ABSTRACT
Hepatitis B virus (HBV) DNA integration into the host cell genome is reportedly a major cause of liver cancer, and a source of hepatitis B surface antigen (HBsAg). High HBsAg levels can alter immune responses which therefore contributes to the progression of HBV-related disease. However, to what extent integration leads to the persistent circulating HBsAg is unclear. Here, we aimed to determine if the extent of HBV DNA integration is associated with the persistence of circulating HBsAg in people exposed to HBV. We established a digital droplet quantitative inverse PCR (dd-qinvPCR) method to quantify integrated HBV DNA in patients who had been exposed to HBV (anti-HBc positive and HBeAg-negative). Total DNA extracts from both liver resections (n = 32; 14 HBsAg-negative and 18 HBsAg-positive) and fine-needle aspirates (FNA, n = 10; 2 HBsAg-negative and 8 HBsAg-positive) were analysed. Using defined in vitro samples for assay establishment, we showed that dd-qinvPCR could detect integrations within an input of <80 cells. The frequency of integrated HBV DNA in those who had undergone HBsAg loss (n = 14, mean ± SD of 1.514 × 10−3 ± 1.839 × 10−3 integrations per cell) was on average 9-fold lower than those with active HBV infection (n = 18, 1.16 × 10−2 ± 1.76 × 10−2 integrations per cell; p = 0.0179). In conclusion, we have developed and validated a highly precise, sensitive and quantitative PCR-based method for the quantification of HBV integrations in clinical samples. Natural clearance of HBV is associated with fewer viral integrations. Future studies are needed to determine if dynamics of integrated HBV DNA can inform the development of curative therapies.
KEYWORDS: HBV DNA integration, functional cure, fine needle aspiration, digital droplet PCR, HBsAg
Introduction
Chronic infection with the Hepatitis B virus is the single most common driver of liver cancer. Worldwide, over 360 million people are chronically infected with HBV, which causes ∼1.1 million deaths each year due to liver cancer or liver failure [1]. The current treatments (reverse transcriptase inhibitors) are taken for indefinite periods as they simply suppress virus replication, but do not cure the infection [2]. The risks of life-threatening disease and the lifelong persistence of HBV infection lead to considerable stigma for people living with chronic hepatitis B, causing impacts to quality of life [3]. Thus, one of the main goals of HBV research is developing a finite-term treatment strategy that eliminates both liver disease progression and viral persistence.
In a consensus statement from the HBV research community, the loss of serum HBV surface antigen (HBsAg) has been selected as one of the primary endpoints for novel therapies (defining as functional cure) [4]. The main rationale for this state is drawn from the natural history of chronic HBV infection (i.e. HBsAg loss), where a dramatic reduction in liver disease progression and hepatocellular carcinoma (HCC) risk accompanies the loss of serum HBsAg [5,6].
This loss of serum HBsAg reflects both an increase in the integration rate of neutralizing antibodies against HBsAg as well as a reduction of HBsAg-secreting cells in the liver. However, despite the loss of detectable HBsAg in the blood, HBV forms that encode HBsAg persist in the liver. First, covalently closed circular DNA (cccDNA, the episomal virus template for all HBV products) has been detected in both animal models of acute HBV infection (in woodchucks and chimpanzees) [7–9], as well as in patients who have spontaneously cleared chronic HBV infection [10–13]. Some of these cccDNA molecules are “defective” and contain mutations rendering them replication incompetent [14]. However, a subset of replication-competent cccDNA forms are still present and can re-initiate HBV infection in cases of immunosuppression [15].
In addition, integrated HBV DNA can be detected in animal models and in humans after HBsAg loss. These forms of HBV DNA inserted into the host cell genome are replication-defective but can maintain HBsAg expression [16,17] (indeed becoming the main source of HBsAg in latter stages of disease [17–20]). At initial infection, integrations occur in ∼1 in 104–105 cells [21–23], but can become at least 10-fold more frequent through the hepatocyte clonal expansion that occurs during immune-mediated cell death and subsequent liver turnover [24–27].
Given this, an immune response directed against both cccDNA and integrated forms of HBV DNA may be necessary to induce HBsAg loss [28–30]. While levels of cccDNA have been quantified in HBsAg-negative patients [31], the frequency of integrated HBV DNA has not been well characterized due to technical difficulties in quantifying this form.
Various approaches have been used to characterize HBV integration sites in liver tissues, including Southern blot hybridization [32–34], Alu-PCR [35–38] and recent high-throughput sequencing technology such as whole genome sequencing [39–42] and RNA sequencing [43,44]. These methods have unique capabilities but do not have the sensitivity to detect single copies of integrations [16]. Inverse nested PCR (invPCR) is a specialized technique that allows the amplification of unknown human DNA regions adjacent to integrated HBV sequence [22,27,45]. invPCR involves a first step of DNA digestion using specific restriction enzymes, followed by DNA circularization of cleavage products through self-ligation, then amplification using specific primers that can amplify the cellular DNA sequences proximal to the HBV integration [46,47], Our team has demonstrated invPCR has high sensitivity, allowing absolute quantification of integrations down to single copy virus-cell junctions [21,48]. However, quantification with invPCR relies on manual serial dilution and lacks precision. We therefore built on the existing invPCR technique and established a new streamlined and highly quantitative assay based on digital droplet PCR (ddPCR) technology to simplify the study of HBV DNA integration.
Using this digital droplet quantitative inverse PCR (dd-qinvPCR) method, we compared HBV DNA integration rates in people with chronic HBV infection with those who have cleared HBsAg (functional cure) to determine the contribution of integrations to the persistence of circulating HBsAg. As our key result, we found that the natural clearance of HBV (i.e. HBsAg loss after a confirmed chronic HBV infection without any external therapy) is associated with fewer viral integrations.
Methods
Patient liver tissues
Resection liver tissue for this study was sourced from 32 Taiwanese patients previously exposed to HBV who underwent liver surgery (patient details in Table 1). All tissue donors gave written informed consent for the use of liver specimens excess to diagnostic needs, and the protocol was approved by the ethics review committee of the Institutional Review Board (IRB) of National Cheng Kung University Hospital (IRB protocol number: A-ER-105-437). All patients were confirmed as: (1) anti-HBc positive, indicating prior exposure to HBV; and/or (2) HBsAg-positive, indicating current HBV infection. They were split into two groups: Group 1 – HBsAg-positive chronic HBV patients (n = 18); and Group 2 – HBsAg-negative patients (n = 14). For patients in Group 2, HBsAg-negativity had been observed for at least 2 years and all were confirmed to have undetectable plasma HBV DNA. The liver tissue used was collected excess to clinical need after resection for either liver cancer or metastatic colorectal cancers in the liver. All samples analysed were from non-tumour liver collected at surgery, snap-frozen and stored at −80°C prior to analysis.
Table 1.
Profile of clinical participants.
| Sample Type | Case | Sex | Age | Cancer | Fibrosis | HBV DNA (IU/mL) | AVT | AVT duration | HBeAg | HBsAg | anti-HBc | anti-HBe | anti-HBs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Liver resection | 1 | M | 87 | HCC | F1 | N/A | No | N/A | ND | − | + | ND | ND |
| 2 | F | 58 | HCC | F1 | Not detected | No | N/A | ND | − | + | ND | ND | |
| 3 | M | 62 | HCC | F2 | N/A | No | N/A | ND | − | + | ND | + | |
| 4 | F | 59 | HCC | F4 | Not detected | No | N/A | − | − | + | + | + | |
| 5 | F | 70 | HCC | F4 | N/A | No | N/A | − | − | + | − | + | |
| 6 | M | 56 | HCC | F2 | N/A | No | N/A | ND | − | + | ND | ND | |
| 7 | M | 70 | HCC | F1 | N/A | No | N/A | ND | − | + | ND | + | |
| 8 | M | 68 | HCC | F2 | N/A | No | N/A | ND | − | + | ND | + | |
| 9 | M | 59 | HCC | F2 | <20 | No | N/A | − | + | − | + | ND | |
| 10 | M | 45 | HCC | F4 | 5,000,000 | No | N/A | − | + | + | − | − | |
| 11 | F | 56 | HCC | F1 | 63,5000 | Yes & stopped | 3 years | − | + | + | + | − | |
| 12 | M | 56 | HCC | F4 | <20 | Yes | 7 months | − | + | + | + | − | |
| 13 | F | 61 | HCC | F4 | <20 | Yes | 4 years | − | + | ND | + | − | |
| 14 | M | 41 | HCC | F3 | 313 | No | N/A | − | + | ND | + | − | |
| 15 | M | 51 | HCC | F1 | 160,000 | No | N/A | − | + | ND | + | − | |
| 16 | M | 48 | HCC | F4 | 8080 | No | N/A | − | + | ND | + | − | |
| 17 | M | 43 | HCC | F3 | N/A | No | N/A | − | + | ND | + | − | |
| 18 | M | 69 | HCC | F4 | <20 | Yes | 1 year | − | + | ND | + | − | |
| 19 | M | 64 | HCC | F4 | 171,000 | No | N/A | − | + | ND | ND | ND | |
| 20 | M | 59 | HCC | F2 | 60,500 | No | N/A | − | + | ND | ND | − | |
| 21 | M | 50 | HCC | F4 | 6620 | No | N/A | − | + | ND | + | − | |
| 22 | F | 86 | HCC | F3 | 65.7 | No | N/A | − | + | ND | + | − | |
| 23 | M | 51 | HCC | F4 | 52 | Yes | 2 weeks | − | + | ND | + | − | |
| 24 | F | 50 | MCRC | F2 | N/A | No | N/A | ND | − | + | ND | + | |
| 25 | M | 35 | MCRC | F1 | Not detected | No | N/A | ND | − | + | ND | + | |
| 26 | M | 74 | Adeno-carcinoma | F1 | N/A | No | N/A | ND | − | + | ND | ND | |
| 27 | M | 70 | MCRC | F3 | N/A | No | N/A | ND | − | + | ND | ND | |
| 28 | F | 69 | MCRC | F2 | N/A | No | N/A | ND | − | + | ND | + | |
| 29 | M | 76 | MCRC | F1 | N/A | No | N/A | ND | − | + | ND | + | |
| 30 | M | 58 | MCRC | F1 | 94.5 | Yes | 3 months | ND | + | + | + | − | |
| 31 | M | 57 | MCRC | F1 | <20 | Yes | 5 years | − | + | ND | + | − | |
| 32 | F | 59 | MCRC | F2 | 39.3 | No | N/A | − | + | ND | ND | − | |
| Fine needle aspiration | 1 | M | 73 | No cancer | F0 | Not detected | Yes | 2 years | − | + | ND | + | − |
| 2 | M | 43 | F0 | Not detected | Yes | 5 years | − | + | ND | + | − | ||
| 3 | M | 47 | F0 | 22 | Yes | 2 years | − | + | ND | + | ND | ||
| 4 | F | 57 | F0 | 2340 | No | N/A | + | + | ND | − | − | ||
| 5 | F | 55 | F0 | <10 | Yes | 3 years | − | + | ND | + | ND | ||
| 6 | F | 46 | F0 | 1,440,000 | No | N/A | + | + | ND | − | ND | ||
| 7 | M | 35 | F0 | 4060 | No | N/A | − | + | ND | + | ND | ||
| 8 | M | 73 | F1 | Not detected | No | N/A | − | − | ND | + | + | ||
| 9 | M | 62 | ND | Not detected | Yes | > 1 year | − | − | ND | − | − | ||
| 10 | F | 48 | F2 | 13,600 | No | N/A | − | + | ND | + | − |
Liver fibrosis was quantified by histology for resection tissue and fibroscan for FNAs, and is provided as Metavir Fibrosis score [49].
M: male; F: Female; AVT: antiviral treatment; HCC: hepatocellular carcinoma; MCRC: metastatic colorectal adenocarcinoma.
HBeAg: Hepatitis B e Antigen; HBsAg: Hepatitis B Surface Antigen; anti-HBc: anti-Hepatitis B Core Antibody; anti-HBe: anti-Hepatitis B e Antibody; anti-HBs: anti-Hepatitis B Surface Antibody; HCV Ab: Hepatitis C Virus Antibody.
“+”: Positive; “−”: Negative; ND: not done.
Fine needle aspiration (FNA) samples were collected at Westmead Hospital, Australia from people previously exposed to HBV (clinical details in Table 1). All patients gave written informed consent for the use of liver tissue for research, and the protocol was approved by Western Sydney Local Health District Human Research Ethics Committee (2019/ETH01913). FNA was performed under local anaesthesia and ultrasound guidance using aseptic technique. The skin under the ribs on the right side was infiltrated with local anaesthetic, and then a 22-gauge needle attached to a 5 mL syringe was advanced into the liver. The needle was fanned in the liver several times while maintaining gentle negative pressure on the syringe to aspirate liver cells. The aspirate was transferred into ice cold Dulbecco’s Modified Eagle’s Medium (DMEM), centrifuged at 50 × g, washed twice with 1 mL DMEM, and then stored at −80°C until analysis.
All clinical data were collected and deidentified to protect patient privacy.
Generation of clones containing integrations
A novel in vitro model was used to generate cell lines with de novo HBV DNA integrations. HepG2-NTCP (human hepatoma cell line that expresses the NTCP receptor, a gift from Professor Stephan Urban) were maintained in DMEM and infected as per previous protocols [50]. Briefly, HepG2-NTCP cells (seeded at 5 × 105 cells/mL) were infected in 24-well plates, with a replication-deficient reporter HBV encoding for zeocin-resistance (HBV-Zeo), replacing the HBV surface open reading frame. Infection media containing DMEM supplemented with 10% v/v Fetal Bovine Serum (10099, Sigma-Aldrich) and 20 mM L-glutamine (G7513, Sigma-Aldrich), 40% v/v polyethylene glycol (89510, Sigma-Aldrich) and HBV-Zeo were mixed thoroughly. Media was aspirated from the wells and the infection mix (250 mL/well) was applied to cells and left to incubate at 37°C and 5% CO2 for 18–20 h. The cells were washed three times with DPBS (14 040 141, Gibco), and fresh infection media was applied and incubated for 3 days. HepG2-NTCP cells were trypsinized using TrypLE Enzyme (12604021, ThermoFisher), re-seeded into 6-well plates and incubated for approximately 25 days to select for reporter HBV-infected cells, using 0.5 mg/mL Zeocin Selection Reagent (R25001, Invitrogen). As cccDNA is cleared through mitosis [51], cells in the expanded colonies eliminate cccDNA, leaving only those with integrations. The HBV integration-containing clones were expanded and frozen down until further analysis. In total, 118 single cell-derived clones were isolated.
DNA extraction and invPCR
Total cellular DNA was extracted from cell pellets of HepG2-NTCP and HepG2-NTCP HBV-Zeo clones using the QIAGEN DNeasy Blood and Tissue Kit (69506, Germany) according to the manufacturer’s instructions and eluted in 50 μL of elution buffer. For clinical samples, total DNA was either extracted from three liver tissue fragments (5–10 mg) from each patient, using the QIAGEN DNeasy Blood and Tissue Kit, or from one FNA pass, using QIAGEN AllPrep Kit (80004, Germany). DNA concentration was quantified using NanoDrop™ 2000 spectrophotometer and stored at −30°C for further experiments. Total cellular DNA was analyzed for integrated HBV DNA using invPCR, as previously described [48]. The integration rate was determined by dividing the number of virus-cell junctions detected at the highest dilution by the number of cell equivalents within that dilution of DNA template.
Low molecular weight HBV replicative intermediate DNA was suspected of producing artefacts during invPCR. To test this, high molecular weight genomic DNA (≥10–20 kbp) was isolated through agarose gel purification. 1–5 µg of total liver DNA extract was separated by electrophoresis through a 1% low melt agarose gel (161-3111, Biorad) at 60 V for 2 h [27]. High molecular weight genomic DNA was excised with a disposable plastic drinking straw and placed in 1 mL New England Biolabs (NEB) CutSmart buffer (B6004) with 0.1% Triton X-100 (HFH10, Invitrogen). The agarose was equilibrated with CutSmart buffer at 4°C overnight. The supernatant was aspirated and 5 U Ncol (R0193, NEB) was added. The high molecular weight DNA was digested with incubation at 37°C for 1 h, then heat inactivated at 70°C for 20 min. The DNA was then purified using Qiagen Quick PCR Purification Kit (28104), as per the manufacturer's protocol. invPCR was then continued as previously described.
Quantification of integrated HBV DNA by dd-qinvPCR
10 μL of the DNA extract (∼1 μg) was digested in a 40 μL restriction digestion reaction containing 1 × CutSmart buffer (B6004, NEB) and 10 U Ncol (R0193, NEB). The combined restriction digestion and exonuclease reaction was incubated for 60 min at 37°C. The enzymes were then heat-inactivated at 80°C for 20 min. To circularize the digested fragments, a 400 μL solution containing 500 U of T4 DNA Ligase (M0202, NEB) in 1 × T4 DNA ligase buffer and was added to the reaction. The reaction was incubated at room temperature for 2 h, followed by an inactivation step of 70°C for 20 min. Then, 10 μL of sodium dodecyl sulphate (10% w/v) was added to ensure complete inactivation of the T4 ligase. NaCl (final concentration of 100 mM) and 0.5 μL GlycoBlue (AM9516, Invitrogen) were added to aid DNA precipitation, followed by 900 μL of 100% ethanol. DNA was precipitated at −20°C overnight. The precipitated DNA was pelleted by centrifugation (14,000 × g for 15 min), washed with 70% ethanol, and air-dried. The DNA pellet was then dissolved in 20 μL of H2O. The final linearization step was performed by adding 20 μL solution containing 5 U BsiHKAI (R0570, NEB), 5 U SphI-HF (R3182, NEB) in 1 × CutSmart buffer and incubating at 37°C for 1 h, 65°C for 1 h, then storing at 20°C until further use.
For ddPCR analysis, 5 μL of the inverted product was put in a 20 μL ddPCR reaction composed of 1 × ddPCR Supermix for Probes (1863010, Biorad), the forward and reverse primers and the probe sequences to quantify the Integrated HBV DNA fragment were listed in Table 2. Droplets were generated according to the manufacturer’s protocol using a QX200 Droplet Generator (Biorad). Intra-droplet PCR was carried out using the following protocol: an initial 10 min denaturation, enzyme activation and droplet stabilization step at 95°C, followed by 40 cycles of a 10 s denaturation step at 95°C, a 30 s annealing step at 54°C and a 120 s elongation step at 68°C, finished with a 10 min enzyme deactivation step at 95°C. Products were then stored at 4°C until droplet reading using a QX200 Droplet Reader (Biorad), quantification using fluorescein amidite (FAM) and hexachloro-fluorescein (HEX) channels, and data analysis using QuantaSoft (Biorad).
Table 2.
PCR primers and Probes used in dd-qinvPCR design.
| Forward primer | Reverse primer | ||
|---|---|---|---|
| Sequence (5′→ 3′)a | Position on HBVb | Sequence (5′→ 3′)a | Position on HBVb |
| TTCGCTTCACCTCTGCACG | 1585–1603 | AAAGGACGTCCCGCGCAG | 1422–1405 |
| Probes | |||
| Sequence (5′→ 3′) | Position on HBVb | ||
| /56-FAM/CCATGGCTG/ZEN/CTAGGCTGTG/3IABkFQ/ | 1372–1390 | ||
| /5HEX/TGCTCGCAGCMGGTCTGG/3BHQ_1/ | 1296–1313 | ||
| /5HEX/AGGCACAGCTTGGAGGCTT/3BHQ_1/ | 1866–1884 | ||
| /5HEX/CTAGCAGCCATGGTGCTGGT/3BHQ_1/ | 1805–1816 | ||
Statistics
Data were analysed using the Mann–Whitney test, ratio paired t-test, or linear regression analysis in Graphpad Prism. A p-value of <0.05 was considered statistically significant.
Results
Principles of the dd-qinvPCR assay
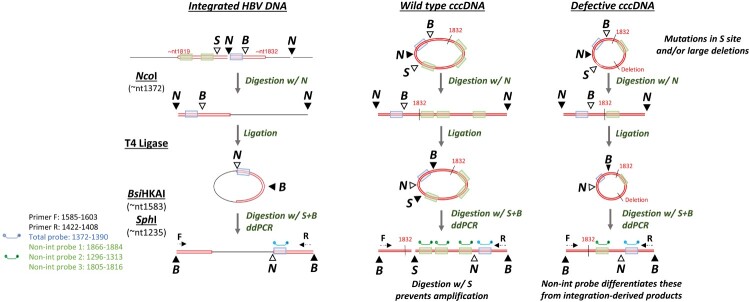
We developed a novel method to expedite the quantification of virus-cell junctions using a ddPCR-dependent approach (Figure 1). Inversion first occurs as per previously published approaches [22,23,47,48]. The genomic DNA is first digested with Ncol that cuts at known sites within the HBV genome. The resulting DNA fragments are then then added to a T4 DNA ligase mediated ligation reaction in a large volume to promote intra-molecular DNA circularization (as opposed to inter-molecular joining of DNA fragments). The circularized product is subsequently cleaved with BsiHKAI to form an inverted product, with viral sequences flanking unknown cellular sequences. The inverted products are also cleaved with SphI to reduce aberrant amplification of HBV cccDNA.
Figure 1.
(A) Schematic diagram of dd-qinvPCR assay HBV cccDNA and integrated HBV DNA sequences (red) and cellular sequences (black) during enzymatic processing over the dd-qinvPCR protocol is shown schematically. The key HBV sequence between nt1372-nt1390 (highlighted in blue), nt1296-nt1313, nt1805-nt1816 and nt1866-nt1884 (highlighted in green). For integrated HBV DNA, the 3’ (right-hand) viral-cellular junction is excised by Ncol (N). Right-hand junction undergoes a T4 DNA ligation reaction, allowing DNA circularization. Circularized product is then cleaved by BsiHKAI (B) to form an inverted product, with viral sequences flanking cellular sequences. Finally, inverted products are cleaved by SphI (S) to minimize cccDNA-derived products. For wild-type cccDNA, the S site prevents the inverted HBV DNA amplifiable due to a double-stranded DNA break. A minority of defective cccDNA (contain mutations in S site and/or large deletions, shown as dashed lines) are consequently lack the S site, which allows the inverted HBV DNA amplifiable. The inverted product is then quantified using ddPCR and its forward primer (F) and reverse primer (R). The probe (blue) binds to sequences (highlighted in blue) that exist in both integrated and cccDNA forms; the probes (green) bind to sequences only present in cccDNA form (absent in integrated forms, highlighted in green). RE sites are shown as triangles (solid colouring indicates the reaction at this step); RE digestions during the steps are indicated as arrows; nt: nucleotide. Figure adapted from [21], based on the nucleotide numbering of the HBV DNA sequence outlined in GenBank accession number U95551.1. (B). ddPCR reading use drop-off assay to exclude the products with HBV sequences not expected in integrated DNA. The fluorophore FAM is used to label total probe which binds to the sequences existing in both integrated and cccDNA forms, HEX is used to label non-integrated probes which only bind to the sequences present in cccDNA forms, allowing the simultaneous detection of two different targets in the same reaction. Orange points indicate cccDNA amplicon, and blue points represent HepG2-NTCP Zeo clones. Y-axis represents the FAM signal amplitude (the total HBV DNA probe); X-axis represents the HEX signal amplitude (the cccDNA specific probe).
Products are amplified using specific HBV-specific forward and reverse primers [48], which are oriented in a way that they amplify across the junction of the integrated viral and host DNA. The total HBV DNA probe (FAM) binds to sequences that exist in both integrated and cccDNA forms, while “non-integrated” probes (HEX) only bind to the sequences present in cccDNA forms (due to being outside of the expected excised HBV DNA junction derived from double stranded linear forms). Multiple “non-integrated” probes were used to detect the broad range of products derived from defective cccDNA templates sequenced by invPCR in our previous studies [22,23,47,48]. The ddPCR drop-off assay differentiated cccDNA from integrated DNA derived products based on HBV sequences not expected in integrated DNA.
Sensitivity, linearity and specificity of the dd-qinvPCR assay
Each HBV DNA integration event occurs at a random site in the human genome [21]. Due to the distribution of restriction enzyme sites used for the inversion and efficiency of circularization of the excised chimeric DNA fragment, not all integrations are detected. Our previous modelling suggested that invPCR could detect ∼10% of all HBV DNA integrations [21]. We empirically tested this assumption using 106 single-cell-derived clones containing unique integrations. These were generated by infecting HepG2-NTCP cells with a reporter HBV virus (encoding a Zeocin resistance gene in place of the surface open reading frame). Cells with HBV DNA integrations were selected for using Zeocin and resultant colonies were isolated for DNA analysis.
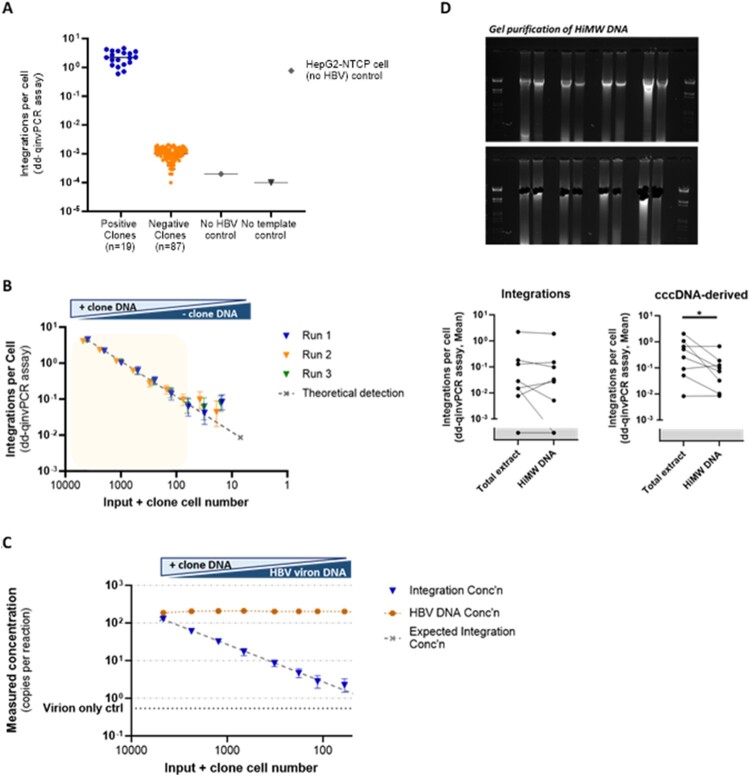
dd-qinvPCR reactions with DNA from parental cells and no template reactions were negative as expected. By analysing this bank of clones with single integrations, we detected viral-cellular junction in 19 of 106 clones (18%) (Figure 2A), relatively consistent with the in-silico estimations [21]. Thus, the actual integration rate is likely ∼5 times higher than what is detected by the invPCR and dd-qinvPCR assays.
Figure 2.
The sensitivity and specificity for dd-qinvPCR assay to quantify integrated HBV DNA. (A) The sensitivity of dd-qinvPCR assay integrated HBV DNA was quantified in DNA extracted from 106 HepG2-NTCP Zeo clones by dd-qinvPCR, these were performed in duplex with reference gene RNaseP (2 copies/cell) for normalization. Inverted DNA that tested positive for integrations among 19 clones (blue points), and significantly above 87 negative clones, uninfected HepG2-NTCP cells (no HBV) and no template control. (B) The linearity of dd-qinvPCR assay was determined by titrating down the positive clone DNA input from 1 μg to 4 nanogram (ng), while DNA from a negative clone was increased to maintain constant DNA input. The integration rates of three repeated runs (triangles in blue, orange and green), the expected integration rate per serial dilution is shown as dashed line, X-axis shows the equivalent input positive clone cell number (C). The specificity of dd-qinvPCR assay for integrated HBV DNA in relation to HBV DNA replicative intermediates DNA from a positive clone was titrated from 1 μg to 4 ng, while supplemented with virion-derived HBV DNA to maintain constant HBV DNA integration rates. dd-qinvPCR was used to quantify the integrated HBV DNA concentration (blue triangles) and total HBV DNA concentration (orange squares). X-axis shows the equivalent input positive clone cell number. The expected concentration (in absolute copies per microliter) assuming 100% detection efficiency is shown as dashed line. False positive signals for integration came up at ratios of 1 copy per ∼300 copies of virion DNA (virion only control in dash line). (D) The specificity of dd-qinvPCR assay by analysing both total and high molecular weight (HiMW) DNA. HiMW DNA was isolated through agarose gel electrophoresis of 1–5 μg of total liver DNA extract. Using dd-qinvPCR assay to quantify the integration rates of HBV DNA in paired total and HiMW preparations of the same DNA extract, there was no significant difference (left, p = 0.349, ratio paired t-test); the HiMW DNA samples showed ∼10 times lower integration rates of cccDNA (right, p = 0.032, ratio paired t-test).
DNA from these positive clones were analysed by standard invPCR, and products were detectable in all of them (and not negative clone controls). We were able to isolate PCR products from DNA agarose gels for 13 clones and sequence them by Sanger sequencing, confirming that they are true integrations (the sequence of human-HBV junction of the 13 clones provided in Supplementary Table 1).
We then tested the linearity of the assay by titrating down the input of DNA from a detectable clone (clone 1, the sequence of human-HBV junction in Supplementary Table 2), while keeping the total input constant with a non-detectable clone (clone 14, the sequence of human-HBV junction provided in Supplementary Table 2) (Figure 2B). We found that the dd-qinvPCR assay could detect integrated HBV DNA in a linear fashion and hewed closely to the expected theoretical value, down to ∼60 cell equivalents in an input of 4800 cells per inversion reaction.
Moreover, we measured the specificity of the dd-qinvPCR assay and its ability to discriminate integrated HBV DNA from other HBV DNA forms (Figure 2C). To test this, we mixed DNA extracted from heparin-affinity purified HBV with DNA from a cell clone that was dd-qinvPCR positive (clone 1). We altered the ratio of these two sources, maintaining a constant copy number of total HBV DNA per reaction. Consistent with the previously determined lower limit of detection, we found linear quantification of the integrated forms down to an input of 72 cells per inversion reaction. The virion only control showed a false positive rate of <1 copy of integrated HBV DNA per 300 copies of HBV DNA.
We then determined if the HBV replicative intermediate DNA within total cellular DNA extracts was at an integration rate sufficient to produce false-positive artefacts. In separate patient samples, we specifically isolated and analysed high molecular weight genomic DNA (thereby depleting intracellular single and double stranded HBV DNA). When we compared the dd-qinvPCR assay analysis of paired total and high molecular weight samples from the same tissue, no significant difference in integration rate was observed (Figure 2D). In contrast (and as expected), significantly lower integration rates of cccDNA-derived forms (∼10 fold) were observed by the dd-qinvPCR assay in the high-molecular weight DNA extracts. Thus, we show that our assay is likely to be highly specific for integrated HBV DNA and that the impact of intracellular replicative intermediates on the signal is minimal.
HBsAg-loss is associated with significantly lower total HBV DNA integration rates and fewer integrations
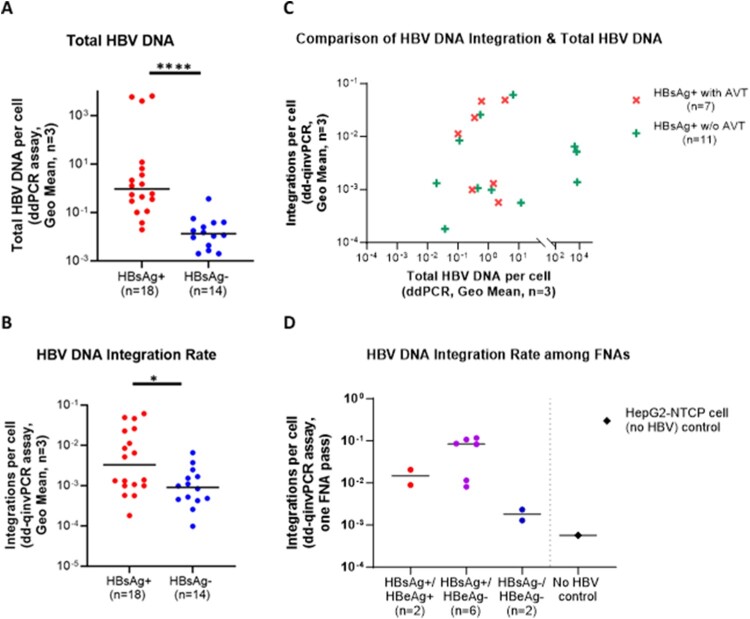
We then analysed intrahepatic total HBV DNA and integrated HBV DNA integration rates in liver tissue from people with chronic HBV infection (n = 18) or those who had cleared HBsAg (i.e. functional cure, n = 15). Three fragments of liver tissue (non-tumour) from each patient were analysed by ddPCR for total HBV DNA and by dd-qinvPCR for integrated HBV DNA. We also quantified integration rates in these tissues using our previously published assay (invPCR) and found high correlation between the two methods (r2 = 0.0228, Supplemental Figure 1). However, many samples positive for dd-qinvPCR were not positive using invPCR. The likely cause for this is the necessitation that integrated forms must be more frequent than the defective cccDNA forms to be detected by invPCR (due to its dependence on end-point titration to isolate single-copy templates for nested PCR). Discrepancies between the quantification of integrations by invPCR and dd-qinvPCR may also be due to the inherent poor precision of end-point titration for quantification. Given these likely explanations, we suggest that the figures provided by dd-qinvPCR are more accurate than those from invPCR.
As expected, total HBV DNA copies per cell were significantly lower in the HBsAg-loss group compared to those with chronic HBV infection (Figure 3A; p < 0.0001, Mann–Whitney test), which is consistent with prior studies [52,53]. Importantly, the integration rate in the HBsAg loss group was significantly lower than in patients with active HBV infection (mean ± SD of 1.514 × 10−3 ± 1.839 × 10−3 vs. 1.365 × 10−2 ± 1.986 × 10−2 integrations per cell; p = 0.0179, Mann–Whitney test), supporting our hypothesis and indicating that natural clearance of HBV is associated with fewer viral integrations (Figure 3B).
Figure 3.
Intrahepatic total HBV DNA and integration rates in patients. (A) Quantified total HBV DNA in HBsAg-positive (n = 18) or HBsAg-negative (n = 14, undergone functional cure) by ddPCR, significantly lower integration rates (p < 0.0001) of total HBV DNA were observed in the HBsAg-loss group compared to those with chronic HBV infection. (3) Quantified integration rates using dd-qinvPCR, the integrated HBV DNA in HBsAg loss group (n = 14) was significantly lower (p = 0.0179) than active HBV infection (n = 18). (C) Comparison of intrahepatic copies of integrated HBV DNA per cell and total HBV qPCR among HBsAg-positive patients that under antiviral treatment (n = 7) or those without treatment (n = 11). No significant association was observed between total HBV DNA and the integration rates of integrated HBV DNA in antiviral treatment group (r2 = 0.1083; p = 0.471), or treatment naïve group (r2 = 0.0447; p = 0.533). HBsAg-positive patients under antiviral treatment did not have significantly different integration rates of HBV DNA integrations compared to those not under treatment (1.90 × 10−2 ± 2.13 × 10−2 vs 1.03 × 10−2 ± 1.85 × 10−2, p = 0.479). Integrations per cell and total HBV DNA represent the Geometric Mean of those 3 fragments. (D) Comparison of copies of integrated HBV DNA per cell in patient FNAs among 10 patients underwent FNA, 2 patients are active hepatitis B infection (HBsAg-positive and HBeAg-positive), 6 patients are chronic hepatitis B infection (HBsAg-positive and HBeAg-negative), 2 patients are no active hepatitis B infection (HBsAg-negative and HBeAg-negative). The copies of detected integrated DNA per cell are 5-fold higher (median 0.8431 vs 0.01478) in chronic infection patients (n = 6) than active infection (n = 2). Compared with uninfected HepG2-NTCP cells (no HBV), the integrated HBV DNA in HBsAg-negative patients (n = 2) is detectable and significantly lower (p = 0.0444, Mann–Whitney test) than HBsAg-positive(n = 8) group.
In HBsAg-positive people, there was no significant association between total HBV DNA and integration rate (p = 0.375) (Figure 3C). Moreover, HBsAg-positive patients receiving antiviral treatment did not have significantly different integration rates compared to those not receiving treatment (1.90 × 10−2 ± 2.13 × 10−2 vs 1.03 × 10−2 ± 1.85 × 10−2, p = 0.479, Mann–Whitney test). This shows that replicative intermediates were not likely to be interfering with the quantification of integrated HBV DNA using our assay.
As our assay displayed high sensitivity and required only low DNA input (Figure 2C), we determined if HBV DNA integrations could be quantified in clinical FNA samples (Figure 3D). Liver FNA is a minimally invasive procedure used to obtain tissue samples, offering a balance between diagnostic accuracy and patient safety [54]. It has been demonstrated to be a reliable method for obtaining serial hepatic tissue samples and can obtain small numbers of liver cells (104–105 cells) [55]. By analysing DNA extracted from 10 FNA samples, we found that dd-qinvPCR could quantify integrations rates that were similar to those observed in liver resection tissues. Two of the patients were HBsAg-negative and had lower integration rates compared to patients who were HBsAg-positive.
Integrated HBV DNA is not associated with other clinical features
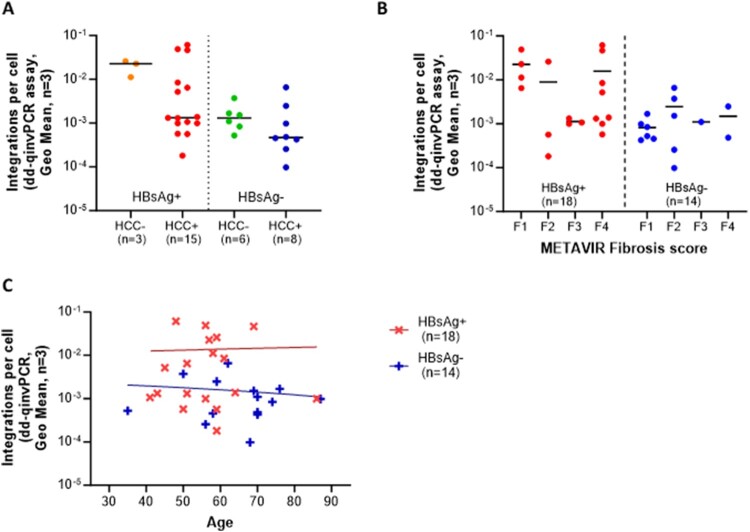
We performed subgroup analyses to examine whether integration rates were modified by clinical features (HCC and cirrhosis) or age. We found that integration rates were not statistically associated with the incidence of HCC (Figure 4A), fibrosis stage (Figure 4B), or age (Figure 4C) in these HBeAg-negative cohorts.
Figure 4.
Intrahepatic integrated HBV DNA integration rates among patients with different clinical features. (A) Integration vs. HCC no significant difference in integration rate was observed (p = 0.246) between HCC + (n = 23) and HCC− groups (n = 9). (B) Integration vs. fibrosis stage no correlation with fibrosis stage in HBsAg-positive (n = 18) and HBsAg-negative (n = 14) groups. Fibrosis grades were classified using the METAVIR scoring system [49] determined by liver histology: F1 means portal fibrosis without septa, F2 means few septa, F3 means numerous septa without cirrhosis, F4 means cirrhosis. (C) Integration vs. age no significant relationship between age and integration rates were observed by linear regression in HBsAg-positive (n = 18, p = 0.888) or HBsAg-negative (n = 14, p = 0.648) cohorts. Integrations per cell are represented by the geometric mean of 3 liver fragments per patient.
Discussion
In this study, we established a highly specific, sensitive, and precise method (dd-qinvPCR) to detect and quantify HBV DNA integrations, even in challenging and limited samples such as FNAs. Using this assay, we found that HBsAg loss (but not other demographic or clinical features) in HBeAg-negative patients is linked with lower integration rates.
The underlying mechanism of this association is not known, as we lack key clinical data to make this distinction (e.g. the time of seroconversion relative to surgery). On one hand, if clearance of HBsAg occurred in the distant past (i.e. decades prior), this result might be due to the reduction in ongoing integration events. Moreover, the decreased inflammation from clearance would reduce the clonal expansion of hepatocytes that is known to drive integration rate increases (e.g. due to HBeAg seroconversion) [27,56]. On the other hand, if HBsAg-seroconversion occurred in the recent past, then this would suggest that the reduction of integrated HBV DNA is necessary to induce HBsAg loss or is caused by the antiviral immune response associated with it (e.g. clearance of HBsAg-producing cells). Distinguishing between these two possibilities will be difficult, given the general lack of clinical justification to perform liver biopsies in patients who have cleared HBsAg. However, our novel assay provides one potential approach: the high sensitivity of dd-qinvPCR allows it to be used to quantify integrations from liver fine needle aspirates, which are less invasive than core needle biopsies.
Nevertheless, we found that HBV DNA integrations in HBsAg-negative patients were not completely eliminated and were still detectable, consistent with animal models [20] and clinical data [57,58]. Our approach was not able to define the full sequence of integrations remaining in the liver after HBsAg loss. Defining the structures of the persisting integrations (e.g. whether they maintain the HBsAg open reading frame) or their epigenetic state (e.g. whether they have been transcriptionally silenced) could provide understanding of the intrahepatic landscape after functional cure of chronic HBV infection and inform therapeutic approaches to induce it.
While several recent studies [59–64] showed nucleoside analogue treatment may reduce several aspects associated with integrated HBV DNA over time (e.g. transcriptionally active integrations), our data did not support the reduction of integrated HBV DNA with antiviral treatment. Reductions of integrated HBV DNA with antiviral treatment have observed in previous studies, but they are only limited changes (∼1 log) over long periods of time (10 years) [60]. It is possible that our small retrospective study was not powered enough to show these small differences. Moreover, the clinical information from the participants in the current study was not detailed enough to provide the length of antiviral treatment prior to surgery, limiting further interpretation over the effect of antiviral therapy on integration rates.
Nonetheless, we believe it is important to understand the impact of new curative therapeutics on HBV DNA integration. As integrations have been shown to be major contributors to HBsAg secretion in HBeAg-negative patients, if reductions of HBV DNA integrations can be achieved, such approaches may be valuable to induce functional cure, in combination with other agents. In particular, quantifying integrations before and after immunotherapies could help understand whether integrations are specifically targeted by therapeutically-activated immune responses. Given that our dd-qinvPCR assay appears to be one to the most sensitive and quantitative approaches available, we believe that it could facilitate this analysis by providing a way to quantify HBV DNA integrations from liver fine needle aspirates (which could be incorporated into clinical trial monitoring).
However, this assay does have limitations. Not only does it detect only a proportion (∼18%) of large HBV DNA integrations (i.e. containing enough of the HBV DNA to include the zeocin selection cassette replacing the HBsAg ORF), but it is likely to miss integrations with large terminal deletions (due to missing primer binding sites). It is still unclear to what extent these sorts of integrations contribute to carcinogenesis or HBsAg expression.
In summary, our work provides a new approach to study HBV DNA integrations and shows that HBsAg loss is associated with fewer integrations. These results highlight the potential importance of targeting HBV DNA integrations with curative therapies and provide a tool to quantify them. Our novel method could be used to inform future HBV cure research, which would benefit the 300 million people worldwide living with chronic hepatitis B.
Supplementary Material
Acknowledgements
We are grateful for the support from the Human Biobank, Research Centre of Clinical Medicine and the Cancer Data Bank of National Cheng Kung University Hospital.
Funding Statement
This work was supported by Australian Centre for HIV and Hepatitis Virology Research; Cancer Institute NSW [grant number 2021/ATRG2028]; China Medical University Hospital [grant number DMR-HHC-113-12]; Deutsche Forschungsgemeinschaft [grant number SFB-TRR 179/2 2020-272983813]; Deutscher Akademischer Austauschdienst [grant number Post-Doctoral Research Fellowship 57407594]; Deutsches Zentrum für Infektionsforschung [grant number TI 07.001-80235CLJOW]; National Health and Medical Research Council [grant number APP1053206, APP2001692, APP1107178]; Paul and Valeria Ainsworth [grant number Precision Medicine Fellowship]; Robert W. Storr [grant number Bequest to the Sydney Medical Foundation].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Global hepatitis report 2024: action for access in low- and middle-income countries. States News Service. 2024.
- 2.Lampertico P, Agarwal K, Berg T, et al. EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67(2):370–398. doi: 10.1016/j.jhep.2017.03.021 [DOI] [PubMed] [Google Scholar]
- 3.Tu T, Block JM, Wang S, et al. The lived experience of chronic hepatitis B: a broader view of its impacts and why we need a cure. Viruses. 2020;12(5):515. doi: 10.3390/v12050515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornberg M, Lok ASF, Terrault NA, et al. Guidance for design and endpoints of clinical trials in chronic hepatitis B—report from the 2019 EASL-AASLD HBV treatment endpoints conference. Hepatology (Baltimore. Md). 2020;71(3):1070–1092. doi: 10.1002/hep.31030 [DOI] [PubMed] [Google Scholar]
- 5.Fattovich G, Olivari N, Pasino M, et al. Long-term outcome of chronic hepatitis B in Caucasian patients: mortality after 25 years. Gut. 2008;57(1):84–90. doi: 10.1136/gut.2007.128496 [DOI] [PubMed] [Google Scholar]
- 6.Yang H-I, Lu S-N, Liaw Y-F, et al. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med. 2002;347(3):168–174. doi: 10.1056/NEJMoa013215 [DOI] [PubMed] [Google Scholar]
- 7.Murray JM, Wieland SF, Purcell RH, et al. Dynamics of hepatitis B virus clearance in chimpanzees. Proc Natl Acad Sci. 2005;102(49):17780–17785. doi: 10.1073/pnas.0508913102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wieland SF, Spangenberg HC, Thimme R, et al. Expansion and contraction of the hepatitis B virus transcriptional template in infected chimpanzees. Proc Natl Acad Sci. 2004;101(7):2129–2134. doi: 10.1073/pnas.0308478100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moraleda G, Saputelli J, Aldrich CE, et al. Lack of effect of antiviral therapy in nondividing hepatocyte cultures on the closed circular DNA of woodchuck hepatitis virus. J Virol. 1997;71(12):9392–9399. doi: 10.1128/jvi.71.12.9392-9399.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mason AL, Xu L, Guo L, et al. Molecular basis for persistent hepatitis B virus infection in the liver after clearance of serum hepatitis B surface antigen. Hepatology (Baltimore. Md). 1998;27(6):1736–1742. doi: 10.1002/hep.510270638 [DOI] [PubMed] [Google Scholar]
- 11.Loriot MA, Marcellin P, Walker F, et al. Persistence of hepatitis B virus DNA in serum and liver from patients with chronic hepatitis B after loss of HBsAg. J Hepatol. 1997;27(2):251–258. doi: 10.1016/S0168-8278(97)80168-7 [DOI] [PubMed] [Google Scholar]
- 12.Rehermann B, Ferrari C, Pasquinelli C, et al. The hepatitis B virus persists for decades after patients’ recovery from acute viral hepatitis despite active maintenance of a cytotoxic T-lymphocyte response. Nat Med. 1996;2(10):1104–1108. doi: 10.1038/nm1096-1104 [DOI] [PubMed] [Google Scholar]
- 13.Michalak TI, Pasquinelli C, Guilhot S, et al. Hepatitis B virus persistence after recovery from acute viral hepatitis. J Clin Invest. 1994;93(1):230–239. doi: 10.1172/JCI116950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei L, Ploss A.. Hepatitis B virus cccDNA is formed through distinct repair processes of each strand. Nat Commun. 2021;12(1):1591–1591. doi: 10.1038/s41467-021-21850-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64(12):1972–1984. doi: 10.1136/gutjnl-2015-309809 [DOI] [PubMed] [Google Scholar]
- 16.Tu T, Zhang H, Urban S.. Hepatitis B virus DNA integration: in vitro models for investigating viral pathogenesis and persistence. Viruses. 2021;13(2):180. doi: 10.3390/v13020180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Podlaha O, Wu G, Downie B, et al. Genomic modeling of hepatitis B virus integration frequency in the human genome. PLoS One. 2019;14(7):e0220376–e0220376. doi: 10.1371/journal.pone.0220376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meier M-A, Calabrese D, Suslov A, et al. Ubiquitous expression of HBsAg from integrated HBV DNA in patients with low viral load. J Hepatol. 2021;75(4):840–847. doi: 10.1016/j.jhep.2021.04.051 [DOI] [PubMed] [Google Scholar]
- 19.Freitas N, Lukash T, Gunewardena S, et al. Relative abundance of integrant-derived viral RNAs in infected tissues harvested from chronic hepatitis B virus carriers. J Virol. 2018;92(10):e02221–17. doi: 10.1128/JVI.02221-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wooddell CI, Yuen M-F, HL-Y C, et al. RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci Transl Med. 2017;9(409):eaan0241. doi: 10.1126/scitranslmed.aan0241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tu T, Budzinska MA, Vondran FWR, et al. Hepatitis B virus DNA integration occurs early in the viral life cycle in an in vitro infection model via sodium taurocholate cotransporting polypeptide-dependent uptake of enveloped virus particles. J Virol. 2018;92(11):e02007–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mason WS, Low H-C, Xu C, et al. Detection of clonally expanded hepatocytes in chimpanzees with chronic hepatitis B virus infection. J Virol. 2009;83(17):8396–8408. doi: 10.1128/JVI.00700-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Summers J, Jilbert AR, Yang W, et al. Hepatocyte turnover during resolution of a transient hepadnaviral infection. Proc Natl Acad Sci. 2003;100(20):11652–11659. doi: 10.1073/pnas.1635109100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tu T, Mason WS, Clouston AD, et al. Clonal expansion of hepatocytes with a selective advantage occurs during all stages of chronic hepatitis B virus infection. J Viral Hepat. 2015;22(9):737–753. doi: 10.1111/jvh.12380 [DOI] [PubMed] [Google Scholar]
- 25.Mason WS, Jilbert AR, Summers J.. Clonal expansion of hepatocytes during chronic woodchuck hepatitis virus infection. Proc Natl Acad Sci. 2005;102(4):1139–1144. doi: 10.1073/pnas.0409332102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, Summers J.. Integration of hepadnavirus DNA in infected liver: evidence for a linear precursor. J Virol. 1999;73(12):9710–9717. doi: 10.1128/JVI.73.12.9710-9717.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mason WS, Gill US, Litwin S, et al. HBV DNA integration and clonal hepatocyte expansion in chronic hepatitis B patients considered immune tolerant. Gastroenterology (New York, NY 1943). 2016;151(5):986–998.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim JH, Ghosh A, Ayithan N, et al. Circulating serum HBsAg level is a biomarker for HBV-specific T and B cell responses in chronic hepatitis B patients. Sci Rep. 2020;10(1):1835–1835. doi: 10.1038/s41598-020-58870-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chua CG, Mehrotra A, Mazzulli T, et al. Optimized ex vivo stimulation identifies multi-functional HBV-specific T cells in a majority of chronic hepatitis B patients. Sci Rep. 2020;10(1):11344–11344. doi: 10.1038/s41598-020-68226-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrari C. HBV and the immune response. Liver Int. 2015;35(s1):121–128. doi: 10.1111/liv.12749 [DOI] [PubMed] [Google Scholar]
- 31.Hayashi S, Isogawa M, Kawashima K, et al. Droplet digital PCR assay provides intrahepatic HBV cccDNA quantification tool for clinical application. Sci Rep. 2022;12(1):2133–2133. doi: 10.1038/s41598-022-05882-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takada S, Gotoh Y, Hayashi S, et al. Structural rearrangement of integrated hepatitis B virus DNA as well as cellular flanking DNA is present in chronically infected hepatic tissues. J Virol. 1990;64(2):822–828. doi: 10.1128/jvi.64.2.822-828.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen JY, Harrison TJ, Lee CS, et al. Detection of hepatitis B virus DNA in hepatocellular carcinoma: analysis by hybridization with subgenomic DNA fragments. Hepatology (Baltimore. Md). 1988;8(3):518–523. doi: 10.1002/hep.1840080315 [DOI] [PubMed] [Google Scholar]
- 34.Edman JC, Gray P, Valenzuela P, et al. Integration of hepatitis B virus sequences and their expression in a human hepatoma cell. Nature (London). 1980;286(5772):535–538. doi: 10.1038/286535a0 [DOI] [PubMed] [Google Scholar]
- 35.Pollicino T, Vegetti A, Saitta C, et al. Hepatitis B virus DNA integration in tumour tissue of a non-cirrhotic HFE-haemochromatosis patient with hepatocellular carcinoma. J Hepatol. 2013;58(1):190–193. doi: 10.1016/j.jhep.2012.09.005 [DOI] [PubMed] [Google Scholar]
- 36.Murakami Y, Saigo K, Takashima H, et al. Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut. 2005;54(8):1162–1168. doi: 10.1136/gut.2004.054452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paterlini-Brechot P, Saigo K, Murakami Y, et al. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene. 2003;22(25):3911–3916. doi: 10.1038/sj.onc.1206492 [DOI] [PubMed] [Google Scholar]
- 38.Gozuacik D, Murakami Y, Saigo K, et al. Identification of human cancer-related genes by naturally occurring hepatitis B virus DNA tagging. Oncogene. 2001;20(43):6233–6240. doi: 10.1038/sj.onc.1204835 [DOI] [PubMed] [Google Scholar]
- 39.Ye R, Wang A, Bu B, et al. Viral oncogenes, viruses, and cancer: a third-generation sequencing perspective on viral integration into the human genome. Front Oncol. 2023;13:1333812–1333812. doi: 10.3389/fonc.2023.1333812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujimoto A, Furuta M, Totoki Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48(5):500–509. doi: 10.1038/ng.3547 [DOI] [PubMed] [Google Scholar]
- 41.Kan Z, Zheng H, Liu X, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23(9):1422–1433. doi: 10.1101/gr.154492.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujimoto A, Totoki Y, Arai Y, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44(7):760–764. doi: 10.1038/ng.2291 [DOI] [PubMed] [Google Scholar]
- 43.Péneau C, Imbeaud S, La Bella T, et al. Hepatitis B virus integrations promote local and distant oncogenic driver alterations in hepatocellular carcinoma. Gut. 2022;71(3):616–626. doi: 10.1136/gutjnl-2020-323153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svicher V, Salpini R, Piermatteo L, et al. Whole exome HBV DNA integration is independent of the intrahepatic HBV reservoir in HBeAg-negative chronic hepatitis B. Gut. 2021;70(12):2337–2348. doi: 10.1136/gutjnl-2020-323300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mason WS, Liu C, Aldrich CE, et al. Clonal expansion of normal-appearing human hepatocytes during chronic hepatitis B virus infection. J Virol. 2010;84(16):8308–8315. doi: 10.1128/JVI.00833-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hui EK, Wang PC, Lo SJ.. Strategies for cloning unknown cellular flanking DNA sequences from foreign integrants. Cell Mol Life Sci. 1998;54(12):1403–1411. doi: 10.1007/s000180050262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsuei D-J, Chen P-J, Lai M-Y, et al. Inverse polymerase chain reaction for cloning cellular sequences adjacent to integrated hepatitis B virus DNA in hepatocellular carcinomas. J Virol Methods. 1994;49(3):269–284. doi: 10.1016/0166-0934(94)90142-2 [DOI] [PubMed] [Google Scholar]
- 48.Tu T, Jilbert AR.. Detection of hepatocyte clones containing integrated hepatitis B virus DNA using inverse nested PCR. Hepatitis B Virus. 2017;1540:97–118. doi: 10.1007/978-1-4939-6700-1_9 [DOI] [PubMed] [Google Scholar]
- 49.Bedossa P, Poynard T.. An algorithm for the grading of activity in chronic hepatitis C. Hepatology (Baltimore. Md). 1996;24(2):289–293. doi: 10.1002/hep.510240201 [DOI] [PubMed] [Google Scholar]
- 50.Ni Y, Lempp FA, Mehrle S, et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology (New York, NY 1943). 2014;146(4):1070–1083.e6. [DOI] [PubMed] [Google Scholar]
- 51.Tu T, Zehnder B, Wettengel JM, et al. Mitosis of hepatitis B virus-infected cells in vitro results in uninfected daughter cells. JHEP Reports. 2022;4(9):100514–100514. doi: 10.1016/j.jhepr.2022.100514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeo YH, Ho HJ, Yang H-I, et al. Factors associated With rates of HBsAg seroclearance in adults With chronic HBV infection: A systematic review and meta-analysis. Gastroenterology (New York, NY 1943). 2019;156(3):635–646.e9. [DOI] [PubMed] [Google Scholar]
- 53.Sonneveld MJ, Chiu S-M, Park JY, et al. Lower pretreatment HBV DNA levels are associated with better off-treatment outcomes after nucleo(s)tide analogue withdrawal in patients with HBeAg-negative chronic hepatitis B: a multicentre cohort study. JHEP Rep. 2023;5(8):100790–100790. doi: 10.1016/j.jhepr.2023.100790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chhieng DC. Fine needle aspiration biopsy of liver - an update. World J Surg Oncol. 2004;2(1):5–5. doi: 10.1186/1477-7819-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lejnine S, Marton MJ, Wang IM, et al. Gene expression analysis in serial liver fine needle aspirates. J Viral Hepat. 2015;22(1):64–76. doi: 10.1111/jvh.12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Budzinska MA, Shackel NA, Urban S, et al. Sequence analysis of integrated hepatitis B virus DNA during HBeAg-seroconversion. Emerging Microbes Infect. 2018;7(1):1–12. doi: 10.1038/s41426-018-0145-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gan W, Gao N, Gu L, et al. Reduction in intrahepatic cccDNA and integration of HBV in chronic hepatitis B patients with a functional cure. J Clin Transl Hepatol. 2023;11(2):314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao N, Guan G, Xu G, et al. Integrated HBV DNA and cccDNA maintain transcriptional activity in intrahepatic HBsAg-positive patients with functional cure following PEG-IFN -based therapy. Aliment Pharmacol Ther. 2023;58(10):1086–1098. doi: 10.1111/apt.17670 [DOI] [PubMed] [Google Scholar]
- 59.Wen X, Wu X, Sun Y, et al. Long-term antiviral therapy is associated with changes in the profile of transcriptionally active HBV integration in the livers of patients with CHB. J Med Virol. 2024;96(6):e29606–e29n/a. doi: 10.1002/jmv.29606 [DOI] [PubMed] [Google Scholar]
- 60.Chow N, Wong D, Lai C-L, et al. Effect of antiviral treatment on hepatitis B virus integration and hepatocyte clonal expansion. Clin Infect Dis. 2023;76(3):e801–e809. doi: 10.1093/cid/ciac383 [DOI] [PubMed] [Google Scholar]
- 61.Zhang M, Zhang H, Cheng X, et al. Liver biopsy of chronic hepatitis B patients indicates HBV integration profile may complicate the endpoint and effect of entecavir treatment. Antiviral Res. 2022;204:105363–105363. doi: 10.1016/j.antiviral.2022.105363 [DOI] [PubMed] [Google Scholar]
- 62.Hsu Y-C, Suri V, Nguyen MH, et al. Inhibition of viral replication reduces transcriptionally active distinct hepatitis B virus integrations with implications on host gene dysregulation. Gastroenterology (New York, NY 1943). 2022;162(4):1160–1170.e1. [DOI] [PubMed] [Google Scholar]
- 63.Grudda T, Hwang HS, Taddese M, et al. Integrated hepatitis B virus DNA maintains surface antigen production during antiviral treatment. J Clin Invest. 2022;132(18):1–10. doi: 10.1172/JCI161818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Erken R, Loukachov V, Dort K, et al. Quantified integrated hepatitis B virus is related to viral activity in patients with chronic hepatitis B. Hepatology (Baltimore. Md). 2022;76(1):196–206. doi: 10.1002/hep.32352 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.