Abstract
In budding yeast, the Cdc14p phosphatase activates mitotic exit by dephosphorylation of specific cyclin-dependent kinase (Cdk) substrates and seems to be regulated by sequestration in the nucleolus until its release in mitosis. Herein, we have analyzed the two human homologs of Cdc14p, hCdc14A and hCdc14B. We demonstrate that the human Cdc14A phosphatase is selective for Cdk substrates in vitro and that although the protein abundance and intrinsic phosphatase activity of hCdc14A and B vary modestly during the cell cycle, their localization is cell cycle regulated. hCdc14A dynamically localizes to interphase but not mitotic centrosomes, and hCdc14B localizes to the interphase nucleolus. These distinct patterns of localization suggest that each isoform of human Cdc14 likely regulates separate cell cycle events. In addition, hCdc14A overexpression induces the loss of the pericentriolar markers pericentrin and γ-tubulin from centrosomes. Overproduction of hCdc14A also causes mitotic spindle and chromosome segregation defects, defective karyokinesis, and a failure to complete cytokinesis. Thus, the hCdc14A phosphatase appears to play a role in the regulation of the centrosome cycle, mitosis, and cytokinesis, thereby influencing chromosome partitioning and genomic stability in human cells.
INTRODUCTION
In the budding yeast Saccharomyces cerevisiae exit from mitosis requires the down-regulation of mitotic cyclin-dependent kinase (Cdk) activity (Murray et al., 1989; Noton and Diffley, 2000). The Cdc14 phosphatase is a member of the mitotic exit network (MEN), a group of proteins that inactivates mitotic Cdk activity and is required for mitotic exit. The MEN includes the GTPase Tem1p and its exchange factor Lte1p; the kinases Cdc5p, Cdc15p, Dbf2p, and Dbf20p; and Mob1p, which binds Dbf2p and Dbf20p (McCollum and Gould, 2001). Genetic and biochemical analyses indicate that Cdc14p activation requires all of the MEN components and lies at the bottom of this pathway (Jaspersen et al., 1998; Visintin et al., 1998).
The activity of Cdc14p seems to be regulated by its subcellular localization (Shou et al., 1999; Visintin et al., 1999). Throughout most of the cell cycle, Cdc14p is sequestered in the nucleolus in an inactive state, bound to a complex consisting of Net1p and Sir2p (the RENT complex) (Shou et al., 1999; Visintin et al., 1999). During early anaphase a regulatory network called the Cdc Fourteen Early Anaphase Release (FEAR) network, which includes Cdc5, the separase Esp1, the kintetochore-associated protein Slk19, and Spo12, promotes Cdc14 release from the nucleolus (Stegmeier et al., 2002). Subsequently, in late anaphase, the MEN network is required to prevent Cdc14p from relocalizing to the nucleolus. The released Cdc14p promotes mitotic cyclin destruction and Cdk inactivation by dephosphorylating the Cdk inhibitor p40Sic1, the APC activator Cdh1p, and the transcription factor Swi5p (Visintin et al., 1998).
Although the Schizosaccharomyces pombe homolog of Cdc14p, flp1p/clp1p, is highly conserved with S. cerevisiae Cdc14p (36% identity) and functionally replaces S. cerevisiae Cdc14p, it behaves quite differently. First, flp1/clp1 is not required for mitotic exit but instead regulates septum formation and cytokinesis as part of the septation-inducing network (SIN) (Cueille et al., 2001; Trautmann et al., 2001). Flp1/clp1 may also regulate the G2/M transition, because its overexpression results in a G2/M arrest (Cueille et al., 2001; Trautmann et al., 2001). Second, flp1/clp1 does not dephosphorylate the same substrates as Cdc14p, for example, the Cdh1p homolog ste9p, nor is it required for the accumulation of the Sic1p homolog rum1p or the degradation of the cyclin cdc13p (Cueille et al., 2001; Trautmann et al., 2001). Finally, flp1p/clp1p localizes to both the spindle pole body (SPB) and the nucleolus during interphase, and early in mitosis it is released from the nucleolus and localizes to the SPB, mitotic spindle, and medial ring (Cueille et al., 2001; Trautmann et al., 2001). During septum synthesis, flp1/clp1 localizes to the contractile ring at the leading edge of the division septum, both SPBs, and the nucleolus. Based on these differences between the Cdc14p homologs in the two yeasts, it is difficult to predict which processes human Cdc14 might regulate.
Intriguingly, nearly all the genes from the S. cerevisae MEN have homologs in the S. pombe SIN (Balasubramanian et al., 2000; Nigg, 2001). Thus, the MEN and SIN seem to control distinct cell cycle events despite sharing a conserved biochemical pathway. Several components of the MEN and SIN, including CDC5 (a Polo kinase), MOB1, and CDC14, are conserved in higher eukaryotes; however, other MEN/SIN components are not readily identifiable in public databases.
The Cdc14 family of phosphatases contains a highly conserved N-terminal catalytic domain and a nonconserved C-terminal domain. Interestingly, two different Cdc14 genes exist in humans, termed Cdc14 A and B, whereas only one isoform is present in Drosophila, Caenorhabditis elegans, S. cerevisiae, and S. pombe. Although hCdc14A and B share high sequence homology (50% identity), they demonstrate two notable differences; hCdc14B possesses a unique 54 amino acid N-terminal extension, and the two proteins have divergent C-terminal domains.
In this study, we demonstrate that hCdc14A localizes to the centrosome and appears to be an important centrosomal regulator. Overexpression of hCdc14A leads to defects in centrosome structure, chromosome segregation, cytokinesis, and nuclear reformation. Furthermore, hCdc14A efficiently and specifically dephosphorylates substrates phosphorylated by cyclin-dependent kinases in vitro. We also examine the localization of hCdc14B and show that it localizes to the nucleolus. Because hCdc14A and B localize to distinct regions of the cell, they are likely to regulate different cell cycle processes.
MATERIALS AND METHODS
Phosphatase Assays
In Vitro Phosphatase Reactions.
The following Cdk substrates were expressed and purified from bacteria as fusion proteins: GST-p27Kip1, MBP-p27Xic1, GST-XCdc6, and GST-Xenopus cyclin E. 6xHis-tagged human Cdh1 (kind gift from J.M. Peters) was purified from baculovirus as described previously (Kramer et al., 2000) and Histone H1 was purchased from Sigma-Aldrich (St. Louis, MO). Each substrate (2.5 μg) was incubated with 1 U of either mitogen-activated protein (MAP) kinase, cyclin B-Cdc2 (New England Biolabs, Beverly, MA), GST-Plk, or baculovirus-expressed cyclin E-Cdk2 in kinase buffer (50 mM Tris, pH 7.5, 10 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol [DTT], and 50 μM ATP) in the presence of 0.15 μCi of γ[32P]ATP for 30 min at 30°C. Samples were then centrifuged through a G-25 spin column (Harvard Apparatus, Holliston, MA) preequilibrated with phosphatase buffer (50 mM imidazole, pH 6.9, 1 mM EDTA, and 1 mM DTT). Bacterially expressed GST-hCdc14A was added to 40 nM, reactions were stopped at various times with sample buffer, and the proteins were resolved by SDS-PAGE and visualized by autoradiography. The peptide substrates Autocantide 3 and Syntide 2 were phosphorylated by αCaM kinase II (Ca2+/calmodulin-dependent protein kinase) (kind gift from Roger Rich, Stanford University). Phosphatase reactions with the peptide substrates were stopped by acid precipitation on P81 Whatman paper and analyzed by scintillation counting. To phosphorylate GSK-3 peptide with Akt, activated Myr-Akt-HA was immunoprecipitated from NIH 3T3 cells stimulated with insulin for 30 min. The extent of radiolabel incorporated into GSK-3 peptide was determined by densitometry. For p-nitrophenylphosphate (pNPP) phosphatase reactions, pNPP was used at 20 mM. pNPP reactions were performed in a volume of 50 μl, stopped by the addition of 200 μl of 0.25 N NaOH, and read at OD405 nm on a spectrophotometer.
Immunoprecipitation (IP) Phosphatase Assays.
For each time point, 1 μg of affinity-purified antibodies raised against hCdc14A(344–623) or Cdc14B(1–54) was coupled to 7 μl of protein A-Sepharose beads for 2 h at 4°C, washed three times in lysis buffer (20 mM HEPES, pH 7.7, 150 mM NaCl, 0.3% Triton X-100, 60 mM β-glycerophosphate, 1 mM EDTA, 1 mM DTT, 10 μg/ml each of leupeptin, pepstatin, and chymostatin, and 1 mM pheylmethylsulfonyl fluoride), and incubated with 100 μg of HeLa lysate for 2 h at 4°C. Complexes were washed four times in lysis buffer followed by four washes in IP phosphatase buffer (50 mM imidazole, pH 7.5, 50 mM NaCl, 1 mM DTT, and 1 mM EDTA). γ[32P]ATP-labeled hCdh1 (see below) was then added to the beads (100–200 ng of hCdh1 per time point) and incubated at 30°C with frequent agitation. At each time point, the reaction was centrifuged in a Nanofuge (Hoeffer, San Francisco, CA) for 5 s to pellet the beads and a sample of the supernatant was removed, followed by addition of SDS-PAGE sample buffer. Reactions were then loaded on an SDS-PAGE gel and proteins were visualized by autoradiography. Baculovirus-purified human Cdh1 was labeled with γ[32P]ATP as follows: crude sera raised against Xenopus cyclin B was coupled to protein A-Sepharose (20 μl of crude sera per 10 μl of packed beads) and used to immunoprecipitate cyclin B/Cdc2 from cytostatic factor-arrested Xenopus egg extracts. Cytostatic factor extract was diluted 1:10 in IP buffer (50 mM β-glycerophosphate, 0.1% Triton X-100, 5 mM EDTA, 100 mM NaCl, 1 mM DTT, and 10 μg/ml each leupeptin, pepstatin, and chymostatin), immunoprecipitated for 2 h at 4°C, washed four times in IP buffer, washed four times in kinase buffer (see above), and then incubated with hCdh1 for 30 min at 30°C on a rotating platform. The supernatant was then spun over a G25 spin column equilibrated with phosphatase buffer. For a typical reaction, 100 μl of Xenopus egg extract was immunoprecipitated by 30 μl of coupled Xcyclin-B beads and used to label 5 μg of Cdh1.
Cell Cycle Arrest, Lysates, Growth Media, and Transfections
HeLa cells were synchronized at the G1/S boundary by a double thymidine block. Briefly, cells were incubated 18 h in complete media containing 2 mM thymidine, washed with PBS and incubated for 8 h in fresh media, and finally incubated for 18 h in media containing 2 mM thymidine. To release the cells from the arrest, the cells were washed in phosphate-buffered saline (PBS) and released into prewarmed fresh media. To synchronize cells in mitosis, cells were treated for 18 h in media supplemented with 100 ng/ml nocodazole. Mitotic cells were isolated by mitotic shake-off and plated into fresh media prewarmed to 37°C. The cell cycle stage of each time point was determined by quantifying DNA stained with propidium iodide using flow cytometry.
To prepare lysates, cells were trypsinized, collected by centrifugation, washed twice in PBS, and the cell pellets were flash frozen in liquid nitrogen and stored at −80°C. The pellets were resuspended in 1 volume of lysis buffer, incubated on ice for 10 min, and then centrifuged at 14,000 rpm in an Eppendorf centrifuge for 10 min at 4°C. Protein concentrations were assayed using Bio-Rad reagent and absorbance measured at 595 nm.
U2OS cells were cultured in McCoy's 5A medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine, and incubated at 37°C in 10% CO2. HeLa cells were grown in DMEM with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine, and incubated at 37°C in 10% CO2.
Transfections were carried out using FuGENE 6 (Roche Applied Science, Indianapolis, IN) according to the manufacturer's instructions. Typically, 100,000 U2OS cells were plated the day before transfections in six-well dishes containing 12-mm coverslips treated with fibronectin.
Antibody Production and Purification
GST fusion proteins of hCdc14A (1–380) and hCdc14A (344–623) were expressed and purified in bacteria for antisera production in rabbits (Josman Immunoresearch). hCdc14B(1–54) was purified as a GST fusion in pGEX6P1 and cleaved with Prescission protease (Pharmacia, Peapack, NJ). The peptide was then coupled to keyhole limpet hemocyanin (Imject Immunogen EDC kit; Pierce Chemical, Rockford, IL) for injection into rabbits. Affinity purification of antisera was performed by acid elution (100 mM glycine, pH 2.5) from MBP fusion proteins coupled to cyanogen bromide-activated Sepharose (Amersham Biosciences, Piscataway, NJ). For blocking experiments, antibodies were incubated at room temperature for 1 h with a fivefold molar excess of cleaved antigen, spun at top speed in an Eppendorf centrifuge for 10 min, and used for either immunofluorescence or Western blotting analysis. The following antibodies were used for Western blot analysis: cyclin B1 (rabbit, catalog no. sc-752; Santa Cruz Biotechnology, Santa Cruz, CA), cyclin A (rabbit, catalog no. sc-751; Santa Cruz Biotechnology), and actin (goat, catalog no. sc-1616; Santa Cruz Biotechnology).
Immunofluorescence Microscopy
Cells were grown on 12-mm coverslips treated with fibronectin and fixed for 5 min in 100% methanol at −20°C. Endogenous hCdc14A was detected with affinity-purified antibodies raised in rabbits against either the N- or C-terminal domain of hCdc14A. Centrosomes were identified by costaining with mouse monoclonal antibodies raised against γ-tubulin (clone GTU-88; Sigma-Aldrich). Centrioles were stained with the centrin mouse monoclonal antibody 20H5 (Salisbury laboratory). Rat antibodies against α-tubulin were from Serotec (MCAP77; Oxford, United Kingdom). For localization experiments with green fluorescent protein (GFP)-hCdc14A, mouse monoclonal antibodies against GFP (catalog no. 8362-1; CLONTECH, Palo Alto, CA) and rabbit antibodies against γ-tubulin (a gift from Tim Stearns laboratory) or pericentrin (a gift from Steve Doxsey) were used. For GFP-hCdc14B localization experiments, rabbit antibodies raised against the N-terminal 1–54 amino acids of hCdc14B were used to detect hCdc14B, and mouse monoclonal antibodies were used to detect nucleolin (clone 4E2; Research Diagnostics, Flanders, NJ). All secondary antibodies were raised in donkeys, conjugated to Texas Red or fluorescein isothiocyanate, and used at 1:150 (Jackson Immunoresearch Laboratories, West Grove, PA).
Molecular Biology
For GFP expression studies, variants of hCdc14A or B were cloned into pEGFP-N1 and pEGFP-C1 vectors (CLONTECH) by using standard molecular biology techniques. Most of the hCdc14A and B variants were tested as both N- and C-terminal fusion to GFP and in each case gave identical localization results. Point mutations were generated using QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA). Accession numbers are as follows: full-length hCdc14A is referred to as hCdc14A2 in GenBank (AF064102.1), and hCdc14B is referred to as hCdc14B2 (AF064104.1).
Microtubule Regrowth Assay
U2OS cells were treated with 10 μg/ml nocodazole (Calbiochem, San Diego, CA) in complete media for 2 h at 37°C to depolymerize microtubules. The cells were washed with PBS and allowed to recover in complete media for 10 min at 37°C and then processed for immunofluorescence as described above with antibodies against α-tubulin.
RESULTS
hCdc14A Dephosphorylates Substrates of Cyclin-dependent Kinases
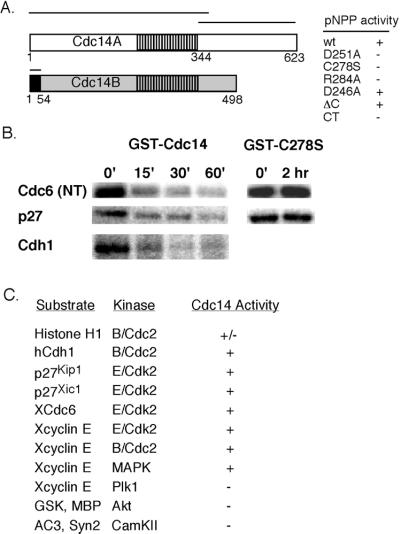
To test whether the human Cdc14 phosphatase (Li et al., 1997) is specific for substrates of cyclin-dependent kinases, we examined the ability of bacterially expressed hCdc14A to dephosphorylate substrates of cyclin B/cdc2 and cyclin E/cdk2 kinases vs. other kinases. As a control, we constructed mutations in hCdc14A at active site residues characteristic of dual-specificity phosphatases, including yeast Cdc14p. These mutations include an active site aspartic acid (D251A), cysteine (C278S), and an arginine (R284A). Each of these mutations caused a complete loss of in vitro phosphatase activity (Figure 1A, right), supporting the idea that hCdc14A has similar structural and catalytic requirements to S. cerevisiae Cdc14 (Taylor et al., 1997). Mutation of the conserved aspartic acid (D246A) or deletion of the C-terminal domain of hCdc14A did not affect its phosphatase activity, using either pNPP or phosphorylated protein substrates (Figure 1A; our unpublished data). Importantly, hCdc14A efficiently dephosphorylated known substrates of cyclin B/Cdc2 and cyclin E/Cdk2, and also of MAP kinase, but not substrates of the Polo-like kinase Plk1, Ca2+/calmodulin-dependent protein kinase kinase II, or Akt (Figure 1, B and C). We also found that Histone H1 was less efficiently dephosphorylated by hCdc14A than other Cdk substrates (Figure 1C). Together, these results suggest that hCdc14A does not broadly dephosphorylate serine/threonine phosphorylation sites, but is selective for serine-proline or threonine-proline-directed Cdk and MAP kinase consensus phosphorylation sites. Previous studies demonstrated that hCdc14A dephosphorylates the mitotically phosphorylated Cdk substrate cyclin E to allow its rebinding to interphase chromatin in Xenopus (Furstenthal et al., 2001), and that hCdc14A and B can dephosphorylate Cdk-phosphorylated p53 (Li et al., 2000), providing further examples that hCdc14A can directly oppose Cdk-directed phosphorylation events.
Figure 1.
Human Cdc14 phosphatase is specific for Cdk substrates. (A, left) Schematic of the hCdc14A and B phosphatases. hCdc14A and B represent highly related (61% identity within the catalytic domain), but independent genes with a 246 amino acid C-terminal domain specific to hCdc14A, and a 54 amino acid N-terminal peptide specific to hCdc14B. Bars indicate peptides used for immunizing rabbits. Right, bacterially expressed hCdc14A was purified as a GST fusion and used to dephosphorylate the chromogenic phosphatase substrate pNPP. Mutations in conserved residues within the active site aspartic acid (D251A), cysteine (C278S), and arginine (R284A) residues abrogated activity against pNPP. Mutation of another aspartic acid (D246A in hCdc14A), conserved in Cdc14 homologs, but not other dual specificity phosphatases, or deletion of the C-terminal domain, did not affect pNPP dephosphorylation activity. The C-terminal domain of hCdc14A had no phosphatase activity. (B) hCdc14A phosphatase dephosphorylates Cdk substrates. Validated Cdk substrates Xenopus Cdc6, human p27Kip1, and human Cdh1 were phosphorylated in vitro with cyclin E/Cdk2 (Cdc6 and p27) or cyclin B/Cdc2 (Cdh1), repurified, and mixed with 40 nM hCdc14A in phosphatase buffer for the indicated times. (C) Selectivity of the hCdc14A phosphatase. As in B, various proteins or peptides were phosphorylated by the indicated kinases, repurified, and assayed as substrates for hCdc14A. +, indicates >90% dephosphorylation; −, indicates <5% dephosphorylation; and ±, indicates <50% dephosphorylation.
Endogenous hCdc14A and B Protein Levels and Activity Vary Modestly during Cell Cycle
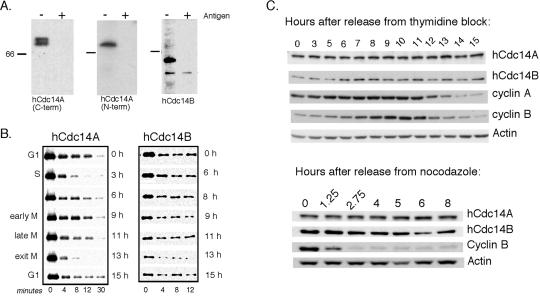
To examine the endogenous isoforms of human Cdc14, we raised antisera to unique determinants of each protein: the hCdc14A C-terminal domain (amino acids 344–623), and the first 54 amino acids of hCdc14B (Figure 1A; see MATERIALS AND METHODS). We also raised an antibody to the N-terminal and central regions of hCdc14A (amino acids 1–380). Affinity-purified antibodies raised against hCdc14A specifically recognized a 69-kDa protein, and anti-Cdc14B antibodies specifically recognized a 60-kDa protein (Figure 2A). Two forms of hCdc14A were resolved using a gradient gel system, which most likely represents phosphorylated forms (Figure 2A, left).
Figure 2.
hCdc14A and B protein levels and phosphatase activity fluctuate modestly through the cell cycle. (A) Antibodies specific to hCdc14A or B recognize endogenous protein. Antibodies raised against the hCdc14A-specific C terminus (residues 344–623) (left) and hCdc14A N terminus (residues 1–380) (middle) and to the specific N-terminal 54 amino acid peptide of hCdc14B (right) were affinity-purified and used to probe immunoblots of asynchronous HeLa cell lysates with (+) or without (−) prebinding to their respective antigens (see MATERIALS AND METHODS). The gradient gel system used only in the left panel was necessary to resolve the hCdc14A doublet. (B) In vitro phosphatase activities of hCdc14A and B vary modestly during the cell cycle. HeLa cells were synchronized in late G1 phase by a double thymidine block and then released from the block into fresh medium. Extracts were prepared at the indicated times (right) and DNA content analyzed by flow cytometry (major cell cycle phase shown at left). hCdc14A and B were immunoprecipitated with specific antibodies coupled to protein A-Sepharose beads, and the immune complexes were used to dephosphorylate the phosphorylated form of the APC activator human Cdh1 as a substrate (see MATERIALS AND METHODS). (C) hCdc14A protein levels are constant, whereas hCdc14B protein levels oscillate modestly during the cell cycle. HeLa cells were synchronized by double thymidine block-and-release (top) and nocodazole block-and-release (bottom) procedures and analyzed by Western blot for the abundance of endogenous hCdc14A and B protein.
To measure the phosphatase activity of endogenous hCdc14A and B, we developed an immunoprecipitation assay using antibodies specific to hCdc14A and B. Phosphatase assays of immunoprecipitated hCdc14A from synchronized HeLa lysates showed that the activity of hCdc14A varied slightly during the cell cycle (Figure 2B), increasing slightly in S phase and late mitosis. Parallel immunoblots showed little cell cycle variation in hCdc14A abundance (Figure 2C). Similar analyses of hCdc14B showed that hCdc14B phosphatase activity (Figure 2B) and the amount of hCdc14B protein (Figure 2B) varied slightly throughout the cell cycle (Figure 2B), with hCdc14B protein levels peaking in mitosis. We do not currently understand how the in vitro phosphatase activity of either Cdc14 isoform relates to its in vivo activity. Because the amount of hCdc14A and B protein levels or phosphatase activity does not seem to fluctuate significantly during the cell cycle, we next wished to determine whether their localization is cell cycle regulated.
Cell Cycle-regulated Localization of hCdc14A to Centrosomes and hCdc14B to Nucleoli
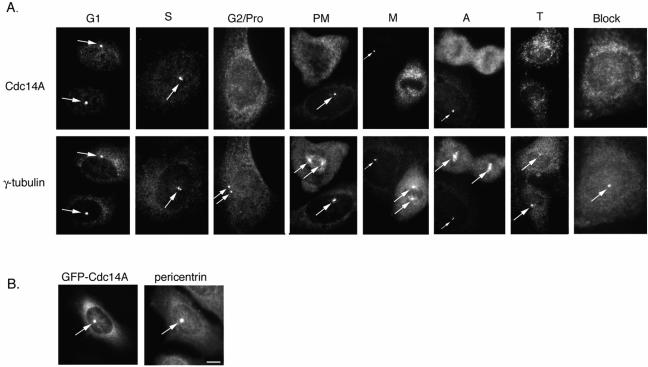
Examination of the subcellular localization of hCdc14A by indirect immunofluorescence revealed the protein to be specifically localized to centrosomes in interphase cells (Figure 3A, leftmost panel). hCdc14A centrosomal staining was apparent in cells with single or duplicated centrosomes during G1, S, and G2 phases (>98% positive), but was displaced from centrosomes by early prophase and throughout mitosis (<2% positive) (Figure 3A). Similar results were observed in both U2OS and HeLa cells. To determine when the loss of hCdc14A from centrosomes occurred, we examined hCdc14A staining in late G2/early prophase cells with newly separated centrosomes (determined by γ-tubulin staining). We found that in 22% (33/147) of cells with separated centrosomes hCdc14A appeared at both centrosomes; in 14% of cells (21/147) hCdc14A staining appeared on only one centrosome; and in 64% of cells hCdc14A staining was absent. We suspect this heterogeneity reflects that hCdc14A is displaced from one then the other centrosome as cells enter mitosis. Thus, hCdc14A staining disappears from centrosomes shortly after centrosome separation (when cells are entering prophase), and is restored upon mitotic exit. GFP-hCdc14A also localized to the centrosome and cytoplasm in U2OS (Figure 3B) or HeLa cells (our unpublished data), recapitulating the staining of endogenous hCdc14A. Brief treatment of cells with the microtubule-depolymerizing drug nocodazole did not affect endogenous hCdc14A centrosomal localization (our unpublished data), demonstrating that the protein is an integral centrosomal protein and not simply recruited by microtubules (Chang and Stearns, 2000).
Figure 3.
hCdc14A associates with the interphase, but not mitotic, centrosome. (A) Endogenous hCdc14A staining. U2OS cells were synchronized by double thymidine block-and-release and stained with affinity-purified antibodies specific to hCdc14A and mouse antibodies to γ-tubulin. Representative images of cells in various stages of the cell cycle are shown. Arrows indicate centrosomes. Note the absence of hCdc14A staining in mitotic stages and in interphase cells where the hCdc14A antibody was preblocked with antigen (“block”). Pro, prophase; PM, prometaphase; M, metaphase; A, anaphase; T, telophase. Also note that in PM, M, and A, hCdc14A stains the centrosome of an adjacent interphase cell (indicated by an arrow). (B) GFP-hCdc14A targets to the centrosome. Full-length hCdc14A was fused to the enhanced variant of green fluorescent protein (eGFP) and transiently transfected into human U2OS cells, fixed with methanol, and costained with antibodies to GFP and pericentrin. Bar, 5 μm.
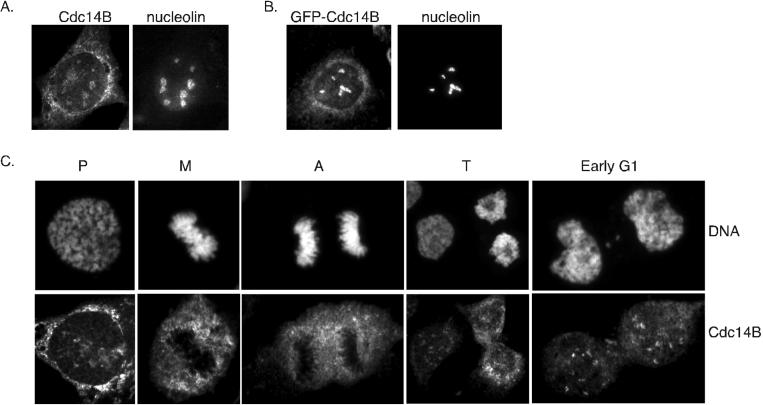
In contrast to hCdc14A, endogenous hCdc14B localized to the nucleolus in U2OS and HeLa cells, as demonstrated by colocalization with the nucleolar marker nucleolin (Figure 4A). Preincubation of Cdc14B antibodies with the immunizing peptide blocked hCdc14B nucleolar staining, indicating that the hCdc14B staining of the nucleolus is specific (our unpublished data). GFP-hCdc14B expressed in U2OS cells also localized to the nucleolus (Figure 4B). Examination of mitotic cells showed that endogenous hCdc14B staining was lost from the nucleolus after prophase until late telophase and was again seen in early G1 cells (Figure 4C); this pattern of localization was nearly identical to that of nucleolin (our unpublished data). Thus, the localization of hCdc14B closely parallels the disassembly and reassembly of the nucleolus, which breaks down at the onset of mitosis and reassembles during telophase (Dundr et al., 2000). It is currently unclear whether the localization of hCdc14B to the nucleolus regulates its activity in a manner akin to S. cerevisiae Cdc14p. A notable difference is the timing in which the two homologs are released from the nucleus; hCdc14B seems to be released in prophase, whereas S. cerevisiae Cdc14p is released in early anaphase.
Figure 4.
hCdc14B localizes to the nucleolus in interphase cells. (A) Endogenous staining of interphase U2OS cell with hCdc14B antibodies. Asynchronously growing U2OS cells were fixed in methanol and costained with antibodies raised against hCdc14B and nucleolin. (B) GFP-hCdc14B localizes to the nucleolus. U2OS cells were transiently transfected with a fusion between eGFP and hCdc14B, fixed in methanol and costained with antibodies against GFP and nucleolin. (C) Nucleolar staining of endogenous hCdc14B is absent during mitosis. Representative images of U2OS cells during various stages of mitosis were fixed in methanol and stained with hCdc14B antibodies. Cells were costained with Hoechst dye to visualize chromosomes. Pro, prophase; M, metaphase; A, anaphase; T, telophase.
Transfection of GFP fusions to various domains of hCdc14A and B facilitated mapping of localization determinants of each isoform (Figure 5, A and B). The hCdc14A N terminus (amino acids 1–377) localized to the centrosome and cytoplasm in the same pattern as full-length hCdc14A (Figure 5B). In addition, the C terminus of hCdc14A localized diffusely throughout the cell (Figure 5B). Thus, the centrosomal localization determinants of hCdc14A reside in the N-terminal region. Interestingly, the first 54 amino acids of hCdc14B (Figure 5A, top) were sufficient to localize the GFP fusion to the nucleolus. Removal of these amino acids (hCdc14BΔ1–54) resulted in localization to the cytoplasm rather than the nucleolus, with 20% of cells also showing localization to the centrosome (Figure 5A). Furthermore, a fusion of the first 54 amino acids of hCdc14B to the N terminus of full-length hCdc14A localized to the nucleolus and centrosome (Figure 5A, bottom), although in a fraction of cells only nucleolar localization was seen; a fusion between hCdc14B(1–54) and hCdc14A localized in the same manner. Therefore, the first 54 amino acids of hCdc14B encode a nucleolar targeting domain.
Figure 5.
Mapping localization determinants of hCdc14A and B. (A) U2OS cells were transfected for 24 h with the indicated GFP fusion constructs and immunostained with antibodies against nucleolin (top) or pericentrin (bottom). Arrows in the lower panels indicate centrosomes. (B) Summary of the localization of various hCdc14A or B constructs fused to GFP. Cells were fixed in methanol and stained with antibodies against GFP, and costained with either pericentrin or nucleolin as in A. NES in hCdc14A (aa 352–367) and in hCdc14B (aa 390–405) are depicted.
Both hCdc14A and B contain putative consensus nuclear export sequences (NES) (hCdc14A, amino acids 352–367; hCdc14B, amino acids 390–405). Mutation of the NES in hCdc14A, or treatment of cells with the nuclear export inhibitor leptomycin B, caused hCdc14A to relocalize to the nucleolus (Mailand et al., 2002). hCdc14A also contains a consensus nuclear localization sequence (amino acids 78–91), which from the analysis presented herein seems to actively transport hCdc14A to the nucleus. Thus, active nuclear export seems to be a prerequisite for hCdc14A localization to the centrosome. These analyses show that hCdc14A and B seem to have the ability to dynamically distribute between nucleolus, centrosome, and possibly other compartments through specific sequences within each isoform. The dynamic restructuring of the centrosome (Hinchcliffe and Sluder, 2001; Stearns, 2001) and the disassembly of the nucleolus (Dundr et al., 2000) during mitosis may play important roles in the partitioning of hCdc14A and B.
Overexpression of hCdc14A Disrupts Centrosome Structure and at High Levels Inhibits Microtubule Regrowth from Centrosomes
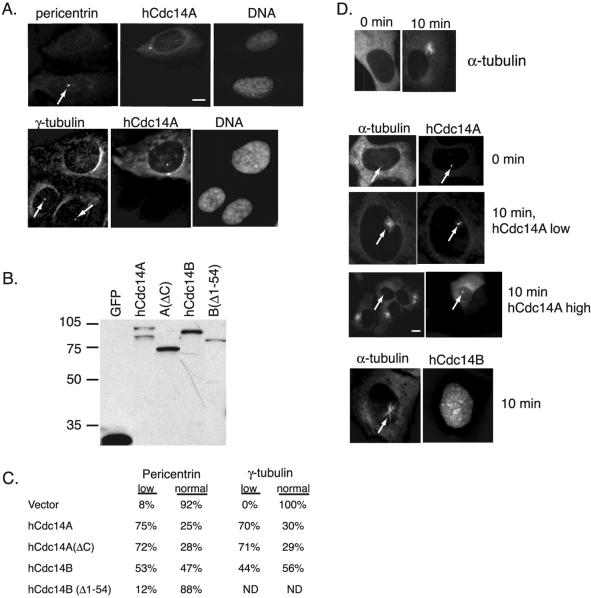
Because hCdc14A is a centrosomal component and potential centrosomal regulator, we tested whether overexpression of hCdc14A affected centrosomal function. hCdc14A overexpression strongly reduced the abundance of the pericentriolar markers pericentrin and γ-tubulin at centrosomes (Figure 6A). Twenty-four hours after transfection, 75% of hCdc14A-expressing cells contained low or undetectable levels of pericentrin staining, and 70% of hCdc14A-cells contained low or undetectable levels of γ-tubulin staining (Figure 6C). Only 8% of GFP-vector control cells contained low pericentrin staining, whereas all vector control cells possessed normal levels of γ-tubulin staining (Figure 6C). Examination of the hCdc14A and B variants demonstrated that hCdc14A had a stronger effect on the abundance of both centrosomal markers than hCdc14B (Figure 6C; see expression levels in 6B). The modest effect of hCdc14B on the centrosome (53 and 44% of positive cells had low pericentrin and γ-tubulin staining, respectively) may be due to an intermediate ability to target to the centrosome. Deletion of the N-terminal 54 amino acids of hCdc14B resulted in only 12% of cells containing low pericentrin staining. Thus, the centrosome-specific form of human Cdc14, hCdc14A, has the strongest influence on centrosome integrity.
Figure 6.
Overexpressed hCdc14A induces loss of pericentriolar material and inhibits microtubule regrowth from centrosomes. (A) hCdc14A overexpression causes loss of pericentriolar material from centrosomes. U2OS cells transiently transfected with pEGFP-hCdc14A were fixed and stained with the pericentriolar markers pericentrin (top) and γ-tubulin (bottom). Arrows indicate centrosomes in untransfected cells. Note that cells overexpressing hCdc14A have low levels of both pericentrin and γ-tubulin compared with untransfected cells. (B) Western blot of GFP-hCdc14 variants expressed in U2OS cells. Extracts from cells transfected with GFP (lane 1), GFP-hCdc14A (lane 2), GFP-hCdc14A(ΔC) (lane 3), GFP-hCdc14B (lane 4), and GFP-hCdc14B(Δ1–54) were resolved by SDS-PAGE and analyzed by Western blotting with antibodies to GFP. (C) Quantitation of pericentrin and γ-tubulin levels in cells transfected with variants of hCdc14A and B. GFP fusions to the indicated hCdc14A or B constructs were transfected in U2OS cells and positive cells were scored for either “low” or “normal” staining of pericentrin and γ-tubulin levels as in A. In each case, at least 150 GFP-positive cells were counted. (D) High levels of GFP-hCdc14A overexpression inhibit microtubule regrowth from centrosomes. U2OS cells transfected with GFP fusions to hCdc14A or B were treated with 10 μg/ml nocodazole at 24 h posttransfection for 2 h to depolymerize microtubules then grown in fresh media for 10 min. Coverslips were then fixed in methanol and processed for immunofluorescence with antibodies against α-tubulin to monitor regrowth of microtubules. Top, cells after treatment for 2 h with nocodazole (0 min) and after 10 min in media without nocodazole (10 min). Middle, GFP-hCdc14A-positive cells at 0 min, and at 10 min with either low or high levels of hCdc14A. Arrows indicate location of centrosomes. Bottom, a GFP-hCdc14B-positive cell after 10 min. Note that hCdc14B, even at high levels, did not inhibit microtubule regrowth. Bars 5 μm.
Because microtubules emanating from the centrosome are nucleated by the pericentriolar material (Stearns, 2001), we tested whether hCdc14A overexpression impaired the ability of centrosomes to nucleate microtubules. To address this question we treated U2OS cells that had been transfected with GFP variants of hCdc14 with nocodazole for 2 h to depolymerize microtubules, then washed the cells in fresh media for 10 min before fixing and staining the cells with α-tubulin to visualize microtubules. After the 2-h treatment with nocodazole microtubules were depolymerized (Figure 6D, top), and after 10 min in fresh media microtubules were readily visible radiating from the centrosomes (Figure 6D, top). Low amounts of hCdc14A overexpression did not impair the ability of centrosomes to nucleate microtubules; however, high levels of hCdc14A expression completely abolished microtubule nucleation from centrosomes, as did hCdc14A(ΔC) (Figure 6D, middle). hCdc14B, even at high levels of expression, had no effect on microtubule nucleation from centrosomes (Figure 6D, bottom). Thus, hCdc14A overexpression inhibits accumulation of pericentriolar material (Figure 6A) and impairs the ability of the centrosome to nucleate microtubules.
In addition to the effects observed in transiently transfected cells, low levels of hCdc14A overexpression in regulatable stable cell lines caused centrosome amplification, consistent with Cdc14 being an important regulator of the centrosome duplication cycle (Mailand et al., 2002). If hCdc14A simply dephosphorylated Cdk substrates at the centrosome, one might expect that Cdc14 overexpression would oppose centriole splitting, because this is a Cdk-directed event (Hinchcliffe et al., 1999; Lacey et al., 1999). It is possible that hCdc14A overexpression increases the dynamics of Cdk-phosphorylation and hCdc14A-dephosphorylation at the centrosome, which in turn leads to enhanced centriole splitting. Alternatively, hCdc14A may block cytokinesis (see below) or cause a failure to separate centrioles into daughter cells.
To determine whether hCdc14A affected the DNA replication cycle, we assayed the DNA content of 293T cells overexpressing hCdc14A by flow cytometry. hCdc14A overexpression did not cause an increase in the percentage of cells at any stage of the cell cycle, suggesting that the effects of hCdc14A overexpression on centrosome structure were specific and did not reflect global changes in the cell cycle. After 48 h of overexpression, however, cells with less than 2N DNA content began to accumulate (our unpublished data; Mailand et al., 2002). This suggests that hCdc14A overexpression induced cells to undergo abnormal cell divisions in which daughter cells receive less than their normal complement of DNA.
hCdc14A Overexpression Causes Defects in Spindle Morphology, Mitosis, and Cytokinesis
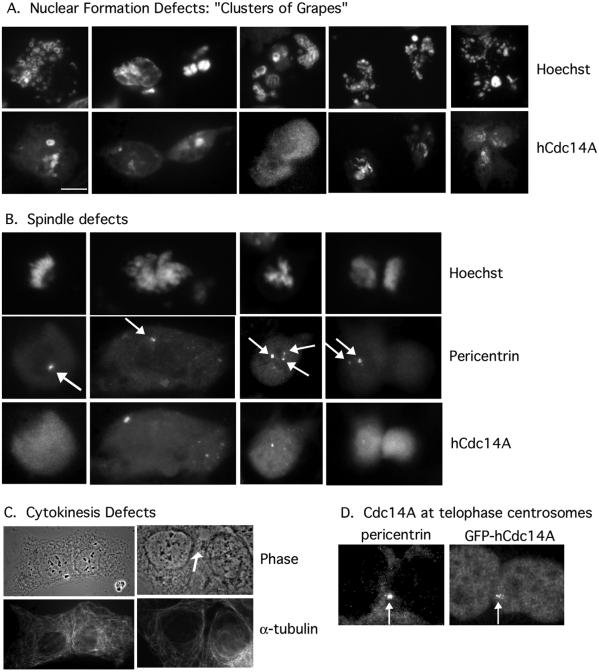
In addition to interfering with the integrity of the pericentriolar material, hCdc14A overexpression might also disrupt subsequent or later events in mitosis. Indeed, we found that overexpression of hCdc14A in U2OS cells caused a variety of mitotic defects, including monopolar or multipolar spindles, missegregated chromosomes, and unequal partitioning of DNA, a failure to reform nuclei after mitosis, and strong cytokinesis defects (Figure 7). In Figure 7, a GFP-hCdc14A fusion was transiently transfected in U2OS cells, and cells were fixed 24 h after transfection. Most hCdc14A-overexpressing cells showed pronounced defects in nuclear formation after mitosis (93% of mitotic cells vs. <1% in control mitotic cells). These defects ranged from fragmented or small nuclei to a large number of karyomeric micronuclei, resembling clusters of grapes (Figure 7A, middle). We also observed cells with fragmented nuclei that seemed to have a tripolar spindle (Figure 7A, right). Karyomeric structures are commonly observed as precursors to complete nuclear formation in embryonic cells (Lemaitre et al., 1998), but may only transiently form in somatic cells.
Figure 7.
Transient overexpression of hCdc14A induces abnormalities in chromosome segregation, karyokinesis, and cytokinesis. U2OS cells were transiently transfected with pEGFP-hCdc14A, fixed in methanol 24 h after transfection, and stained with hCdc14A and Hoechst (A, B, and D), pericentrin (B and D), or α-tubulin (C). (A) Overexpression of hCdc14A induces karyokinesis (“micronuclei”) defects. Bar, 5 μm. (B) Overexpression of hCdc14A induces chromosome abnormalities and missegregation. Arrows indicate centrosomes. (C) Overexpression of hCdc14A induces a block to cytokinesis. Arrow indicates the position of the cytoplasmic bridge. Both cells overexpress GFP-hCdc14A. (D) GFP-hCdc14A colocalizes with centrosomes at the cleavage furrow in telophase cells. Arrows indicate the centrosomes.
hCdc14A overexpression also induced spindle defects, ranging from cells with a single spindle pole (Figure 7B, left) to partially duplicated centrosomes separating to only one daughter cell (Figure 7B, right). Additionally, asymmetric or multipolar DNA masses were frequently seen (Figure 7B, middle). In total, 3% of hCdc14A overexpressing cells displayed clear spindle defects compared with <0.2% of control cells. Because the block in mitosis by hCdc14A may be transient and these aberrant mitotic structures unstable, we suspect that a much higher percentage of cells have spindle abnormalities.
In addition to mitotic and spindle defects, a prominent failure in cytokinesis was observed in a high percentage (94 vs. <2% in control) of hCdc14A overexpressing cells that had recently divided (Figure 7C). Studies of cytokinesis in fission yeast and in budding yeast have supported a role for Cdc14 and the S. cerevisiae MEN or homologous S. pombe SIN in regulating cytokinesis (Balasubramanian et al., 2000; McCollum and Gould, 2001). Herein, we observed blocks to cytokinesis after chromosomes decondensed, characterized by a remaining cytoplasmic bridge. Some cells possessed residual DNA bridges, but many cells showed cytokinesis defects without detectable errors in chromosome segregation or DNA morphology. A recent study by Bornens and colleagues (Piel et al., 2001) suggested that migration of the centriole to the midbody in telophase triggers cytokinesis. Thus, hCdc14A-induced deficiencies in centrosomal structure might affect the ability of the centriole to trigger cytokinesis, or hCdc14A itself may play a role in the triggering event. Consistent with the latter idea, we have observed GFP-hCdc14A colocalizing with centrosomes near the cleavage furrow during telophase (Figure 7D; our unpublished data).
DISCUSSION
We have shown that hCdc14A localizes to the centrosome and hCdc14B localizes to the nucleolus. During mitosis, hCdc14A staining is absent from centrosomes and hCdc14B staining is lost from the nucleolus. The dramatic differences in the localization of hCdc14A and B suggest that despite sharing high sequence homology, these isoforms perform separate functions. Consistent with hCdc14A playing an important role in centrosomal function and controlling various steps in mitosis, misregulation of hCdc14A by transient overexpression leads to defects in centrosome structure (assessed by pericentrin and γ-tubulin staining), progression through mitosis, and cytokinesis.
It is intriguing to speculate that hCdc14A localization to the centrosome may be important for regulating cytokinesis in light of a recent study by Bornens and colleagues in which they show that centrioles play a critical role in triggering cytokinesis (Piel et al., 2001). Consistent with this possibility, we have seen numerous examples of GFP-hCdc14A localized to the centrosome in telophase near the cytokinesis furrow. However, the cytokinesis defects that we see in hCdc14A-overexpressing cells may not be direct, and instead may be caused by an earlier perturbation in the cell cycle, such as incomplete centrosome maturation or defective spindle assembly. Depletion of hCdc14A protein from HeLa cells by siRNA prevented cells from undergoing normal cell division (Mailand et al., 2002). These abnormalities included an abortive anaphase attempt and the failure to undergo cytoplasmic abscission, further supporting a role for hCdc14A in cytokinesis.
Evidence from both yeasts suggests that the MEN from S. cerevisiae and the SIN from S. pombe are important regulators of cytokinesis. Although the SIN has a well established role in regulating cytokinesis, several components of the S. cerevisiae MEN have also been shown to be important for regulating cytokinesis, including Cdc14p, Cdc15p, Cdc5p, and Mob1p (Song et al., 2000; Lee et al., 2001; Luca et al., 2001; Menssen et al., 2001; Song and Lee, 2001). In addition, the Drosophila homolog of Cdc5, the Polo kinase, has been implicated in regulation of cytokinesis (Carmena et al., 1998). Thus, it is quite possible that in higher eukaryotes the conserved homologs of the MEN/SIN pathway are important for regulation of cytokinesis, mitotic exit, and perhaps other cell cycle transitions.
We considered the possibility that hCdc14A controls the G1/S transition by preventing the ubiquitin-mediated proteolysis of the Cdk inhibitor p27Kip1. In S. cerevisiae, Cdc14p may oppose ubiquitin-mediated proteolysis of the related Cdk inhibitor Sic1p (Visintin et al., 1998), whose phosphorylation by Cln/Cdc28 targets it for recognition by the E3 ubiquitin ligase SCFCdc4 (Feldman et al., 1997). Overexpression of Cdc14p in S. cerevisiae also causes a G1 arrest and stabilization of Sic1p (Visintin et al., 1998). In human cells, p27Kip1 is destroyed by ubiquitin-mediated proteolysis, which requires phosphorylation of p27Kip1 by cyclin E/Cdk2 (Carrano et al., 1999; Montagnoli et al., 1999). In vitro, hCdc14A efficiently dephosphorylated p27Kip1 (Figure 1B), however, we found that hCdc14A did not inhibit the SCFSkp2-dependent ubiquitylation of p27Kip1 in HeLa lysate (Kaiser and Eldridge, unpublished data). In addition, transient overexpression of hCdc14A in human embryonic kidney 293T cells did not cause a G1 arrest in the cell cycle (our unpublished data). Therefore, we do not believe that hCdc14A regulates the stability of p27Kip1.
Localization Requirements in hCdc14A and B
GFP fusions of both hCdc14A and hCdc14B recapitulated the localization of the endogenous protein. By fusing different domains of hCdc14A and B to GFP, we were able to map their localization requirements. Based on these studies, the N terminus of hCdc14A (amino acids 1–377) is necessary and sufficient for localization to the centrosome. It remains unclear what role the C-terminal domain of hCdc14A performs; hCdc14A(ΔC) dephosphorylated substrates as efficiently as the full-length protein in vitro, localized in an identical manner, and when overexpressed, caused similar defects in the accumulation of pericentrin and γ-tubulin at centrosomes, mitotic progression, and cytokinesis. Both hCdc14A and B contain consensus nuclear export sequences that seem to actively transport both isoforms out of the nucleus. Mutating the NES of hCdc14A caused the fusion to localize to the nucleolus (Mailand et al., 2002). In hCdc14B, removal of the first 54 amino acids relocalized the GFP fusion to the cytoplasm, indicating that the putative NES of hCdc14B likely exported the full-length protein from the nucleus. Conversely, a fusion of GFP with only the first 54 amino acids of hCdc14B localized to the nucleolus. Thus, hCdc14B localization to the nucleolus requires the first 54 amino acids, and in their absence hCdc14B is exported from the nucleolus. These GFP mapping studies suggest that endogenous hCdc14A and B dynamically distribute between the nucleus and cytoplasm. In addition, hCdc14A seems to actively distribute between the cytoplasm and centrosome as demonstrated by fluorescence recovery after photobleaching analysis (Mailand et al., 2002). The GFP mapping studies demonstrated that the first 54 amino acids are required for localizing hCdc14B to the nucleolus; however, it is not known whether other proteins are required for this localization. The majority of endogenous hCdc14B is part of a large complex (>600 kDa) by gel filtration analysis of HeLa interphase lysate (our unpublished data), suggesting that other proteins may be important for targeting hCdc14B to the nucleolus. In S. cerevisiae, Net1p tethers Cdc14p to the nucleolus in a complex (the RENT complex) that includes Sir2p (Shou et al., 1999; Straight et al., 1999; Visintin et al., 1999). In S. pombe, there is no known homolog of Net1p; however, there is a conserved homolog of Sir2p that could be responsible for localizing flp1p/clp1p to the nucleolus (Cueille et al., 2001). In human cells there is also no clear homolog of Net1p, however, there is a homolog of Sir2p (Brachmann et al., 1995). Thus, the human homolog of Sir2p is an interesting candidate protein that could be required for hCdc14B nucleolar localization.
Regulation of Cdc14
We examined several factors that might indicate how hCdc14A and B are regulated throughout the cell cycle, including steady-state protein levels, intrinsic phosphatase activity, and localization. The protein levels of hCdc14A seem constant throughout the cell cycle, and hCdc14B protein levels fluctuate only slightly (Figure 2C). The intrinsic phosphatase activity of both isoforms also varies little throughout the cell cycle (Figure 2B). However, the immunoprecipitation assay we used to measure Cdc14A and B phosphatase activity may not reflect the in vivo activity. Localization thus seems likely to be the predominant mechanism regulating hCdc14A and B.
The changes in localization may reflect a means of regulating hCdc14A and B activity or may bring both isoforms into proximity with their substrates at the correct time in the cell cycle. hCdc14A may be important for regulating the centrosome cycle only during interphase and must be displaced during mitosis. It is possible that a fraction of hCdc14A remains at the centrosomes in mitosis, but is not detectable by our immunofluorescence methods. Indeed, at lower levels of expression, GFP-hCdc14A is present on mitotic centrosomes, although at much lower levels than in interphase (Mailand et al., 2002).
Another intriguing possibility is that Cdk phosphorylation of Cdc14 is important for its regulation. We have identified seven possible Cdk phosphorylation sites in hCdc14A (serine-proline or threonine-proline dipeptides) and four such sites in hCdc14B. In vitro, hCdc14A is an efficient substrate for cyclin E/Cdk2 and hCdc14A is able to autodephosphorylate (Kaiser and Jackson, unpublished data); we have not yet tested whether hCdc14B is a substrate for cyclin E/Cdk2. Furthermore, we have observed at least two forms of hCdc14A on SDS-PAGE that likely represent phosphorylated species of hCdc14A (Figure 2A). Similar to hCdc14A, flp1p/clp1p protein levels in S. pombe are constant throughout the cell cycle, but flp1p/clp1p becomes hyperphosphorylated during mitosis (Cueille et al., 2001). Whether this phosphorylation is important for flp1p/clp1p localization or for regulating flp1/cp1 activity is not known.
We considered that hCdc14A and B might heterodimerize in vivo, which could offer an additional means of regulation. Cdc14p in S. cerevisiae homodimerizes (Taylor et al., 1997) and bacterial expressed hCdc14A also seems to dimerize based on gel filtration analysis (our unpublished data). We have no reason to believe, however, that hCdc14A and B associate in vivo because hCdc14A specific antibodies never detected a nucleolar signal and hCdc14B specific antibodies never detected a centrosomal signal in immunofluorescence studies.
Substrates and Function of Cdc14
The finding that the hCdc14A phosphatase localizes to the centrosome and may be required for centrosome maintenance is interesting in light of the fact that numerous kinases, including cyclin E and A/Cdk2, Mps1, Polo-like kinase (Plk1), and Aurora A, are important regulators of centrosome function (Hinchcliffe et al., 1999; Lacey et al., 1999; Fisk and Winey, 2001; Meraldi et al., 2002). It seems likely that some phosphatase(s) should oppose these kinases, and based on the analyses presented herein hCdc14A seems a likely candidate.
We do not know the relevant physiological substrates of hCdc14A or B. We have shown that hCdc14A efficiently and specifically dephosphorylates substrates of cyclin-dependent kinases in vitro, consistent with substrate specificity of S. cerevisiae Cdc14p. A candidate substrate for hCdc14 is nucleophosmin/B23, which localizes to centrosomes in mitosis and through G1 (Okuda et al., 2000). Phosphorylation by cyclin E/Cdk2 causes nucleophosmin/B23 to dissociate from centrosomes, and this dissociation is required for centrosome duplication (Okuda et al., 2000). Mutation of the nucleophosmin/B23 Cdk phosphorylation site (Thr199Ala) inhibits centrosome duplication (Tokuyama et al., 2001).
Another interesting candidate substrate of hCdc14A is C-Nap1, a validated substrate of centrosomal kinase Nek2 (Fry et al., 1998). C-Nap1, like hCdc14A, is concentrated at centrosomes in interphase cells, but diminishes in abundance at the mitotic centrosome (Fry et al., 1998). Similar to the phenotype we observe with hCdc14A transient overexpression, overexpression of Nek2 in U2OS cells causes loss of pericentriolar material (45% of cells, assayed by γ-tubulin staining) and promotes centrosome splitting (45% of cells) (Fry et al., 1998). An intriguing possibility is that Nek2 may be a target of hCdc14A and that dephosphorylation of Nek2 by hCdc14A activates Nek2, which in turn promotes centrosome duplication.
Identification of bona fide hCdc14A and B substrates is an immediate goal and will help to elucidate which processes each isoform regulates. In addition, identification and characterization of functional homologs of the MEN/SIN, which are likely to exist in higher eukaryotes, is of vital importance to the understanding of hCdc14 and mitotic exit in higher organisms.
ACKNOWLEDGMENTS
We thank Bryan Gardner and Laura Furstenthal for superb technical support and advice; Adam Eldridge for careful reading of the manuscript and consultation as roving molecular biologist; A. Kaiser for help with typing the manuscript; Paul Chang and the Stearn's laboratory for antibodies and advice; and Neils Mailand, Claudia Lukas, Jiri Lukas, and Jiri Bartek for sharing unpublished information. This work was supported by National Institutes of Health grants GM-60439 and GM-54811 (to P.K.J.) and 5T32CA09151 (to Z.Z.), U.S. Public Health Service grant CA-09302, awarded by the National Cancer Institute, Department of Health and Human Services (to B.K.K.), and a Lieberman Fellowship (to B.K.K.).
Footnotes
DOI: 10.1091/mbc.01–11–0535.
REFERENCES
- Balasubramanian MK, McCollum D, Surana U. Tying the knot: linking cytokinesis to the nuclear cycle. J Cell Sci. 2000;113:1503–1513. doi: 10.1242/jcs.113.9.1503. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- Carmena M, Riparbelli MG, Minestrini G, Tavares AM, Adams R, Callaini G, Glover DM. Drosophila Polo kinase is required for cytokinesis. J Cell Biol. 1998;143:659–671. doi: 10.1083/jcb.143.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- Chang P, Stearns T. δ-Tubulin and ε-tubulin: two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat Cell Biol. 2000;2:30–35. doi: 10.1038/71350. [DOI] [PubMed] [Google Scholar]
- Cueille N, Salimova E, Esteban V, Blanco M, Moreno S, Bueno A, Simanis V. Flp1, a fission yeast orthologue of the S. cerevisae CDC14 gene, is not required for cyclin degradation or rum1p stabilization at the end of mitosis. J Cell Sci. 2001;114:2649–2664. doi: 10.1242/jcs.114.14.2649. [DOI] [PubMed] [Google Scholar]
- Dundr M, Misteli T, Olson MO. The dynamics of postmitotic reassembly of the nucleolus. J Cell Biol. 2000;150:433–446. doi: 10.1083/jcb.150.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Fisk HA, Winey M. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106:95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J Cell Biol. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furstenthal L, Kaiser BK, Swanson C, Jackson PK. Cyclin E uses Cdc6 as a chromatin-associated receptor required for DNA replication. J Cell Biol. 2001;152:1267–1278. doi: 10.1083/jcb.152.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Sluder G. “It takes two to tango”: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 2001;15:1167–1181. doi: 10.1101/gad.894001. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Tinker-Kulberg RL, Morgan DO. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey KR, Jackson PK, Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proc Natl Acad Sci USA. 1999;96:2817–2822. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Frenz LM, Wells NJ, Johnson AL, Johnston LH. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr Biol. 2001;11:784–788. doi: 10.1016/s0960-9822(01)00228-7. [DOI] [PubMed] [Google Scholar]
- Lemaitre JM, Geraud G, Mechali M. Dynamics of the genome during early Xenopus laevis development: karyomeres as independent units of replication. J Cell Biol. 1998;142:1159–1166. doi: 10.1083/jcb.142.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ernsting BR, Wishart MJ, Lohse DL, Dixon JE. A family of putative tumor suppressors is structurally and functionally conserved in humans and yeast. J Biol Chem. 1997;272:29403–29406. doi: 10.1074/jbc.272.47.29403. [DOI] [PubMed] [Google Scholar]
- Li L, Ljungman M, Dixon JE. The human Cdc14 phosphatases interact with and dephosphorylate the tumor suppressor protein p53. J Biol Chem. 2000;275:2410–2414. doi: 10.1074/jbc.275.4.2410. [DOI] [PubMed] [Google Scholar]
- Luca FC, Mody M, Kurischko C, Roof DM, Giddings TH, Winey M. Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol Cell Biol. 2001;21:6972–6983. doi: 10.1128/MCB.21.20.6972-6983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Lukas C, Kaiser BK, Jackson PK, Bartek J, Lukas J. Deregulated human Cdc14A phosphatase disrupts centrosome separation and chromosome segregation. Nat Cell Biol. 2002;4:318–322. doi: 10.1038/ncb777. [DOI] [PubMed] [Google Scholar]
- McCollum D, Gould KL. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 2001;11:89–95. doi: 10.1016/s0962-8924(00)01901-2. [DOI] [PubMed] [Google Scholar]
- Menssen R, Neutzner A, Seufert W. Asymmetric spindle pole localization of yeast Cdc15 kinase links mitotic exit and cytokinesis. Curr Biol. 2001;11:345–350. doi: 10.1016/s0960-9822(01)00095-1. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53(−/−) cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A, Fiore F, Eytan E, Carrano AC, Draetta GF, Hershko A, Pagano M. Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes Dev. 1999;13:1181–1189. doi: 10.1101/gad.13.9.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW, Solomon MJ, Kirschner MW. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989;339:280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- Noton E, Diffley JF. CDK inactivation is the only essential function of the APC/C and the mitotic exit network proteins for origin resetting during mitosis. Mol Cell. 2000;5:85–95. doi: 10.1016/s1097-2765(00)80405-0. [DOI] [PubMed] [Google Scholar]
- Okuda M, et al. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- Piel M, Nordberg J, Euteneuer U, Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Charbonneau H, Deshaies RJ. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Song S, Grenfell TZ, Garfield S, Erikson RL, Lee KS. Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures. Mol Cell Biol. 2000;20:286–298. doi: 10.1128/mcb.20.1.286-298.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Lee KS. A novel function of Saccharomyces cerevisiae CDC5 in cytokinesis. J Cell Biol. 2001;152:451–469. doi: 10.1083/jcb.152.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T. Centrosome duplication. a centriolar pas de deux. Cell. 2001;105:417–420. doi: 10.1016/s0092-8674(01)00366-x. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- Taylor GS, Liu Y, Baskerville C, Charbonneau H. The activity of Cdc14p, an oligomeric dual specificity protein phosphatase from Saccharomyces cerevisiae, is required for cell cycle progression. J Biol Chem. 1997;272:24054–24063. doi: 10.1074/jbc.272.38.24054. [DOI] [PubMed] [Google Scholar]
- Tokuyama Y, Horn HF, Kawamura K, Tarapore P, Fukasawa K. Specific phosphorylation of nucleophosmin on Thr (199) by cyclin-dependent kinase 2-cyclin E and its role in centrosome duplication. J Biol Chem. 2001;276:21529–21537. doi: 10.1074/jbc.M100014200. [DOI] [PubMed] [Google Scholar]
- Trautmann S, Wolfe BA, Jorgensen P, Tyers M, Gould KL, McCollum D. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr Biol. 2001;11:931–940. doi: 10.1016/s0960-9822(01)00268-8. [DOI] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]