Abstract
GATA-1 is essential for the generation of the erythroid, megakaryocytic, eosinophilic and mast cell lineages. It acts as an activator and repressor of different target genes, for example, in erythroid cells it represses cell proliferation and early hematopoietic genes while activating erythroid genes, yet it is not clear how both of these functions are mediated. Using a biotinylation tagging/proteomics approach in erythroid cells, we describe distinct GATA-1 interactions with the essential hematopoietic factor Gfi-1b, the repressive MeCP1 complex and the chromatin remodeling ACF/WCRF complex, in addition to the known GATA-1/FOG-1 and GATA-1/TAL-1 complexes. Importantly, we show that FOG-1 mediates GATA-1 interactions with the MeCP1 complex, thus providing an explanation for the overlapping functions of these two factors in erythropoiesis. We also show that subsets of GATA-1 gene targets are bound in vivo by distinct complexes, thus linking specific GATA-1 partners to distinct aspects of its functions. Based on these findings, we suggest a model for the different roles of GATA-1 in erythroid differentiation.
Keywords: chromatin, GATA-1, hematopoiesis, repression, transcription factors
Introduction
Hematopoiesis has served as a model for cellular commitment and differentiation. Hematopoietic stem cells (HSCs) commit to a number of divergent yet narrowly defined lineages, each giving rise to a specific type of blood cell. Lineage commitment involves the upregulation of a particular transcription program with the concomitant suppression of ‘multipotentiality' and transcriptional programs specifying alternative lineages (Orkin, 2000).
GATA-1 is a key regulator of the differentiation of the erythroid, megakaryocytic, eosinophilic and mast cell lineages and is the founding member of the GATA family of zinc-finger factors implicated in the development and differentiation of several cell types. Efforts to understand GATA-1 functions have identified a number of protein interactions with transcription factors, such as TAL-1 (and its associated proteins Ldb1, LMO2 and E2A), EKLF, PU.1 and Sp1 (reviewed by Cantor and Orkin, 2002). GATA-1 is also reported to interact with chromatin remodeling/modification proteins, including the CBP/p300 histone acetyltransferases (HATs) and the SWI/SNF chromatin remodeling complex (Blobel et al, 1998; Kadam and Emerson, 2003). Prominent among the GATA-1 interacting partners is FOG-1, originally identified in a yeast two-hybrid screen (Tsang et al, 1997). A direct interaction between the two factors is required for erythroid differentiation (Crispino et al, 1999) and the GATA-1 and FOG-1 knockout phenotypes are very similar (Tsang et al, 1998).
GATA-1 functions as both an activator and a repressor. The GATA-1/FOG-1 complex has been shown to repress some genes, such as GATA-2, and activate others, such as β-globin or the EKLF gene (Anguita et al, 2004; Letting et al, 2004; Pal et al, 2004). GATA-1 has also been linked to the repression of genes with cell proliferation functions, for example, myc and myb (Rylski et al, 2003), although its protein partners are unknown. The multimeric GATA-1/TAL-1/Ldb1/E2A/LMO2 complex binds to closely spaced GATA and E-box binding motifs and has been associated with the activation of erythroid genes, such as glycophorin A and the α-globin locus (Anguita et al, 2004; Lahlil et al, 2004). The duality of GATA-1 as an activator and repressor has been reinforced by the recent microarray analysis of terminal erythroid differentiation following induction of GATA-1 expression (Welch et al, 2004). Despite all this information, important questions remain as to how can GATA-1 accommodate all these functions and interactions at the same time in erythroid cells? In addressing this, we have undertaken a biotinylation tagging–proteomics approach to characterize GATA-1 complexes from erythroid cells (de Boer et al, 2003). We show that GATA-1 forms distinct complexes with hematopoietic transcription factors and chromatin remodeling and modification complexes. Our findings provide an explanation for a number of previous observations regarding GATA-1 functions and protein interactions. In addition, we provide evidence for distinct GATA-1 complexes performing specific functions in erythroid cells, thus providing a new framework for future work on GATA-1 and its parallel functions in erythroid differentiation.
Results
Identification of GATA-1 complexes from erythroid cells by biotinylation tagging and mass spectrometry
GATA-1 was tagged by fusing of a small (23 aa) peptide sequence to its N-terminus. This tag is efficiently biotinylated by the bacterial BirA biotin ligase, which is coexpressed in stably transfected mouse erythroleukemic (MEL) cells, allowing a single-step purification of biotinylated GATA-1 using streptavidin beads under mild conditions (de Boer et al, 2003). Proteins copurified with GATA-1 from MEL cells chemically induced to undergo terminal differentiation were identified by mass spectrometry, classified according to Gene Ontology terms or by BLAST searches and compared to the background (Supplementary Table 1). Additional experiments employed more stringent conditions and different nuclear extract preparations from induced MEL cells. We rejected proteins that appeared in the background binding experiments (de Boer et al, 2003), or proteins that belonged to a subnuclear compartment from which GATA-1 is excluded, for example, the nucleolus (Elefanty et al, 1996). Streptavidin pull-downs of nuclear extracts under more stringent conditions (Figure 1A–C) and immunoprecipitations of induced nontransfected MEL nuclear extracts provided further validation (Figure 1D–G). The identities of proteins confirmed in this way as copurifying with GATA-1 are shown in Table I.
Figure 1.
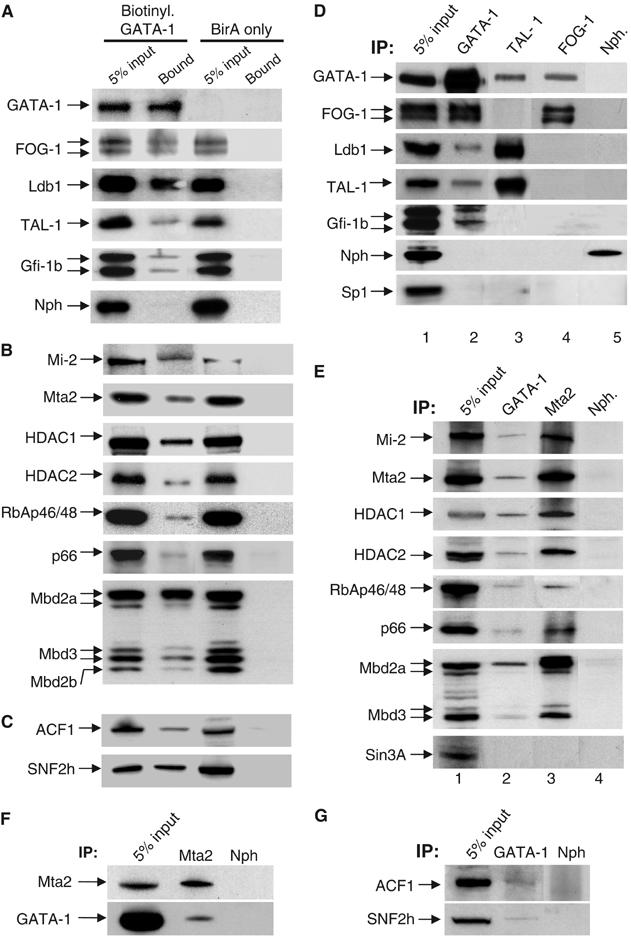
Confirmation by streptavidin pull-downs (A–C) and immunoprecipitations (D–G) of proteins identified copurifying with GATA-1. (A) Streptavidin pull-downs of transcription factors. Biotinylated GATA-1 (top panel) is detected by streptavidin–HRP and is absent from the BirA-only transfected cells. (B) Pull-downs of the MeCP1 complex. (C) Pull-downs of ISWI-containing complexes. SB: streptavidin-bound. (D) Immunoprecipitations (IP) using antibodies against GATA-1, TAL-1, FOG-1 (lanes 2, 3 and 4, respectively) and nucleophosmin as negative control (lane 5). (E) IP of the MeCP1 complex by antibodies against GATA-1 and MTA2 (lanes 2 and 3) and nucleophosmin (lane 4). (F) GATA-1 can be specifically immunoprecipitated by an antibody against MTA2. (G) IP of the ACF/WCRF complex by GATA-1 antibodies. Nuclear extract equivalent to 5% used in each pull-down or IP was loaded as control for input material. IP: immunoprecipitating antibody. Arrows show the detecting antibodies.
Table 1.
Proteins specifically copurifying with biotin-tagged GATA-1 as compared to the control purification (de Boer et al, 2003)
| Protein identity | Number of peptides | Additional purifications |
|---|---|---|
| MeCP1 complex | ||
| Mi-2 | 82 | + |
| HDAC 1 | 10 | + |
| HDAC 2 | 17 | + |
| MTA1 | 47 | + |
| MTA2 | 10 | + |
| MTA3 | 4 | − |
| Mbd2 | 14 | + |
| Mbd3 | 9 | + |
| p66 | 11 | + |
| RbAp46 | 10 | + |
| RbAp48 | 8 | + |
| ACF/WCRF complex | ||
| SNF2h | 21 | + |
| ACF1 | 4 | − |
| Transcription factors | ||
| FOG-1 (Hem.) | 47 | + |
| TAL-1 (Hem.) | 2 | + |
| Gfi-1b (Hem.) | 1 | − |
| Ldb1 (Ubiq.) | 1 | + |
| DNA repair | ||
| Rfc5 | 10 | + |
| XRCC1 | 3 | − |
| Ku70 | 9 | + |
| PARP | 10 | + |
| DNA ligase III-β | 1 | − |
| DNA topological change | ||
| DNA Topo I | 34 | + |
| DNA Topo II α | 64 | + |
| DNA Topo II β |
32 |
− |
| A number of these proteins have been validated by immunoprecipitations and other assays (see text). Hem: hematopoietic transcription factors; Ubiq: ubiquitous transcription factors. | ||
Finding FOG-1, TAL-1 and Ldb1 copurifying with GATA-1 (Tsang et al, 1997; Wadman et al, 1997) validated our approach. The Gfi-1b hematopoietic transcription factor was also identified under moderate stringency conditions and verified by immunoprecipitation (Figure 1D, lane 2) demonstrating an interaction between the two factors. This is in line with the similarities observed in the Gfi-1b and GATA-1 knockout phenotypes, which result in differentiation arrest of the erythroid and megakaryocytic lineages (Pevny et al, 1995; Saleque et al, 2002). Chromatin remodeling and modification proteins also coeluted with GATA-1 (Table I) including the entire MeCP1 complex (Figure 1). MeCP1 consists of the methyl-DNA binding protein MBD2 (Feng and Zhang, 2001), p66/p68 (Feng et al, 2002) and the multi-subunit Mi-2/NuRD complex containing the nucleosome stimulated Mi-2β ATPase, the histone deacetylases HDAC1 and HDAC2 and other subunits of unknown function. The Mi2/NuRD and MeCP1 complexes are associated with epigenetic mechanisms of repression during development (Ahringer, 2000), potentially linking the GATA-1 repressive functions to the MeCP1 complex.
The SNF2h and ACF1 members of mammalian ISWI chromatin remodeling complexes also copurified with GATA-1 (Table I and Figure 1C and G). SNF2h, a homolog of the Drosophila protein ISWI, is the ‘signature' ATPase of this class of complexes and participates in three distinct complexes in human cells: RSF, hACF/WCRF and hCHRAC (reviewed by Corona and Tamkun, 2004). We did not detect by mass spectrometry or immunoprecipitation (not shown) the additional p15 and p17 protein partners present in the hCHRAC complex; hence, GATA-1 appears to interact with SNF2h/ACF1 in the context of the ACF/WCRF complex (Bochar et al, 2000). ISWI/SNF2h-containing chromatin remodeling complexes have been associated with gene activation and repression (reviewed by Corona and Tamkun, 2004). The interaction between SNF2h and GATA-1 may help explain the observation that knocking down SNF2h expression in primary hematopoietic progenitor cells blocked erythroid differentiation (Stopka and Skoultchi, 2003).
Further validation for the GATA-1 interactions was provided by reverse immunoprecipitations using antibodies against TAL-1, FOG-1 (Figure 1D) or MTA2 (Figure 1F). TAL-1 antibodies specifically immunoprecipitated GATA-1 and Ldb1 (Figure 1D), as previously observed (Osada et al, 1995; Visvader et al, 1997). LMO2 or E2A were not detected copurifying with GATA-1 from induced MEL cells, but it cannot be excluded that their absence is due to the very low abundance of the GATA-1/TAL-1/Ldb1/E2A/LMO2 complex (Table I), in agreement with previous reports of a very small fraction of LMO2 being immunoprecipitated by GATA-1 antibodies (Osada et al, 1995). Interestingly, FOG-1 antibodies immunoprecipitated GATA-1 but not TAL-1, Ldb1 or Gfi-1b (Figure 1D, lane 4). The converse was also true using TAL-1 antibodies (Figure 1D, lane 3). Thus, GATA-1 interactions with TAL-1, FOG-1 and Gfi-1b are non-overlapping and must occur in distinct complexes. GATA-1 was also immunoprecipitated by MTA2 antibodies (Figure 1F). By contrast, the Sin3A corepressor, which interacts with HDACs but not in the MeCP1 complex, was not immunoprecipitated by GATA-1 or MTA2 antibodies (Figure 1E), further supporting the specificity of the GATA-1 interactions with MeCP1. The participation of GATA-1 in multiple protein interactions is supported by size fractionation experiments of nuclear extracts, which showed that GATA-1 fractionates with a broad profile overlapping the profiles of the partners identified here (Supplementary Figure 1), also providing evidence for interactions occurring in distinct complexes (e.g., fractionation peaks of MeCP1 versus SNF2h/ACF1).
Other abundant chromatin-associated proteins also copurified with GATA-1 (Table I), including topoisomerases and Ku autoantigen or ADP ribosyltransferase (PARP). We tested the association of these proteins with DNA and GATA-1 by treating nuclear extracts with DNase I. In contrast to MTA2, there were no topoisomerase I or PARP copurifying with GATA-1 after DNase I treatment (Supplementary Figure 2). Although it remains formally possible that interactions of GATA-1 with topoisomerase I or PARP are relevant and require DNA, on the basis of our DNase I results and on previous evidence by other groups describing topoisomerases as a common contaminant (Eberharter et al, 2001), we did not pursue these further.
GATA-1 forms several distinct complexes
To confirm directly the distinct GATA-1 interactions and to assess how the GATA-1 partners may be partitioned in the GATA-1 complexes, we carried out sequential immunodepletion experiments. First, we used an antibody against one of the GATA-1 partners, that is, FOG-1, TAL-1 or MTA2, in order to immunodeplete from a nuclear extract the fraction of GATA-1 that is in complex with this factor (Figure 2A). The remaining GATA-1 in the supernatant was subsequently immunoprecipitated with a GATA-1 antibody and both immunoprecipitates were tested for the presence or absence of GATA-1 and interacting proteins (Figure 2A).
Figure 2.
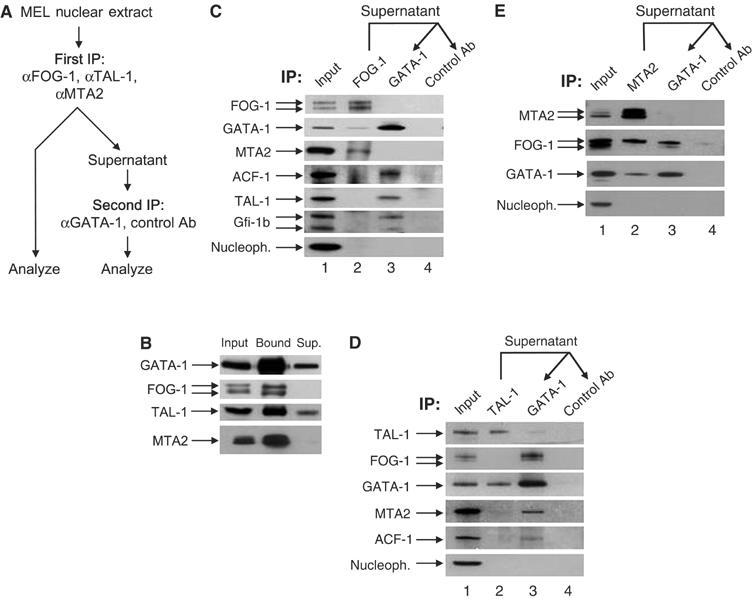
Distinct GATA-1 complexes by sequential immunoprecipitations (IP). (A) Experimental procedure. (B) Efficiency of immunoprecipitating antibodies (also used to detect the immunoprecipitated protein). Sup: supernatant after IP. (C) FOG-1 immunodepletion, and FOG-1 IP (lane 2) followed by IP of supernatant with GATA-1 or control antibodies (lanes 3 and 4). (D) TAL-1 immunodepletion; same as (C), using TAL-1 antibodies in first IP. (E) MTA2 immunodepletion; same as (C) and (D), using MTA2 antibodies in first IP. The MTA2 antibody used in panels B and E is different to that used in (C) and (D) (see Supplementary data). IP: immunoprecipitating antibody. Arrows show the detecting antibodies.
We first established that antibodies against GATA-1, TAL-1, FOG-1 and MTA2 were efficient in immunodepleting most of these proteins from nuclear extracts (Figure 2B). As expected, FOG-1 antibodies immunoprecipitated a fraction of GATA-1 (Figure 2C, lane 2). Surprisingly, MTA2 was also specifically immunoprecipitated by FOG-1 antibodies (Figure 2C, lane 2), suggesting an interaction between FOG-1 and the MeCP1 complex. This was confirmed by the reverse immunoprecipitation of FOG-1 by an MTA2 antibody (Figure 2E). There was no immunoprecipitation of TAL-1, Gfi-1b or ACF1 by FOG-1 antibodies (Figure 2C, lane 2). Importantly, MTA2 could no longer be detected in the subsequent immunoprecipitation of the supernatant with GATA-1 antibodies (Figure 2C, lane 3). Thus, the fraction of GATA-1 that is in complex with MTA2 (and MeCP1) was depleted in the first step by the FOG-1 antibodies, leading us to conclude that FOG-1 and GATA-1 interact together in the same complex with MeCP1. Further confirmation was provided by the immunoprecipitation of MeCP1-associated HDAC activity by GATA-1 and FOG-1 antibodies (Supplementary Figure 3). Following the FOG-1 immunodepletion, GATA-1 antibodies could still immunoprecipitate TAL-1, Gfi-1b and ACF1 (Figure 3C, lane 3). This confirms that GATA-1 participates in a complex with FOG-1 and MeCP1 that is distinct from that with TAL-1, Gfi-1b or ACF/WCRF. Using TAL-1 antibodies in the first immunodepletion step, a small fraction of GATA-1, but not MTA2 or ACF1, was immunoprecipitated (Figure 2D, lane 2).
Figure 3.
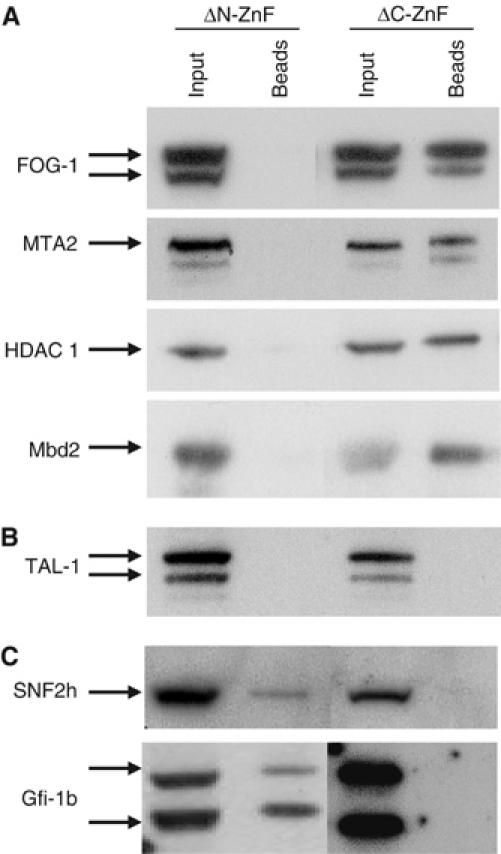
(A–C) Differential interactions mediated by the GATA-1 zinc-fingers. GATA-1 zinc-finger deletions were expressed as biotin-tagged proteins in MEL cells and interactions were assessed by streptavidin pull-downs and Western blots. (A) FOG-1 and the MeCP1 complex require N-ZnF for interactions. (B) TAL-1 requires both zinc-fingers. (C) Gfi-1b and SNF2h require C-ZnF for interactions with GATA-1. The TAL-1 antibody used is different from that used in Figure 1.
We also carried out an MTA2 immunodepletion to determine whether the entire fraction of GATA-1 interacting with FOG-1 does so in the context of the MeCP1 complex. Following the immunodepletion of MTA2 (Figure 2B), an appreciable amount of FOG-1 was subsequently immunoprecipitated by GATA-1 antibodies (Figure 2E, lane 3). Thus, GATA-1 also interacts with FOG-1 independently of the MeCP1 complex. Interestingly, the MTA2 antibody specifically immunoprecipitated the slower migrating of the two bands detected by the FOG-1 antibody, suggesting differential interaction with one of the two FOG-1 isoforms, while GATA-1 can interact with both FOG-1 isoforms (Figure 2E, lane 3). Taken together, these experiments show that GATA-1 forms at least five complexes: first with FOG-1 and MeCP1, second with FOG-1 alone, third with TAL-1 (and Ldb1 since it can be almost completely immunodepleted by TAL-1 antibodies (not shown)), fourth with Gfi-1b and fifth with ACF/WCRF.
The GATA-1 zinc-fingers mediate differential protein interactions
GATA-1 contains two evolutionarily conserved, closely spaced zinc-finger domains. The C-terminal zinc-finger (C-ZnF) is essential for DNA binding, whereas the N-terminal zinc-finger (N-ZnF) is primarily involved in protein–protein interactions, for example with FOG-1, which contribute to the specificity and stability of DNA binding by C-ZnF (reviewed by Blobel and Weiss, 2001). Significantly, C-ZnF is essential for all in vivo GATA-1 functions, whereas N-ZnF is required for definitive, but not primitive, erythropoiesis (Shimizu et al, 2001). We addressed how the GATA-1 zinc-fingers mediated its multiple protein interactions by expressing in MEL cells biotin-tagged mutants lacking N-ZnF or C-ZnF followed by streptavidin pull-downs (Figure 3A–C). As described (Tsang et al, 1998), GATA-1 interaction with FOG-1 requires N-ZnF. Interactions of the MeCP1 members MTA2, Mbd2 and HDAC 1 also occur through the N-ZnF of GATA-1 (Figure 3A). Interestingly, interactions of GATA-1 with TAL-1 require both zinc-fingers (Figure 3B), whereas interactions with SNF2h or Gfi-1b require only C-ZnF (Figure 3C). We tested by immunoprecipitation using Gfi-1b antibodies whether Gfi-1b and SNF2h were in complex but found no evidence of such an interaction (not shown). Thus, the multiple, distinct interactions of GATA-1 are differentially mediated through its zinc-finger domains.
FOG-1 mediates interactions of GATA-1 with the MeCP1 complex in repressing transcription
We next tested whether FOG-1 mediates interactions between GATA-1 and the MeCP1 complex. GATA-1 was transiently expressed in HeLa cells (which express endogenous MeCP1, but not GATA-1 or FOG-1) with or without FOG-1, followed by immunoprecipitation using FOG-1 or MTA2 antibodies (Figure 4A and B). We find that the interaction of GATA-1 with MTA2 occurs only in the presence of FOG-1 (Figure 4B), whereas FOG-1 interacts with MTA2 regardless of the presence or absence of GATA-1 (Figure 4B, upper panel). Expression of the GATA-1 zinc-finger deletion mutants (Figure 4B, lower panel) confirmed these observations. We conclude that interaction of GATA-1 with the MeCP1 complex requires interaction with FOG-1, which thus serves as the bridging factor.
Figure 4.
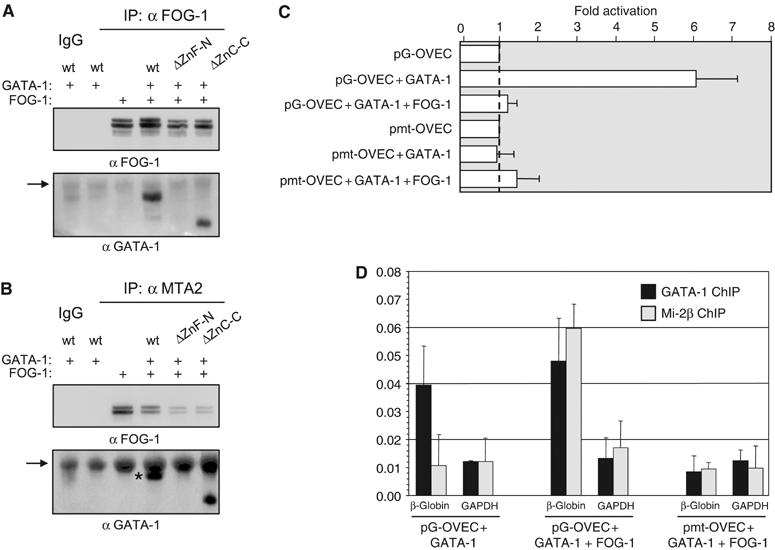
(A–B) FOG-1 bridges GATA-1 and MeCP1. Nuclear extracts from HeLa cells transfected with the FOG-1 and GATA-1 combinations indicated were immunoprecipitated with FOG-1 (A) or MTA2 antibodies (B) and detected with FOG-1 and GATA-1 antibodies. ΔZn-N and ΔZn-C: GATA-1 N- and C-terminal zinc-finger deletion mutants. Arrows: crossreacting IgG. Asterisk (B): GATA-1 signal. (C) Real-time PCR transcription assays in transfected HeLa cells. GATA-1 activates transcription of pG-OVEC, whereas cotransfection of FOG-1 represses to basal levels. (D) Specific recruitment of Mi-2β by cotransfected GATA-1 and FOG-1 by ChIP assays in HeLa cells. Mi-2β recruitment to the repressed gene requires GATA-1 binding to the promoter.
We next tested whether the well-known GATA-1- and FOG-1-mediated repression is due to the recruitment of the MeCP1 complex to a GATA-dependent promoter. To this end, we used a reporter plasmid containing the rabbit β-globin minimal promoter (pOVEC-1; Westin et al, 1987), carrying four copies of an optimal GATA-1 binding sequence, or four copies of a mutated sequence that abolishes GATA-1 binding (Whyatt et al, 1993). The GATA-binding promoter was activated more than six-fold by cotransfection of GATA-1 alone (Figure 4C). As expected, cotransfection of FOG-1 and GATA-1 repressed activation of the GATA-dependent promoter (Figure 4C). Chromatin immunoprecipitation (ChIP) showed that repression by GATA-1 and FOG-1 was due to the specific recruitment of the MeCP1 complex. Binding of Mi-2β to the repressed gene was specifically enriched in GATA-1- and FOG-1-transfected cells (Figure 4D), but not in cells transfected with GATA-1 only. The promoter bearing the mutated GATA binding sites does not bind MeCP1, even in the presence of FOG-1 (Figure 4D). Thus, FOG-1/MeCP1 repression is mediated through GATA-1 binding at its cognate binding sites.
GATA-1, FOG-1, MeCP1 and Gfi-1b are bound to repressed genes in vivo
The results above suggest that GATA-1/FOG-1 interactions can tether MeCP1 to repressed GATA-1 target sequences in vivo. We therefore employed ChIP assays using the GATA-2 locus, which is repressed by GATA-1 in a FOG-1-dependent manner (Grass et al, 2003; Pal et al, 2004). As seen before, the −2.8 kb region upstream of the GATA-2 promoter was enriched for GATA-1 and FOG-1 binding (Figure 5A). The same sequence was also enriched for Mbd2 binding (Figure 5A), with similar results obtained with an antibody against Mi-2β (Supplementary Figure 4A). No binding of TAL-1 or Gfi-1b was observed in the GATA-2 sequences. Interestingly, the −3.4 kb, but not the −4.2 and −2.2 kb flanking sequence used as negative controls for GATA-1 binding, was enriched for FOG-1 and Mbd2 binding (Figure 5A and Supplementary Figure 4A), suggesting that the FOG-1/MeCP1 binding at −3.4 kb may reflect a very localized spreading of these proteins over a few nucleosomes to sequences upstream of the −2.8 kb element, or that they were accidentally crosslinked to neighboring DNA. The latter possibility would suggest that the FOG-1/MeCP1 complex is closer to the upstream sequences around the GATA binding sites (see also below). We found no evidence that the binding of the GATA-1/FOG-1/MeCP1 complex to the −2.8 kb region was mediated by DNA methylation (Supplementary Figure 5); however, this does not exclude the possibility of highly localized methylation to specific CpG residues elsewhere in the GATA-2 locus. Thus, considering that GATA-1 binding is essential for GATA-2 repression (Pal et al, 2004), our findings strongly suggest that GATA-1, FOG-1 and MeCP1 form the repressive complex responsible for GATA-2 silencing (see below).
Figure 5.
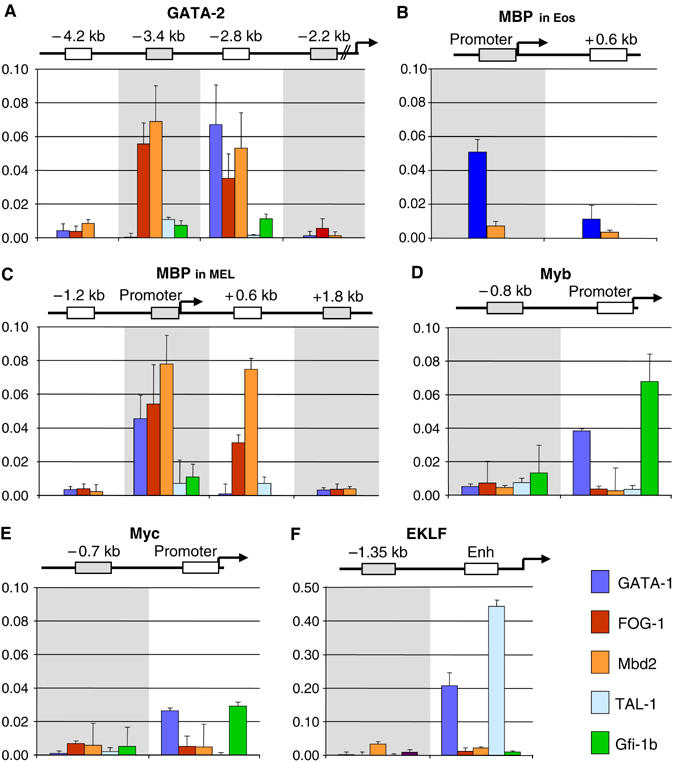
Binding of GATA-1 repressive and activating complexes to target genes by ChIP assays in induced MEL cells. Binding patterns of GATA-1, FOG-1, Mbd2, TAL-1 and Gfi-1b to the −2.8 kb element of the GATA-2 locus (A), the MBP promoter in eosinophils (B) and in MEL cells (C), the myb (D) and myc (E) promoters and at the EKLF upstream enhancer (F). Relative enrichment has been normalized to input and corrected for background binding of species- and isotype-matched immunoglobulins. Antibodies: GATA-1, N6 (Santa Cruz); FOG-1 as in Tsang et al (1997); Mbd2 S923 (Ng et al, 1999); TAL-1 as in Porcher et al (1996); Gfi-1b, D19 (Santa Cruz).
Ectopic expression of FOG-1 in eosinophilic cells results in the downregulation of eosinophilic GATA-1 target genes and the reprogramming of these cells toward an earlier, less-differentiated cell type that may represent a common progenitor for the erythroid/megakaryocytic and eosinophilic lineages (Querfurth et al, 2000). We thus reasoned that eosinophilic GATA-1 target genes, like the major basic protein (MBP) (Du et al, 2002), which is inactive in erythroid cells (Welch et al, 2004), may be suppressed by the GATA-1/FOG-1/MeCP1 complex. We tested this hypothesis by ChIP in induced MEL cells using as control chromatin from mouse eosinophils where MBP is expressed (Guyot et al, 2004). As expected, the promoter of the MBP gene was bound by GATA-1 in eosinophils (Figure 5B). Importantly, GATA-1 was also bound to the inactive MBP promoter in induced MEL cells (Figure 5C). FOG-1 and Mbd2 were also bound to the MBP promoter in MEL cells but not in eosinophils (Figure 5C), consistent with the prediction above. Similar results were also obtained with an antibody against Mi-2β (Supplementary Figure 4). Again, no TAL-1 of Gfi-1b binding was detected in the MBP promoter (Figure 5C). Strikingly, in MEL cells, we again found binding of the FOG-1 and MeCP1 complex, but not of GATA-1, to the +0.6 kb sequence located close to the MBP promoter but not to other sequences located further upstream (−1.8 kb) or downstream (+1.2kb) of the promoter (Figure 5C and Supplementary Figure 4B). This observation is similar to that seen at the GATA-2 −2.8 kb element.
We next tested the myc and myb genes, which are downregulated with MEL differentiation (Lachman and Skoultchi, 1984; Chen and Bender, 2001, and references therein). Repression of the myc and myb genes has been linked to the proliferation arrest that accompanies terminal erythroid differentiation. The myc gene has also been shown to be a GATA-1 target gene in G1E cells (Rylski et al, 2003; Welch et al, 2004). We found GATA-1 binding to both promoters in induced MEL cells but we could not detect binding of FOG-1 or Mbd2 to the same sequences (Figure 5D and E). By contrast, Gfi-1b (absent from all other genes tested) was found binding to both promoters (Figure 5D and E), suggesting a role for the GATA-1/Gfi-1b complex in the repression of genes associated with cell proliferation. This may explain the observations of rapidly proliferating Gfi-1b−/− immature erythroid precursors in colony assays (Saleque et al, 2002) and of Gfi-1b overexpression inducing proliferation arrest and differentiation in erythroid progenitors (Garcon et al, 2005).
Finally, we also tested the EKLF gene as an example of a gene that is activated during erythropoiesis. The EKLF enhancer sequence contains a GATA-E-box motif (Anderson et al, 1998), which is bound in vivo by GATA-1 independently of FOG-1 (Letting et al, 2004). Strong GATA-1 binding and a clear enrichment for TAL-1 binding were indeed detected at the EKLF enhancer (Figure 5F), thus providing a clear demonstration for the alternative (activating) GATA-1 complex with TAL-1 binding to a target gene in vivo. This may be related to the low level of HDAC activity associated with the TAL-1 immunoprecipitate (Supplementary Figure 3A). No significant binding of FOG-1, Mbd2 or Gfi-1b could be detected in the EKLF enhancer sequences (Figure 5F).
Our analysis of the GATA-1/ACF/WCRF complex by ChIP, or any other, assays has been hindered by the quality of ACF/WCRF reagents available to us; hence, it is presently not known whether the GATA-1 and ACF/WCRF complex binds to active or repressed genes.
GATA-1 represses GATA-2 expression through the recruitment of FOG-1 and MeCP1
In order to confirm that GATA-2 repression during erythroid differentiation is specifically due to GATA-1 recruiting FOG-1 and the MeCP1 complex, we took advantage of the GATA-1 null G1E proerythroblastic cell line. These cells are derived from in vitro-differentiated GATA-1 null ES cells and can undergo terminal differentiation only upon restoration of GATA-1 expression (Weiss et al, 1997). We used two G1E cell lines. The first one expresses wild-type GATA-1 fused to an estrogen receptor (ER) ligand binding domain (GATA-1-ER), which can mediate terminal erythroid differentiation upon induction by estradiol (Tsang et al, 1997). The second cell line expresses ER fused to a mutant GATA-1 form bearing a single V205M amino-acid substitution in the GATA-1 N-terminal zinc-finger. While not affecting GATA-1 DNA binding, this mutant abrogates interaction with FOG-1 and fails to rescue differentiation of G1E cells (Crispino et al, 1999; Nichols et al, 2000). We first determined that repression of the GATA-2 gene in G1E cells was absolutely dependent on GATA-1 being capable of interacting with FOG-1 (Figure 6A). We next tested by ChIP whether interaction of GATA-1 with FOG-1 binding at −2.8 kb was responsible for the recruitment of MeCP1 to this sequence and to the neighboring −3.4 kb sequence (Figure 5A). As control, we also tested the more distal −4.2 kb sequence, which did not show binding for any of these factors (Figure 5A). In agreement with the MEL data, GATA-1, FOG-1 and Mi-2β were bound to the −2.8 kb and to the −3.4 kb sequence (for FOG-1 and Mi-2β) in differentiated GATA-1-ER cells (24 h after induction with estradiol), albeit at lower levels compared to MEL cells (Figure 6B, left panels). By contrast, in the GATA-1(V205M)-ER expressing cells, GATA-1 was bound to the −2.8 kb sequence, but no binding of FOG-1 and Mi-2β to the −2.8 or −3.4 kb sequences was detected (Figure 6B, right panels). We conclude that FOG-1 and the MeCP1 complex are specifically recruited by GATA-1 to the GATA-2 locus and are responsible for GATA-2 repression in terminal erythroid differentiation.
Figure 6.
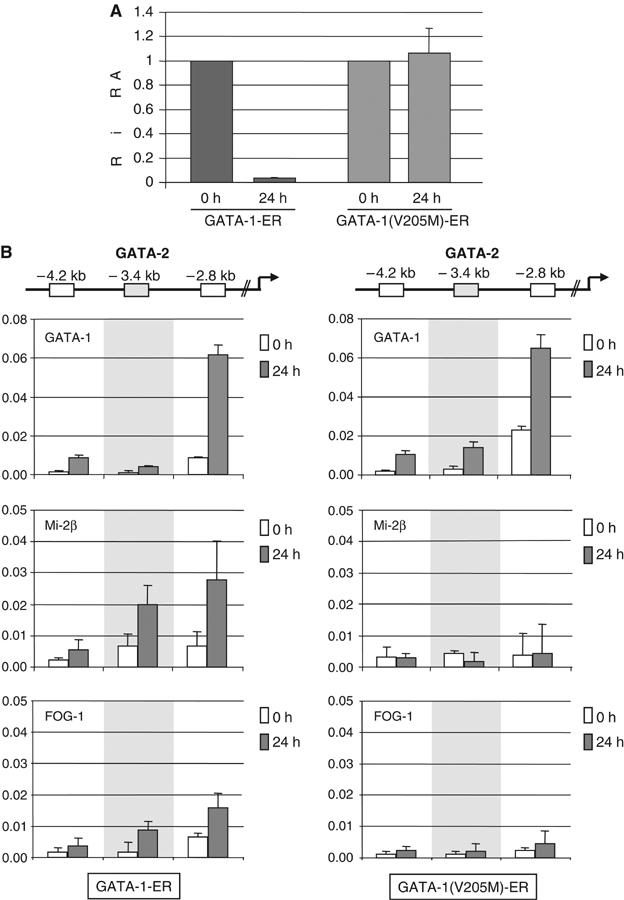
Silencing of GATA-2 requires recruitment of FOG-1 and MeCP1 by GATA-1. (A) The V205M GATA-1 mutation fails to repress GATA-2. GATA-2 mRNA was measured by real-time PCR in G1E GATA-1-ER and G1E GATA-1(V205M)-ER cells before (0 h) and after 24 h of estradiol induction. Expression at 0 h was normalized against GAPDH expression and set as 1. (B) ChIP to show binding of GATA-1, Mi-2β and FOG-1 in G1E GATA-1-ER (left panels) and in G1E GATA-1(V205M)-ER (right panels) at time 0 and 24 h of estradiol induction. Relative enrichment has been normalized to input and corrected for background binding of species- and isotype-matched immunoglobulins. Data represent mean of two independent IPs and three PCRs with duplicate samples. Antibodies used were as in the legend of Figure 5, except Mi-2β antibody (Fujita et al, 2003).
Discussion
We describe here the characterization of GATA-1 complexes from erythroid cells by in vivo biotinylation tagging and purification by streptavidin beads. This work has led to a number of important findings. First, we identified novel GATA-1 partners, including the essential hematopoietic factor Gfi-1b and the chromatin remodeling and modification complexes MeCP1 and ACF/WCRF, in addition to the known GATA-1 interacting factors FOG-1, TAL-1 and Ldb1. Second, we showed that GATA-1 forms several distinct complexes with FOG-1, FOG-1 and MeCP1, TAL-1/Ldb1, Gfi-1b and the ACF/WCRF complex. Third, we found that the most abundant of the GATA-1 complexes are those with FOG-1 and with FOG-1 and MeCP1, with FOG-1 serving as the bridging factor between GATA-1 and the MeCP1 complex. Fourth, we showed that the distinct interactions of GATA-1 with its protein partners are differentially mediated through the two GATA-1 zinc-finger domains. Fifth, we show that the known GATA-1- and FOG-1-mediated repression is due to the recruitment of the MeCP1 complex to the repressed gene(s). Sixth, we present evidence for the in vivo binding of the repressive GATA-1/FOG-1/MeCP1 complex to silenced hematopoietic genes in erythroid cells and of the activating GATA-1/TAL-1 complex to erythroid-specific genes. Significantly, we also showed binding of the GATA-1/Gfi-1b complex to genes associated with cell proliferation functions, which become repressed with erythroid differentiation. Finally, our work demonstrates the utility of biotinylation tagging as an efficient approach for the rapid isolation and identification by mass spectrometry of multiple protein complexes.
Biotinylation tagging and protein complex purification
From our previous work (de Boer et al, 2003) and the work described here, we show that background using biotinylation tagging consists of naturally biotinylated proteins, of abundant nuclear proteins such as splicing factors binding nonspecifically to the beads (de Boer et al, 2003) and, potentially, of abundant chromatin-associated proteins, such as topoisomerase I, which are indirectly pulled down with the tagged transcription factor (Supplementary Table 1). We have validated a number of the remaining proteins as being true GATA-1 partners, some of which represent low abundance or weaker GATA-1 interactions, for example, with TAL-1/Ldb1, Gfi-1b and ACF/WCRF. Importantly, purification required a single capture step.
We cannot be certain that we identified all GATA-1 complexes in differentiated MEL cells. Indeed, some of the size-fractionation profiles (Supplementary Figure 1) suggest that there may be additional protein partners that were not identified perhaps due to their very low abundance or instability. This may be the case for the multimeric GATA-1/TAL-1/Ldb1/E2A/LMO2 complex. Several lines of evidence have suggested the presence of this complex in erythroid cells binding to distinct E-box and GATA motifs spatially arranged 9–12 nucleotides apart (reviewed by Lecuyer and Hoang, 2004). Many erythroid genes identified to date contain such motifs, including GATA-1 itself, EKLF, glycophorin A and 4.2 protein (Lecuyer and Hoang, 2004). Evidence for the multimeric GATA-1/TAL-1 complex binding to erythroid genes in vivo, such as α globin and glycophorin A, has been provided recently by ChIP assays (Anguita et al, 2004; Lahlil et al, 2004). Nevertheless, we did not find any copurification of E2A or LMO2 with GATA-1 from induced MEL cells. The complementary isolation by biotinylation tagging of protein partners, such as TAL-1, will be informative in that respect and may also reveal additional protein partners.
Novel GATA-1 protein partners
We describe here, for the first time, an interaction of GATA-1 with the essential hematopoietic transcription factor Gfi-1b. This factor contains six C-terminal C2H2 zinc-fingers, which bind a defined DNA consensus sequence, and an N-terminal SNAG domain associated with repression (Duan and Horwitz, 2003; Doan et al, 2004). The Gfi-1b knockout is remarkably similar to that of GATA-1, that is, it shows embryonic lethality at E15 due to the developmental arrest of erythroid and megakaryocytic differentiation in the fetal liver (Saleque et al, 2002). Our data that GATA-1 and Gfi-1b (but not FOG-1 or MeCP1) are bound to the myb and to the myc promoters provide a basis for the similarities in the two knockouts (Pevny et al, 1995; Saleque et al, 2002). These data may also be related to the proliferation defects observed in GATA-1-overexpressing mice (Whyatt et al, 2000). It is important to note that although there are also similarities between the FOG-1 and Gfi-1b knockout phenotypes, we did not find FOG-1 and Gfi-1b to interact directly in induced MEL cells (Figure 2). Possibly, the two factors regulate common gene targets through distinct complexes and binding sites. Alternatively, the functions of GATA-1 with FOG-1 or Gfi-1b could be separate, for example, differentiation (FOG-1) versus proliferation arrest (Gfi-1b), with each function being essential for erythropoiesis.
We also describe, for the first time, interactions of GATA-1 with the MeCP1 and ACF/WCRF complexes, linking GATA-1 to repressive functions (with MeCP1) and chromatin structure. Previous evidence linking GATA-1 to chromatin structure involved interactions with the HATs CBP and p300 (Blobel et al, 1998) and in vitro experiments where GATA-1 cooperated with the SWI/SNF remodeling complex in transcriptional activation (Kadam and Emerson, 2003). However, we did not observe these interactions in our GATA-1 purification from induced MEL cells or in immunoprecipitations (data not shown).
Our observations on the interactions of GATA-1 (and FOG-1) with the MeCP1 complex add to previous reports linking MeCP1 (and the closely related NuRD complex) to transcription factors in hematopoiesis (Kim et al, 1999; O'Neill et al, 2000; Hutchins et al, 2002). The conditional knockout of Mi-2β in thymocytes revealed a requirement in different stages of T-cell maturation (Williams et al, 2004). In addition, the characterization of the MTA3 member of NuRD in B lymphocytes showed an interaction with BCL-6, a key repressor of the mature plasma cell transcription program (Fujita et al, 2004). It was thus suggested that MTA3 and the NuRD complex play a role in the maintenance of a population of less-differentiated ‘poised' B lymphocytes (Fujita et al, 2004). In contrast to these data, our data in erythroid cells suggest that the MeCP1 complex works with tissue-specific transcription factors to effect terminal differentiation by shutting down transcription programs associated with early multipotential (‘poised') states.
GATA-1 and FOG-1 interactions
Considerable evidence has linked GATA-1 functions to FOG-1 (reviewed by Blobel and Weiss, 2001; Cantor and Orkin, 2002). A single amino-acid change in the N-terminal zinc-finger of GATA-1, which abolishes interaction with FOG-1 (Crispino et al, 1999), resulted in lethality in mice due to severe anemia (Chang et al, 2002) and is associated with dyserythropoietic anemia in patients (Nichols et al, 2000). Our work suggests that the overlapping functions of GATA-1 and FOG-1 in erythropoiesis occur in the context of two distinct complexes, a GATA-1/FOG-1/MeCP1 complex and a GATA-1/FOG-1 complex. Clearly, the association of GATA-1 and FOG-1 with the MeCP1 complex provides the molecular basis for the well-documented repressive properties of GATA-1 and FOG-1 interactions (Crispino et al, 1999; Fox et al, 1999; Letting et al, 2004; Pal et al, 2004). Only the slower migrating isoform of FOG-1 (Figure 3) interacts with the GATA-1/MeCP1 complex, providing a potential mechanism for the selective formation of the GATA-1/FOG-1/MeCP1 complex.
We suggest that the separate GATA-1/FOG-1 complex without MeCP1 links GATA-1 with FOG-1 to transcriptional activation. For example, disruption of GATA-1 and FOG-1 interactions downregulates erythroid genes such as α and β globin, Band 3, DC11 and HD2 genes (Crispino et al, 1999; Letting et al, 2004). ChIP assays have also shown GATA-1 and FOG-1 to be bound in vivo to active genes such as the α globin locus and the GATA-1 gene itself (Anguita et al, 2004; Pal et al, 2004). Significantly, in the α globin locus, the GATA-1/FOG-1 complex occupies sites distinct from those occupied by the GATA-1/TAL-1/Ldb1 complex (Anguita et al, 2004), in agreement with our findings of distinct GATA-1 complexes.
Our finding that FOG-1 bridges GATA-1 to the repressive MeCP1 complex partly explains the common features of the GATA-1 and FOG-1 knockouts and the phenotypes caused by the single amino-acid change in the N-terminal zinc-finger of GATA-1 in mice and patients. In the GATA-1 knockout, FOG-1/MeCP1 cannot be tethered to target genes, whereas in the FOG-1 knockout, the interaction between GATA-1 and the MeCP1 complex cannot take place. In patients, the lack of interaction between GATA-1 and FOG-1 would also fail to tether the MeCP1 complex to some of their target genes.
GATA-1 complexes and erythropoiesis
An important aspect in hematopoietic development to a particular lineage is the suppression of alternative ‘primed' lineage transcription programs and of genes that maintain multipotentiality, while upregulating genes associated with the differentiated cell type (Enver et al, 1998; Orkin, 2000). In addition, erythroid terminal differentiation is accompanied by cell cycle arrest. GATA-1 has been implicated in the regulation of most of these aspects (Blobel and Weiss, 2001). In fact, a recent microarray analysis of GATA-1-dependent erythroid terminal maturation revealed an early wave of repression of genes like GATA-2, myc and myb, followed by the upregulation of erythroid-specific genes (Welch et al, 2004). Here, we identified two GATA-1 repressive complexes acting on distinct sets of genes. Thus, we suggest that the GATA-1/Gfi-1b complex acts early and suppresses genes involved in cell proliferation, for example, myc and myb, while the GATA-1/FOG-1/MeCP1 complex also acts early to suppress genes required to maintain the ‘primed' multipotential state, for example, GATA-2 and alternative hematopoietic lineage genes, for example, MBP (Figure 7). In contrast, the GATA-1/FOG-1 and the GATA-1/TAL-1/Ldb1 complexes would play a major role in the later upregulation of erythroid genes (Figure 7). The role of the GATA-1/ACF/WCRF complex remains to be established. Thus, GATA-1 provides specific early versus late differentiation functions in the context of distinct complexes (Figure 7). The model of different GATA-1 complexes executing specific tasks in different stages of erythroid differentiation suggests a dynamic aspect in the GATA-1 complex interactions during differentiation and also raises the prospect of dissecting the contribution of distinct GATA-1 interactions in erythropoiesis (i.e. essential versus dispensable) by selectively manipulating a specific GATA-1 complex at a time.
Figure 7.
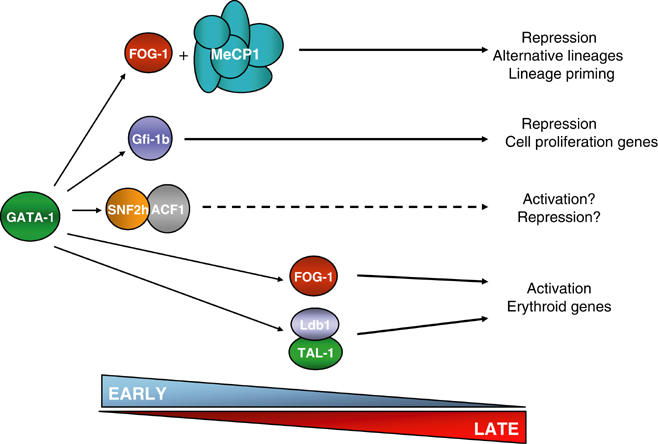
Model for the distinct GATA-1 complexes and their role in erythropoiesis. The broken arrow indicates unknown function and timing. See text for explanation.
Materials and methods
Constructs, nuclear extract preparation, streptavidin binding, mass spectrometry and immunoblot analysis
Tagged constructs and procedures involving MEL cells, biotinylated proteins and mass spectrometry were previously described (de Boer et al, 2003). The GATA-1 zinc-finger deletions have been described (Whyatt et al, 1993). G1E cells and induction were described (Tsang et al, 1997; Weiss et al, 1997).
Superose 6 gel filtration
Size fractionation of protein complexes was carried out on an AKTA FPLC apparatus with a Superose 6 10/30 column (Amersham Biosciences, Piscataway, NJ). Fractions were precipitated with 100% trichloroacetic acid and analyzed by Western immunoblotting, as described (de Boer et al, 2003). Molecular size standards were thyroglobulin (670 kDa) and albumin (66 kDa) (Amersham Biosciences, Piscataway, NJ).
Immunoprecipitations
Nuclear extracts were precleared at 4°C using Protein G Sepharose beads and affinity-purified IgG (rat (Santa Cruz, CA, sc-2026), rabbit (Santa Cruz, sc-2027), goat (Santa Cruz, sc-2028)) in HENG150 buffer. GATA-1 and TAL-1 antibodies were crosslinked to beads using dimethyl pimelimidate. Immunoprecipitations were performed in HENG150/0.3% NP-40 buffer overnight at 4°C using protein-G Sepharose beads. Washes were carried out at room temperature in HENG250/0.3% NP-40 buffer. Bound material was eluted by boiling in 1 × Laemmli buffer.
HeLa transient transfection and transcription assays
GATA-1 and FOG-1 cDNAs cloned in pCDNA 3.1 (Invitrogen, Carlsbad, CA) were transiently transfected using 2 μg DNA and Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Cells were harvested after 24 h and nuclear extracts were used for immunoprecipitations as above. pEGFP-N1 (Invitrogen, Carlsbad, CA) was included as transfection efficiency control. Transcription was assayed by real-time PCR with primers for exon 2 of the pOVEC reporter plasmid. ChIP assays were performed as below using GATA-1 and Mi-2β antibodies. The endogenous human GAPDH gene was used as control.
ChIP assays
Preparation of crosslinked chromatin (2 × 107 induced MEL cells treated with 0.4% formaldehyde for 10 min at room temperature), sonication to 300–800 bp fragments and immunoprecipitations were as described in the Upstate protocol (http://www.upstate.com). Anti-GATA-1 protein–DNA immunocomplexes were immunoprecipitated in an additional step with an AffiniPure rabbit anti-rat antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). Eosinophilic chromatin was prepared as previously described (Guyot et al, 2004). At least two independent ChIP assays were carried out per experiment. Antibodies used were as follows: GATA-1, N6 (Santa Cruz); Mbd2, S923 sheep polyclonal (Ng et al, 1999) and rabbit polyclonal anti-Mbd2/3 antibody (Upstate 07-199); FOG-1 rabbit polyclonal (Tsang et al, 1997); TAL-1 rabbit polyclonal (Porcher et al, 1996); Gfi-1b D19 goat polyclonal (Santa Cruz sc-8559).
Real-time PCR
Quantitative real-time PCR (Opticon I, MJ Research) was performed using SYBR Green I. PCR primers were designed by Primer Express 2.0 (PE Applied Biosystems). The qPCR Core Kit (Eurogentec, Belgium) was used with 400 nM of each primer under the following cycling conditions: 95°C for 10 min, 40 cycles of 30 s at 95°C, 60 s at 60°C and 15 s at 75°C. Enrichment for a specific DNA sequence was calculated using the comparative CT method (Litt et al, 2001). PCR primer sequences are provided in Supplementary data.
Antibodies
See Supplementary data.
Supplementary Material
Supplementary Information
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Table 1
Acknowledgments
We are grateful to our many colleagues cited in Supplementary data for generously sharing reagents. We are also grateful to Karim Bouazoune and Alexander Brehm (Adolf Butenandt Institut, Munich) for help with the HDAC assays, Mitch Weiss (Children's Hospital of Philadelphia) for G1E cells, Sjaak Philipsen and Marieke von Lindern (Erasmus MC) for comments on the manuscript and Raymond Poot (CSC, London) for helpful discussions. We are grateful to Gerd Blobel (Children's Hospital of Philadelphia) for exchanging prepublication data. This work has been supported by the Netherlands Research Organization (NWO), the European Union (grant HPRN-CT-2000-00078), the NIH (grant RO1 HL 073455-01) and the Netherlands Proteomics Center.
References
- Ahringer J (2000) NuRD and SIN3 histone deacetylase complexes in development. Trends Genet 16: 351–356 [DOI] [PubMed] [Google Scholar]
- Anderson KP, Crable SC, Lingrel JB (1998) Multiple proteins binding to a GATA-E box-GATA motif regulate the erythroid Kruppel-like factor (EKLF) gene. J Biol Chem 273: 14347–14354 [DOI] [PubMed] [Google Scholar]
- Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR (2004) Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J 23: 2841–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA, Nakajima T, Eckner R, Montminy M, Orkin SH (1998) CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci USA 95: 2061–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA, Weiss MJ (2001) Nuclear factors that regulate erythropoiesis. In Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management, Steinberg MH, Forget BG, Higgs DR, Nagel RL (eds) pp 72–94. Cambridge: Cambridge University Press [Google Scholar]
- Bochar DA, Savard J, Wang W, Lafleur DW, Moore P, Cote J, Shiekhattar R (2000) A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc Natl Acad Sci USA 97: 1038–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH (2002) Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 21: 3368–3376 [DOI] [PubMed] [Google Scholar]
- Chang AN, Cantor AB, Fujiwara Y, Lodish MB, Droho S, Crispino JD, Orkin SH (2002) GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc Natl Acad Sci USA 99: 9237–9242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bender TP (2001) A novel system to identify Myb target promoters in friend murine erythroleukemia cells. Blood Cells Mol Dis 27: 429–436 [DOI] [PubMed] [Google Scholar]
- Corona DF, Tamkun JW (2004) Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim Biophys Acta 1677: 113–119 [DOI] [PubMed] [Google Scholar]
- Crispino JD, Lodish MB, MacKay JP, Orkin SH (1999) Use of altered specificity mutants to probe a specific protein–protein interaction in differentiation: the GATA-1:FOG complex. Mol Cell 3: 219–228 [DOI] [PubMed] [Google Scholar]
- de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J (2003) Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci USA 100: 7480–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan LL, Porter SD, Duan Z, Flubacher MM, Montoya D, Tsichlis PN, Horwitz M, Gilks CB, Grimes HL (2004) Targeted transcriptional repression of Gfi1 by GFI1 and GFI1B in lymphoid cells. Nucleic Acids Res 32: 2508–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Stankiewicz MJ, Liu Y, Xi Q, Schmitz JE, Lekstrom-Himes JA, Ackerman SJ (2002) Novel combinatorial interactions of GATA-1, PU.1, and C/EBPepsilon isoforms regulate transcription of the gene encoding eosinophil granule major basic protein. J Biol Chem 277: 43481–43494 [DOI] [PubMed] [Google Scholar]
- Duan Z, Horwitz M (2003) Gfi-1 oncoproteins in hematopoiesis. Hematology 8: 339–344 [DOI] [PubMed] [Google Scholar]
- Eberharter A, Ferrari S, Langst G, Straub T, Imhof A, Varga-Weisz P, Wilm M, Becker PB (2001) Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J 20: 3781–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefanty AG, Antoniou M, Custodio N, Carmo-Fonseca M, Grosveld FG (1996) GATA transcription factors associate with a novel class of nuclear bodies in erythroblasts and megakaryocytes. EMBO J 15: 319–333 [PMC free article] [PubMed] [Google Scholar]
- Enver T, Heyworth CM, Dexter TM (1998) Do stem cells play dice? Blood 92: 348–351, discussion 352 [PubMed] [Google Scholar]
- Feng Q, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Zhang Y (2002) Identification and functional characterization of the p66/p68 components of the MeCP1 complex. Mol Cell Biol 22: 536–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Zhang Y (2001) The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev 15: 827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Liew C, Holmes M, Kowalski K, Mackay J, Crossley M (1999) Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J 18: 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, Wade PA (2004) MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell 119: 75–86 [DOI] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Kajita M, Geigerman C, Moreno CS, Wade PA (2003) MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell 113: 207–219 [DOI] [PubMed] [Google Scholar]
- Garcon L, Lacout C, Svinartchouk F, Le Couedic JP, Villeval JL, Vainchenker W, Dumenil D (2005) Gfi-1B plays a critical role in terminal differentiation of normal and transformed erythroid progenitor cells. Blood 105: 1448–1455 [DOI] [PubMed] [Google Scholar]
- Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH (2003) GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci USA 100: 8811–8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyot B, Valverde-Garduno V, Porcher C, Vyas P (2004) Deletion of the major GATA1 enhancer HS 1 does not affect eosinophil GATA1 expression and eosinophil differentiation. Blood 104: 89–91 [DOI] [PubMed] [Google Scholar]
- Hutchins AS, Mullen AC, Lee HW, Sykes KJ, High FA, Hendrich BD, Bird AP, Reiner SL (2002) Gene silencing quantitatively controls the function of a developmental trans-activator. Mol Cell 10: 81–91 [DOI] [PubMed] [Google Scholar]
- Kadam S, Emerson BM (2003) Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell 11: 377–389 [DOI] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, Kingston R, Georgopoulos K (1999) Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10: 345–355 [DOI] [PubMed] [Google Scholar]
- Lachman HM, Skoultchi AI (1984) Expression of c-myc changes during differentiation of mouse erythroleukaemia cells. Nature 310: 592–594 [DOI] [PubMed] [Google Scholar]
- Lahlil R, Lecuyer E, Herblot S, Hoang T (2004) SCL assembles a multifactorial complex that determines glycophorin A expression. Mol Cell Biol 24: 1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Hoang T (2004) SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp Hematol 32: 11–24 [DOI] [PubMed] [Google Scholar]
- Letting DL, Chen YY, Rakowski C, Reedy S, Blobel GA (2004) Context-dependent regulation of GATA-1 by friend of GATA-1. Proc Natl Acad Sci USA 101: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Recillas-Targa F, Prioleau MN, Felsenfeld G (2001) Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J 20: 2224–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A (1999) MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet 23: 58–61 [DOI] [PubMed] [Google Scholar]
- Nichols KE, Crispino JD, Poncz M, White JG, Orkin SH, Maris JM, Weiss MJ (2000) Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet 24: 266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill DW, Schoetz SS, Lopez RA, Castle M, Rabinowitz L, Shor E, Krawchuk D, Goll MG, Renz M, Seelig HP, Han S, Seong RH, Park SD, Agalioti T, Munshi N, Thanos D, Erdjument-Bromage H, Tempst P, Bank A (2000) An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol Cell Biol 20: 7572–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH (2000) Diversification of haematopoietic stem cells to specific lineages. Nat Rev Genet 1: 57–64 [DOI] [PubMed] [Google Scholar]
- Osada H, Grutz G, Axelson H, Forster A, Rabbitts TH (1995) Association of erythroid transcription factors: complexes involving the LIM protein RBTN2 and the zinc-finger protein GATA1. Proc Natl Acad Sci USA 92: 9585–9589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Cantor AB, Johnson KD, Moran TB, Boyer ME, Orkin SH, Bresnick EH (2004) Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc Natl Acad Sci USA 101: 980–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L, Lin CS, D'Agati V, Simon MC, Orkin SH, Costantini F (1995) Development of hematopoietic cells lacking transcription factor GATA-1. Development 121: 163–172 [DOI] [PubMed] [Google Scholar]
- Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH (1996) The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell 86: 47–57 [DOI] [PubMed] [Google Scholar]
- Querfurth E, Schuster M, Kulessa H, Crispino JD, Doderlein G, Orkin SH, Graf T, Nerlov C (2000) Antagonism between C/EBPbeta and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev 14: 2515–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylski M, Welch JJ, Chen YY, Letting DL, Diehl JA, Chodosh LA, Blobel GA, Weiss MJ (2003) GATA-1-mediated proliferation arrest during erythroid maturation. Mol Cell Biol 23: 5031–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleque S, Cameron S, Orkin SH (2002) The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev 16: 301–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu R, Takahashi S, Ohneda K, Engel JD, Yamamoto M (2001) In vivo requirements for GATA-1 functional domains during primitive and definitive erythropoiesis. EMBO J 20: 5250–5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka T, Skoultchi AI (2003) The ISWI ATPase Snf2h is required for early mouse development. Proc Natl Acad Sci USA 100: 14097–14102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang AP, Fujiwara Y, Hom DB, Orkin SH (1998) Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev 12: 1176–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH (1997) FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90: 109–119 [DOI] [PubMed] [Google Scholar]
- Visvader JE, Mao X, Fujiwara Y, Hahm K, Orkin SH (1997) The LIM-domain binding protein Ldb1 and its partner LMO2 act as negative regulators of erythroid differentiation. Proc Natl Acad Sci USA 94: 13707–13712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH (1997) The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J 16: 3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MJ, Yu C, Orkin SH (1997) Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol 17: 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ (2004) Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104: 3136–3147 [DOI] [PubMed] [Google Scholar]
- Westin G, Gerster T, Muller MM, Schaffner G, Schaffner W (1987) OVEC, a versatile system to study transcription in mammalian cells and cell-free extracts. Nucleic Acids Res 15: 6787–6798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyatt D, Lindeboom F, Karis A, Ferreira R, Milot E, Hendriks R, de Bruijn M, Langeveld A, Gribnau J, Grosveld F, Philipsen S (2000) An intrinsic but cell-nonautonomous defect in GATA-1-overexpressing mouse erythroid cells. Nature 406: 519–524 [DOI] [PubMed] [Google Scholar]
- Whyatt DJ, deBoer E, Grosveld F (1993) The two zinc finger-like domains of GATA-1 have different DNA binding specificities. EMBO J 12: 4993–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, Qi X, Keables P, Von Andrian UH, Georgopoulos K (2004) The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity 20: 719–733 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Table 1


