Abstract
Nuclear Rad51 focus formation is required for homology-directed repair of DNA double-strand breaks (DSBs), but its regulation in response to non-DSB lesions is poorly understood. Here we report a novel human SQ/TQ cluster domain-containing protein termed ASCIZ that forms Rad51-containing foci in response to base-modifying DNA methylating agents but not in response to DSB-inducing agents. ASCIZ foci seem to form prior to Rad51 recruitment, and an ASCIZ core domain can concentrate Rad51 in focus-like structures independently of DNA damage. ASCIZ depletion dramatically increases apoptosis after methylating DNA damage and impairs Rad51 focus formation in response to methylating agents but not after ionizing radiation. ASCIZ focus formation and increased apoptosis in ASCIZ-depleted cells depend on the mismatch repair protein MLH1. Interestingly, ASCIZ foci form efficiently during G1 phase, when sister chromatids are unavailable as recombination templates. We propose that ASCIZ acts as a lesion-specific focus scaffold in a Rad51-dependent pathway that resolves cytotoxic repair intermediates, most likely single-stranded DNA gaps, resulting from MLH1-dependent processing of base lesions.
Keywords: FHA domain, KIAA0431, MMS, MNNG, oxidative damage
Introduction
Checkpoints play critical roles in maintaining genome integrity and thereby prevent the onset of cancer by delaying cell division in the presence of damaged DNA and by regulating the repair of distinct lesions by appropriate DNA repair pathways (Kastan and Bartek, 2004). A critical part of the DNA damage response at the cellular level is the organization of various checkpoint complexes into dynamic subnuclear domains that can be visualized as DNA damage-induced foci by fluorescence microscopy. These subnuclear domains seem to form in the vicinity of damaged chromatin in the early phase of the DNA damage response (within minutes) to set up efficient checkpoint signaling networks, and then diversify over hours into pathway-specific DNA repair centers coincident with the processing of original lesions into repair intermediates (Lisby et al, 2004).
ATM-dependent phosphorylation of histone H2AX on Ser139 (γH2AX) near double-strand breaks (DSBs) is among the earliest responses to DNA damage. γH2AX foci are therefore a very sensitive cytological marker for checkpoint activation in response to DSBs, but γH2AX can also be formed in response to DNA damage that does not involve DSBs, in which case Ser139 is phosphorylated by the ATM-related kinase ATR (Stojic et al, 2004). In the repair phase, two mutually exclusive classes of DSB-induced foci can be distinguished: Mre11–Rad50–Nbs1 complex-containing foci that are believed to be sites of DSB repair by the nonhomologous end-joining (NHEJ) pathway, and Rad51-containing foci that are believed to be sites of homologous recombination (HR) (Paull et al, 2000; Mirzoeva and Petrini, 2001).
The recombinase Rad51 is essential for HR and coats single-stranded DNA (ssDNA) 3′-tails resulting from processing of primary DSBs by 5′–3′ exonucleases to form a nucleoprotein filament required for invasion of homologous double-stranded DNA sequences (Sung et al, 2003; West, 2003). Loading of Rad51 onto ssDNA tails in vitro depends on recombination mediators, such as Rad52 in yeast and the Rad51 paralogs Rad51B–D and XRCC2–3 in vertebrates (Sung et al, 2003). Interestingly, the same mediators are also required for the formation of DSB-induced Rad51 foci in vivo (Bishop et al, 1998; O'Regan et al, 2001; Takata et al, 2001; Lisby et al, 2004). DSB-induced Rad51 focus formation in mammalian cells also depends on BRCA2 (Chen et al, 1999; Yuan et al, 1999; Yu et al, 2000), and it is likely that the impairment of this function contributes significantly to increased genome instability and cancer predisposition associated with BRCA2 mutations in familial breast cancer and D1-type Fanconi's anemia (D'Andrea and Grompe, 2003; West, 2003). In yeast, only few (usually one to two) repair foci are formed in response to a much larger number of DSBs, and distinct DSBs can be recruited to the same focus (Lisby et al, 2003). It was proposed that focus formation increases the local concentration of repair proteins to promote their efficient recycling for consecutive repair of multiple DSBs (Lisby et al, 2003).
In addition to its role in DSB repair, Rad51 also modulates the progression of stalled replication forks that encounter DNA lesions during S phase (Henry-Mowatt et al, 2003). This feature may be involved in the spontaneous formation of Rad51 foci during S phase in the absence of exogenous DNA-damaging agents (Scully et al, 1997). Interestingly, in contrast to defective DSB-induced Rad51 focus formation, spontaneous Rad51 focus formation during S phase is unaffected in BRCA2-mutated pancreatic carcinoma cells (Tarsounas et al, 2003), demonstrating the existence of alternative lesion-specific Rad51 focus formation pathways. This notion is also supported by morphological differences between Rad51 foci resulting from methylating agents that do not give rise to primary DSBs and B-cell activation in the same cells (Li and Maizels, 1997), but proteins involved in such alternative Rad51 focus forming pathways have so far remained elusive.
Here, we report a novel human protein termed ASCIZ that forms Rad51-containing foci in response to DNA methylating agents, but not in response to DSBs. ASCIZ is required for Rad51 focus formation only under conditions where it forms foci itself, and we have identified an ASCIZ core domain that can organize Rad51 into focus-like structures in the absence of DNA-damaging agents. We propose that ASCIZ functions as a scaffold in a novel lesion-specific Rad51 focus formation pathway that also involves the mismatch repair (MMR) protein MLH1 as an upstream component.
Results
Identification of ASCIZ as a candidate DNA damage response protein
Forkhead-associated (FHA) domains have important protein–protein interaction functions in DNA damage checkpoints (Durocher and Jackson, 2002; Hammet et al, 2003), which makes them useful as baits in yeast two-hybrid screens to identify novel DNA damage response proteins (Pike et al, 2004). We therefore used the human CHK2 kinase FHA domain (Matsuoka et al, 1998) as bait to screen a human placental cDNA library (∼3 × 106 clones). The 25 most strongly interacting clones isolated encoded the same protein (16 full-length, nine truncated; Supplementary Figure S1A) corresponding to the uncharacterized KIAA0431 cDNA (Ishikawa et al, 1997). This protein (Figure 1A) contains an N-terminal double C2H2 Zn2+-finger domain, a nuclear localization signal and a total of 18 SQ/TQ motifs (13 TQ, five SQ), 17 of which are clustered in an SQ/TQ cluster domain (SCD; residues 265–656). SCDs are hallmarks of DNA damage response proteins and potential substrates for the checkpoint kinases ATM and ATR (Matsuoka et al, 1998; Traven and Heierhorst, 2005). Based on these properties, we have termed this protein ASCIZ (ATM/ATR-substrate CHK2-interacting Zn2+-finger protein) to avoid confusion with unrelated KIAA proteins. Details of the ASCIZ/CHK2 interaction and ASCIZ phosphorylation by ATM/ATR-like kinases are shown in Supplementary Figure S1.
Figure 1.
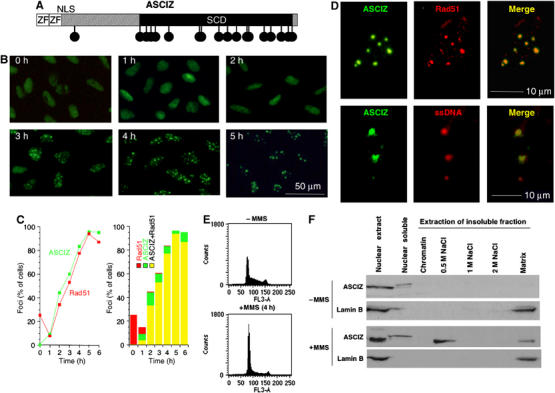
DNA damage-induced ASCIZ focus formation. (A) Schematic diagram of ASCIZ domain organization. Circles indicate SQ/TQ motifs; ZF, Zn2+-finger; NLS, nuclear localization signal. (B) Time course of ASCIZ focus formation in a stable GFP-ASCIZ-expressing U2OS cell line at the indicated times after addition of 0.02% MMS. (C) Quantification of ASCIZ focus formation (green) compared to Rad51 focus formation (red). In the right panel, red bars denote cells that contain only Rad51 foci, green bars cells that contain only ASCIZ foci and yellow bars cells that contain both ASCIZ and Rad51 foci. Data are the mean of two independent experiments with >200 cells scored per sample. Similar data were obtained in other experiments. (D) Colocalization of MMS-induced ASCIZ foci with RAD51 (top) and BrdU-labeled ssDNA (bottom) in single nuclei of GFP-ASCIZ-expressing U2OS cells. (E) Flow cytometry of stable GFP-ASCIZ cells without and after 4 h treatment with 0.02% MMS, stained for DNA content with propidium iodide. (F) Immunoblots of endogenous ASCIZ in subnuclear fractions prepared from untransfected U2OS cells as described (Conlan et al, 2004), and after extraction with increasing salt buffers. Chromatin fraction indicates proteins eluted after DNase treatment of the insoluble fraction.
ASCIZ forms Rad51-containing foci in response to methylating DNA damage
To evaluate DNA damage response functions of ASCIZ, we generated stable human U2OS osteosarcoma cell lines that express ASCIZ as a GFP fusion protein. Interestingly, GFP-ASCIZ redistributed from a diffuse nuclear localization to a predominantly focal pattern after DNA damage treatment with the methylating agent methylmethane sulfonate (MMS) (Figure 1B). Over a 5 h time course, ASCIZ focus formation occurred in >90% of cells (Figure 1C) with 10±2 foci per cell. Similar results were obtained in transiently transfected U2OS cells and other cell lines (Supplementary Figure S2A). An antibody raised against ASCIZ crossreacted strongly with two more abundant bands of ∼50–60 kDa on Western blots, limiting its use in immunofluorescence microscopy analyses to overexpressed ASCIZ (Supplementary Figure S3). Using this antibody for microscopic analysis of cells transfected with the ASCIZ cDNA lacking GFP, we found that the GFP-free protein formed similar MMS-induced foci (Supplementary Figure S2B), demonstrating that ASCIZ focus formation is not restricted to the GFP fusion protein. To corroborate these findings for the endogenous protein, we performed cell fractionation analyses of untransfected U2OS cells. In the absence of DNA-damaging agent, endogenous ASCIZ was found exclusively in the soluble fraction of nuclear extracts, but following MMS treatment it mostly redistributed into the insoluble fraction of nuclear extracts, from where it could be partially extracted by high-salt buffer (Figure 1F). These biochemical data are consistent with the incorporation of ASCIZ into DNA damage-induced macromolecular structures such as foci.
The relatively slow time course of focus formation suggested that ASCIZ foci play a role in DNA repair rather than primary DNA damage detection and signaling. Consistent with this hypothesis, we found that MMS-induced ASCIZ foci also contained Rad51 (Figure 1D, upper panel). If the presence of Rad51 in ASCIZ foci reflects a role in DNA repair, ASCIZ foci should also be associated with ssDNA. We therefore combined MMS treatment with ssDNA-specific nondenaturing BrdU-labeling procedures and found that ASCIZ foci indeed contained ssDNA (Figure 1D, lower panel), demonstrating that they represent genuine DNA damage structures.
In contrast to ASCIZ, Rad51 formed foci in ∼25% of cells under basal conditions (Figure 1C), which are most likely S-phase foci caused by stalled DNA replication (in control experiments under similar conditions, ∼28% of parental U2OS cells contained spontaneous Rad51 foci and >98% of these were in S phase based on BrdU pulse-labeling for 40 min; data not shown). However, in response to MMS treatment, Rad51 foci initially dispersed and then increased in number with an apparently slight delay but otherwise similar kinetics compared to ASCIZ foci (Figure 1C, left panel). At later time points, Rad51 foci were only found in ASCIZ foci-containing cells (and were then almost exclusively associated with ASCIZ foci), whereas some cells contained ASCIZ foci but no Rad51 foci (Figure 1C, right panel). Although it should be stressed that the slightly different time courses of ASCIZ and Rad51 focus formation could reflect different detection limits for GFP-ASCIZ compared to anti-Rad51 immunofluorescence, these results suggest that ASCIZ focus formation may precede Rad51 recruitment into these foci.
MMS-induced ASCIZ focus formation does not depend on DNA replication
Flow cytometry analyses indicated that the majority of cells accumulated in G1 phase under the conditions where ASCIZ foci formed (Figure 1E), consistent with a checkpoint-dependent cell cycle arrest (note enhanced p53 Ser15 phosphorylation as a biochemical marker for G1 checkpoint activation after 4 h MMS treatment; Supplementary Figure S4). Given that ASCIZ foci contain Rad51 and ssDNA, and are therefore likely sites of recombinational repair, this was surprising because HR, at least in response to DSB-inducing agents, is believed to be inefficient during G1 phase. In unsynchronized cells incubated with BrdU during MMS treatment, BrdU-positive and unlabeled cells (most of which should be in G1) formed ASCIZ foci with similar efficiency (Figure 2A). To investigate specifically ASCIZ focus formation during G1, cells were synchronized in mitosis using nocodazole and released into BrdU-containing medium. Under these conditions, cells started to enter S phase from ∼6 h after nocodazole release (Figure 2B and Supplementary Figure S5). MMS treatment at 4 h after release, when >99% of cells had not incorporated BrdU (i.e., were still in G1), led to ASCIZ focus formation in >60% of cells (Figure 2A and C). At 12 h after release, when ∼35% of them had entered S phase, a similar percentage of cells formed MMS-induced ASCIZ foci and the efficiency of ASCIZ focus formation was similar in both BrdU-positive and -negative populations (Figure 2A and C). These results demonstrate that ASCIZ foci form as efficiently in G1 as in S phase, and that MMS-induced ASCIZ focus formation therefore does not depend on DNA replication.
Figure 2.
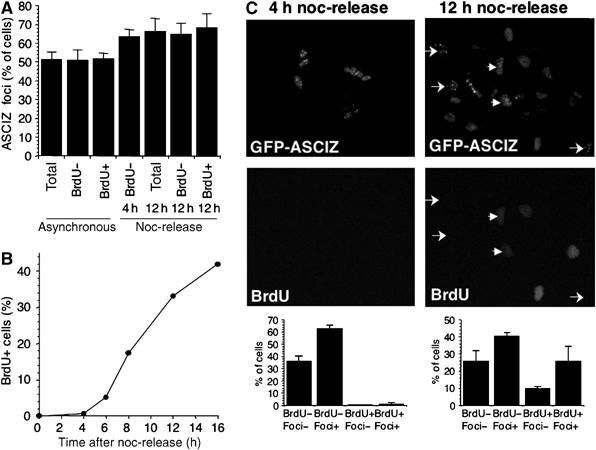
ASCIZ focus formation can occur during G1 phase. (A) Grouped data for ASCIZ focus formation in stable GFP-ASCIZ-expressing U2OS cells after 4 h treatment with 0.02% MMS in unsynchronized cells, or 4 and 12 h after release from nocodazole (noc) arrest. Asynchronous cells were incubated with BrdU during MMS treatment; synchronized cells were released into BrdU-containing medium before MMS treatment. Data are the mean±s.e. of three independent experiments, scoring >250 cells per sample. (B) Kinetics of S-phase entry, detected by BrdU incorporation, in the same cell line after release from nocodazole arrest. Propidium iodide-stained FACS profiles of a similar experiment are shown in Supplementary Figure S5. (C) Examples of micrographs of MMS-treated cells at 4 or 12 h after nocodazole release (top panels), and grouped data of the fractional distribution of BrdU and ASCIZ foci indices, scored as in (A). Arrows indicate examples of G1 cells containing ASCIZ foci and arrowheads indicate post-G1 cells containing ASCIZ foci.
ASCIZ is required for cell survival and Rad51 focus formation in response to MMS
To test if ASCIZ foci are involved in the repair of MMS-induced DNA damage, U2OS cells were treated with two separate synthetic ASCIZ-specific siRNA duplexes. This led to almost complete depletion of the endogenous protein within 48–72 h; however, levels of other proteins including Rad51 were not reduced when compared to actin as a loading control (Figure 3A and Supplementary Figure S6A; only data for one ASCIZ siRNA, si579, are shown in the main figures; similar results for a second ASCIZ siRNA, si226, are shown in Supplementary Figure S6). A control luciferase siRNA (GL2) (Elbashir et al, 2001) had no effect on protein levels (Figure 3A and Supplementary Figure S6A). Interestingly, ASCIZ depletion led to dramatically increased apoptosis (five- to 10-fold) following MMS treatment (Figure 3B and Supplementary Figure S6B), demonstrating that ASCIZ plays a crucial role in the cellular survival of methylating DNA damage.
Figure 3.
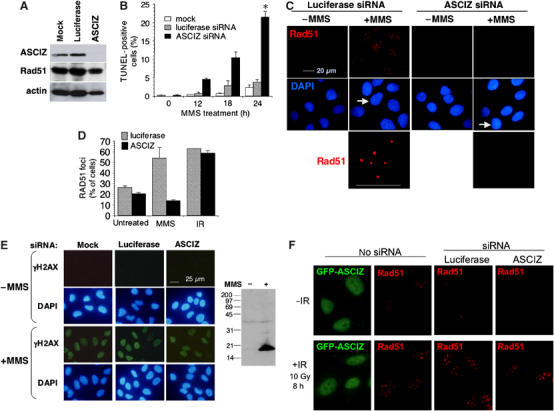
ASCIZ is required for Rad51 focus formation and cell survival in response to MMS treatment. (A) Western blot analysis of ASCIZ, Rad51 and actin from ASCIZ siRNA- and luciferase siRNA-treated U2OS cells. (B) Apoptosis in U2OS cells siRNA-treated as in (A) at the indicated times after 0.005% MMS addition (mean±s.e. of three independent experiments), detected by TUNEL staining. *P<0.01 (paired t-test, two-sided) versus mock and GL2 at 24 h. (C) MMS-induced Rad51 focus formation in siRNA-treated U2OS cells. The bottom panel shows enlargements of nuclei labeled by arrows above. (D) Quantitation of Rad51 focus formation (mean±s.e. of 2–3 independent experiments) in siRNA-treated U2OS cells under basal conditions and in response to MMS or IR as in (B) and (E). Similar results for another ASCIZ siRNA are shown in Supplementary Figure S6C. (E) γH2AX formation in siRNA-treated U2OS cells with or without MMS treatment. The right panel shows an overexposed Western blot demonstrating the specificity of the antibody. In the experiment shown here, 0.14 and 99.5% of control and 0.25 and 97.1% of ASCIZ-depleted cells contained γH2AX in the absence or presence of MMS treatment, respectively (>170 nuclei scored per sample). (F) Formation of Rad51 foci but not GFP-ASCIZ foci in response to IR (left panels), and normal Rad51 focus formation after IR in ASCIZ siRNA-treated U2OS cells (right panels).
As Rad51 foci play important roles in DNA damage repair, we tested if the MMS hypersensitivity of ASCIZ-depleted cells could be due to impaired Rad51 focus formation. Remarkably, MMS-induced Rad51 focus formation was severely impaired in ASCIZ-depleted cells, but unaffected in mock-treated (not shown) and luciferase-treated controls (Figure 3C and D and Supplementary Figure S6C). In contrast, ASCIZ depletion had only minor effects on spontaneous Rad51 focus formation (Figure 3D), and it also did not impair MMS-induced γH2AX formation (Figure 3E) as a sensitive marker for checkpoint activation that can detect methylating DNA damage unrelated to DSBs (Stojic et al, 2004), demonstrating that ASCIZ depletion does not generally interfere with DNA damage detection and signaling. Altogether, these data indicate that ASCIZ is required for efficient DNA repair in response to methylating agents by regulating Rad51 focus formation.
As ASCIZ and Rad51 focus formation is associated with MMS-induced accumulation of cells in G1 phase (Figure 1E), it was important to determine if ASCIZ siRNA treatment leads to accumulation of cells in another cell cycle phase, thereby indirectly preventing Rad51 focus formation. Importantly, ASCIZ siRNA treatment resulted in a marked increase in G1 cells already in the absence of DNA-damaging agents (Figure 4A), which was further increased after MMS treatment (data not shown), demonstrating that ASCIZ depletion impairs MMS-induced Rad51 focus formation in exactly the cell cycle phase where these foci usually form.
Figure 4.
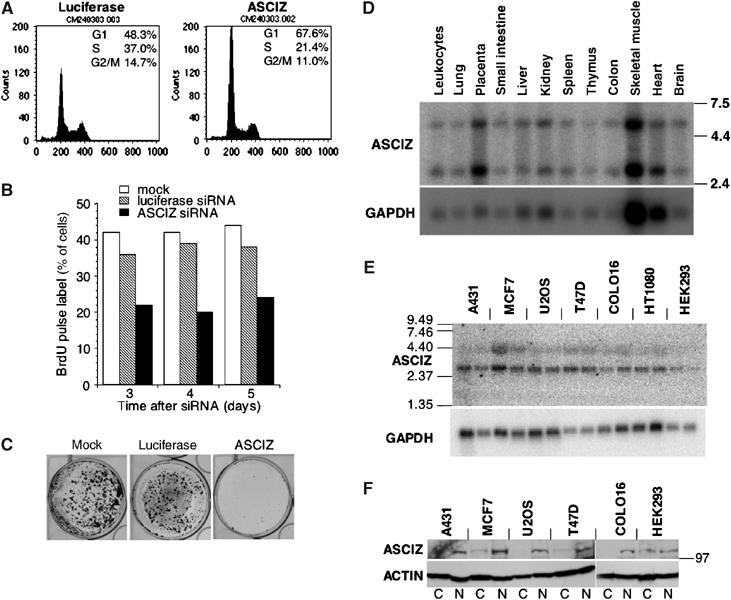
ASCIZ siRNA phenotypes and expression profiles. (A) Flow cytometry of luciferase siRNA- and ASCIZ siRNA-treated U2OS cells, stained with propidium iodide. Cell cycle distribution was analyzed using ModFit LT. (B) S-phase indices of siRNA-treated U2OS cells pulse-labeled for 40 min with 10 μg/ml BrdU. (C) Colony formation of siRNA-treated U2OS cells. (D, E) Northern blot analysis of ASCIZ and GAPDH mRNAs in normal human tissues (D) and human cancer cell lines (E) as indicated. For each cell line, samples were prepared from semiconfluent (left lanes) and confluent (right lanes) cultures. Mass standards (kb) are indicated on the left. The mRNAs correspond to two differently sized ASCIZ cDNAs in GenBank (BC002701 and NM_015251). (F) Western blot analysis of endogenous ASCIZ in cytosol (C) and nuclear (N) fractions of the cell lines indicated above. The position of a 97 kDa marker band is indicated on the right, and actin levels are shown below as a loading control.
Consistent with their G1 accumulation, S-phase indices in ASCIZ-depleted cells were considerably reduced based on flow cytometry (Figure 4A) or BrdU pulse-labeling assays (Figure 4B). This effect was stable for several days (Figure 4B), and as a result ASCIZ depletion also led to markedly impaired cell proliferation in colony formation assays (Figure 4C and Supplementary Figure S6D). It should be noted, however, that the ASCIZ siRNAs were not actually cytotoxic, as they did not lead to increased spontaneous apoptosis detectable by flow cytometry (Figure 4A) or TUNEL staining (Figure 3B and Supplementary Figure S6B), and cells remained viable and eventually formed colonies after longer growth periods (data not shown). Northern and Western blots demonstrated that ASCIZ is ubiquitously expressed at remarkably similar levels (compared to loading controls) in a wide range of normal human tissues (with higher levels in placenta as a highly proliferative tissue) and cancer cell lines (Figure 4D–F). Altogether, these results suggest that ASCIZ has an important function in normal cell proliferation, possibly related to its role in DNA repair.
ASCIZ does not form foci and is not required for Rad51 focus formation in response to DSBs
As Rad51 focus formation is best understood in response to DSBs, we tested if ASCIZ also forms foci in response to ionizing radiation (IR). Surprisingly, ASCIZ did not form foci in response to IR under a range of conditions where Rad51 foci readily formed in the same cells (Figure 3F, ‘no siRNA' panels, and data not shown), indicating that ASCIZ foci form in a lesion-specific manner unrelated to DSBs. This result provided us with an experimental system to test if ASCIZ was also required for Rad51 focus formation under DNA damage conditions where it does not form foci itself. Interestingly, in contrast to the MMS response (Figure 3C and D), IR-induced Rad51 focus formation was normal in ASCIZ-depleted cells (Figure 3D and F, ‘siRNA' panels). These results indicate that ASCIZ acts locally to regulate Rad51 recruitment into foci only in response to DNA lesions that first give rise to ASCIZ foci.
ASCIZ focus formation is highly lesion specific and depends on MLH1
To further explore the lesion specificity of ASCIZ focus formation, we tested additional DNA-damaging agents. Similar to IR (Figure 3F), neither GFP-ASCIZ nor the untagged protein formed foci in response to the DSB-inducing drugs adriamycin and bleomycin, the replication-blocking agent hydroxyurea or the crosslinking agents mitomycin C and UV-B (Supplementary Figure S2B, and data not shown). However, similar to MMS, ASCIZ foci formed in response to another methylating agent, 1-methyl-3-nitro-1-nitrosoguanidine (MNNG; Figure 5A). Although MMS and MNNG are both methylating agents, they produce a somewhat different spectrum of DNA base lesions. The major mutagenic and cytotoxic products of MMS as an SN2 alkylating agent are believed to be N7-methylguanine (N7mG) and N3-methyladenine (N3mA), which are rapidly converted to abasic sites (Glaab et al, 1998, 1999). The SN1 alkylating agent MNNG likewise produces predominantly N7mG and N3mA, but also the biochemically more stable O6-methylguanine (O6mG), which is not converted into abasic sites and is believed to be the major cytotoxic product at lower MNNG doses (Stojic et al, 2004). To resolve which of these lesions could give rise to ASCIZ foci, we compared dose responses of ASCIZ focus formation with cytotoxic effects (half-maximal lethality, LD50, in clonogenic survival assays) of MMS and MNNG in the stable GFP-ASCIZ cell line. Half-maximal ASCIZ focus formation occurred at ∼0.01% MMS (Figure 5B), which was in a similar range to the LD50 of MMS (∼0.006%; data not shown), whereas ASCIZ focus formation in response to MNNG was half-maximal only at 10-fold higher doses (∼8 μM; Figure 5A) than the LD50 of this drug (∼0.8 μM; data not shown). These data suggest that the initiating lesions for ASCIZ focus formation are most likely abasic sites resulting from N7mG and N3mA in response to MMS and high doses of MNNG (note that ASCIZ focus formation was not affected by inhibition of methylguanine methyl-transferase with O6-benzylguanine, supporting that O6mG is not the primary lesion; data not shown).
Figure 5.
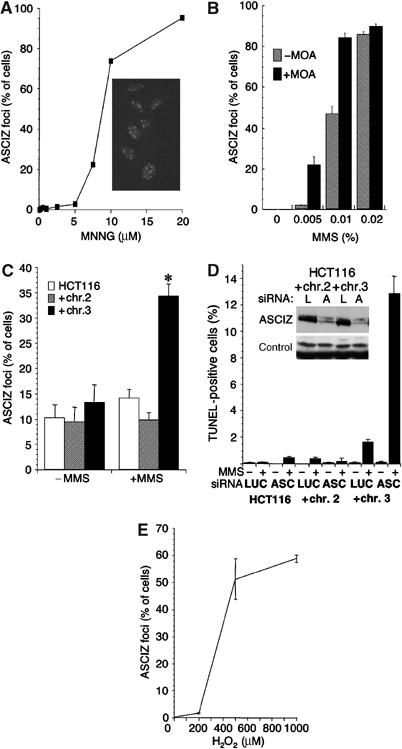
Regulation of ASCIZ focus formation and DNA damage-induced apoptosis. (A) Dose response of ASCIZ focus formation in the stable GFP-ASCIZ-expressing U2OS cell line after 6 h MNNG treatment. The micrograph was taken 6 h after addition of 20 μM MNNG. (B) Dose response of ASCIZ focus formation in the same cell line after 4 h MMS treatment in the presence (filled bars) or absence (hatched bars) of 6 mM methoxyamine (MOA). (C) ASCIZ focus formation in transiently transfected HCT116 cells (open bars), or HCT116 cells complemented with chromosome 2 or 3 without or after 4 h 0.02% MMS treatment. *P<0.01% versus all other samples (paired t-test, two-sided). (D) Apoptosis in the HCT116 cell lines treated with luciferase (LUC) or ASCIZ (ASC) siRNA 24 h after addition of 0.005% MMS. The inset shows an anti-ASCIZ Western blot of the chromosome 2 and 3 complemented HCT116 lines treated with luciferase (L) or ASCIZ (A) siRNAs, using crossreacting bands on the same blot as a loading control. (E) ASCIZ focus formation in stable GFP-ASCIZ-expressing U2OS cells in response to the indicated H2O2 doses (mean±s.e. of 3–4 independent experiments; >200 cells per sample scored).
The finding that ASCIZ foci do not form in response to a variety of DSB-inducing agents indicates that the ssDNA present in ASCIZ foci (Figure 1D) cannot be ssDNA 3′-tails, which lead to Rad51 focus formation after DSB processing. In addition to 3′-tails, Rad51 can also utilize extended ssDNA gaps as recombination substrates (Hatanaka et al, 2005), and we therefore hypothesized that the ssDNA in ASCIZ foci represents gapped DNA. MMS-induced base lesions can be repaired by competing pathways involving base-excision repair (BER) (Cline and Hanawalt, 2003) or MMR proteins (Glaab et al, 1998, 1999). A characteristic feature of DNA damage processing by MMR complexes is that it generates extended ssDNA gaps as repair intermediates (Cline and Hanawalt, 2003). If our hypothesis that ASCIZ foci form in response to ssDNA gaps was true, focus formation should be dependent on functional MMR proteins and antagonized by the BER pathway.
To test this hypothesis, we first inhibited the BER pathway with methoxyamine, which stabilizes abasic sites by preventing access of apurinic/apyrimidinic endonucleases (Liu et al, 2003). As predicted, methoxyamine treatment resulted in enhanced ASCIZ focus formation at lower MMS concentrations (Figure 5B), indicating that ASCIZ and BER act in competing pathways for processing of abasic sites. To test if ASCIZ foci depend on the MMR machinery, we utilized the colon cancer cell line HCT116, where MMR deficiency resulting from biallelic MLH1 mutation can be complemented by an additional chromosome 3 containing wild-type MLH1 (Tindall et al, 1998). Approximately 10% of HCT116 cells contained ASCIZ foci in the absence of exogenous DNA-damaging agents for unknown reasons (Figure 5C; these spontaneous ASCIZ foci were not associated with PML bodies, see below; data not shown). This fraction did not change in response to MMS treatment in the parental cell line or HCT116 cells complemented with an irrelevant extra chromosome 2 (Figure 5C). In contrast, in chromosome 3-complemented HCT116 cells, MMS treatment caused a significant increase (to 35%) in ASCIZ focus formation (Figure 5C and Supplementary Figure S2C), strongly indicating that DNA damage-induced ASCIZ focus formation depends on MLH1 and possibly other MMR components.
Finally, because the methoxyamine experiment (Figure 5B) indicated that abasic sites are substrates for the ASCIZ pathway, we also tested if oxidative DNA damage as another source of abasic sites (Barnes and Lindahl, 2004) can give rise to ASCIZ foci. Figure 5E shows that ASCIZ foci were indeed induced by treatment of cells with H2O2. As base oxidation is a major cause of endogenous DNA lesions, it is tempting to speculate that the basal growth defect of ASCIZ-deficient cells (Figure 4) may reflect slower repair of endogenous DNA damage.
Increased apoptosis in ASCIZ-depleted cells depends on MLH1
Although the mutagenicity of MMS and MNNG is increased in MMR-deficient cells, these cells are paradoxically resistant to the cytotoxic effects of the same agents, indicating that repair intermediates generated by the MMR pathway rather than the primary base lesions are responsible for the cytotoxicity of methylating agents (Glaab et al, 1998; Stojic et al, 2004). Given that ASCIZ focus formation is MLH1 dependent (Figure 5C), we wanted to test if the increased MMS-induced apoptosis in ASCIZ-depleted cells (Figure 3B) also depends on MLH1. As expected, MMS did not cause significant apoptosis in the parental or chromosome 2-complemented lines, but led to ∼10-fold increased apoptosis in chromosome 3-complemented HCT116 cells (Figure 5D). Importantly, ASCIZ depletion had no significant effect on the parental or chromosome 2-complemented lines, but led to a further eight-fold increase in MMS-induced apoptosis in the MLH1-positive chromosome 3-complemented line (Figure 5D). Altogether, these data indicate that ASCIZ foci form in response to, and are required to resolve repair intermediates that result from MLH1-dependent processing of the original lesions and that otherwise lead to increased apoptosis.
Identification of an ASCIZ core domain involved in Rad51 focus formation
We next performed a truncation analysis to identify regions of ASCIZ that are required for focus formation and Rad51 recruitment. A fragment containing the Zn2+-finger region and nuclear localization signal (residues 1–73) did not form foci, while an ASCIZ fragment lacking the Zn2+-finger domain (residues 67–667) behaved similar to the full-length protein in that it formed foci in a strictly DNA damage-dependent manner (Figure 6B). These results indicate that the Zn2+-finger region as a potential DNA-binding domain is not involved in ASCIZ focus formation. Further truncations revealed that residues 67–286 located between the Zn2+-fingers and SCD formed focus-like structures in nuclei of transfected U2OS cells already in the absence of DNA-damaging agents, while the remainder of the protein (residues 285–667, fused to the ASCIZ nuclear localization signal) did not form foci at all (Figure 6B). Taken together, these results suggest that residues 67–286, which we refer to as the ASCIZ core domain, are responsible for ASCIZ focus formation, and that the SCD negatively regulates the incorporation of ASCIZ into focus-like structures under basal conditions.
Figure 6.
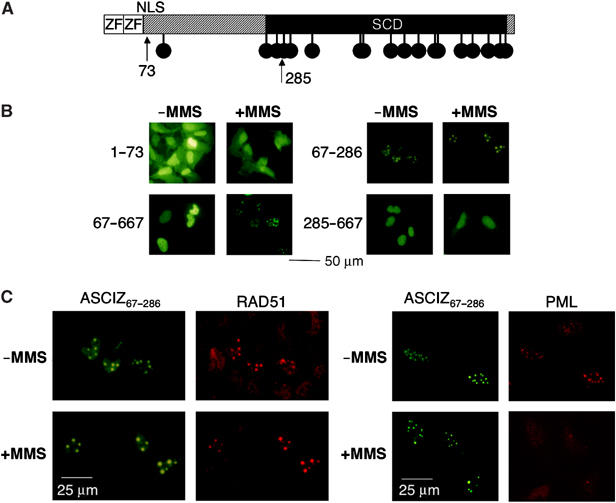
Identification of a focus-forming ASCIZ core domain. (A) Schematic diagram of ASCIZ domains with arrows indicating truncation points. (B, C) Representative micrographs of focus formation by the indicated ASCIZ fragments in the absence or presence of 0.02% MMS for 4 h in transiently transfected U2OS cells, and co-staining of GFP-ASCIZ residues 67–286 with Rad51 (C, left) and PML antibodies (C, right). Note that micrographs were taken from densely grown fields of cells, and that Rad51 foci are considerably more intense in ASCIZ core domain-expressing cells.
Remarkably, co-staining with the Rad51 antibody revealed that overexpression of the ASCIZ core domain led to a dramatic concentration of Rad51 in the same focus-like structures under basal conditions as well as in response to MMS treatment (Figure 6C, left panels). We suspected that the focus-like staining pattern under basal conditions could reflect association of the ASCIZ core domain with PML bodies, which are cytologically similar to DNA damage-induced foci and often contain DNA repair proteins (Conlan et al, 2004). This was confirmed in double-labeling immunofluorescence experiments with an anti-PML antibody (Figure 6C, right panels). However, similar to what we have reported recently (Conlan et al, 2004), the PML bodies largely dispersed after MMS treatment; yet, ASCIZ core domain-Rad51 foci were maintained (Figure 6C), indicating that the core domain retains the ability to concentrate Rad51 in repair sites for MMS-induced DNA lesions independently of PML. Neither full-length ASCIZ nor the core domain interacted directly with Rad51 in yeast two-hybrid assays (data not shown), suggesting that at least one other protein mediates the interaction between ASCIZ and Rad51. Altogether, the simplest explanation for the data presented in Figure 6 would be that ASCIZ functions as a scaffold for the assembly of Rad51 foci.
Discussion
Here, we have identified ASCIZ as a novel DNA damage response protein, and as a key component of a highly lesion-specific Rad51 focus formation pathway. The fundamental importance of ASCIZ is underscored by the fact that its absence leads to dramatically increased apoptosis in response to methylating DNA damage treatment (Figure 3B), most likely because of defective DNA repair resulting from impaired Rad51 focus formation under these conditions (Figure 3C and D).
We propose the following pathway for the formation of ASCIZ-Rad51 foci and their role in DNA repair (Figure 7). First, the original lesions for ASCIZ focus formation are likely abasic sites resulting from conversion of N3mA and N7mG or oxidized bases; the importance of abasic sites for this pathway is indicated by the finding that ASCIZ focus formation is enhanced in methoxyamine-treated cells where abasic sites are shielded from the BER pathway (Figure 5B). Second, abasic sites lead to ASCIZ foci when they are further processed in an MLH1-dependent manner. While the dependence of ASCIZ focus formation on MLH1 (Figure 5C) could be explained by a sensor function of MLH1, the delayed time course of ASCIZ focus formation (5 h to maximum from the onset of methylating DNA damage; Figure 1C) and the requirement of MLH1 for increased MMS-induced apoptosis in ASCIZ-depleted cells (Figure 5D) would be more consistent with an active role in damage processing to yield repair intermediates that are cytotoxic if not resolved in ASCIZ-Rad51 foci. Interestingly, MLH1 interacts directly with exonuclease EXO1 (Jager et al, 2001) and this interaction plays a critical role in the generation of extended ssDNA gaps as the characteristic repair intermediate of the MMR pathway (Cline and Hanawalt, 2003). Together with the fact that ASCIZ foci do not form in response to a range of DSB-inducing agents that give rise to 3′-ssDNA tails (Figure 3D and F, and data not shown), this indicates that the ssDNA present in ASCIZ foci (Figure 1D) represents ssDNA gaps. We therefore propose that ASCIZ foci form in response to persistent ssDNA gaps generated from MLH1-dependent processing of abasic sites. Third, ASCIZ foci then recruit Rad51, which is required for repair of ssDNA gaps and, if repair is successful, prevents apoptosis.
Figure 7.
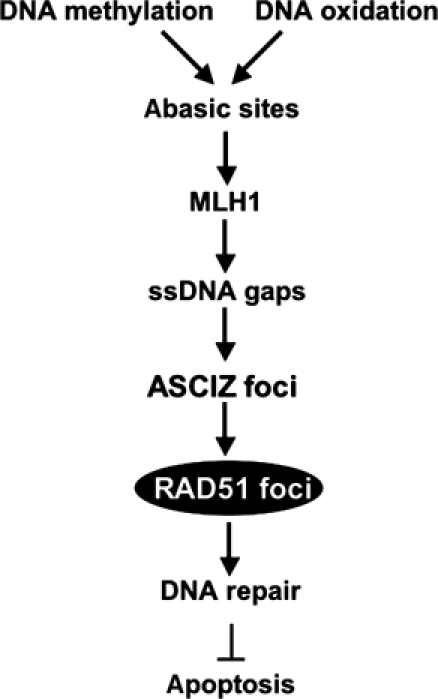
Summary model. Primary methylating or oxidative base damage is converted to abasic sites, which are further processed into ssDNA gaps in an MLH1-dependent manner. ssDNA gaps lead to the formation of ASCIZ foci that recruit Rad51 for DNA repair to prevent DNA damage-induced apoptosis.
This proposed cellular function of ASCIZ foci in a lesion-specific pathway for the formation of Rad51 foci in response to ssDNA gaps is reminiscent of the biochemical function of the RecFOR complex in bacteria (Morimatsu and Kowalczykowski, 2003), which functions as a specialized mediator for recombinase loading onto ssDNA gaps, in contrast to the classical recombination mediator RecBCD for DSB-derived free ssDNA 3′-ends. However, because the ASCIZ core domain is able to concentrate Rad51 in focus-like structures in the absence of appropriate DNA lesions (Figure 6), we believe that ASCIZ is more likely to function as a lesion-specific focus scaffold rather than as a mediator (i.e., a protein that loads recombinases onto DNA substrates). A scaffold function would also be consistent with the findings that ASCIZ is only required for Rad51 focus formation in response to damage that induces ASCIZ foci (MMS; Figures 1 and 3) but not under conditions where ASCIZ foci do not form (i.e., spontaneously during S phase and after IR-induced DSBs; Figures 1 and 3), and that ASCIZ focus formation seems to precede Rad51 recruitment to the same foci (Figure 1).
The most straightforward explanation for the presence of Rad51 and ssDNA in ASCIZ foci (Figure 1D) is that these foci are sites of HR. This conclusion raises a number of issues. Firstly, ASCIZ-Rad51 foci seem to form while the vast majority of cells are checkpoint-arrested in G1 (Figure 1E). The preferred templates for HR repair are sister chromatids, which are only available during late S and G2 phases. However, it is clear that the absence of sister chromatids does not preclude HR, as HR can also utilize corresponding alleles on homologous chromosomes in diploid cells (after all, this is believed to be the major cause for loss of heterozygosity; Richardson and Jasin, 2000) or other homologous sequences as templates (in fact, some of the most frequently used HR assays are based on intrachromosomal gene conversions, e.g. mating type switching in yeast (Ira et al, 2004), I-SceI-induced GFP reporter gene conversion (Richardson and Jasin, 2000) and templated immunoglobulin variable chain conversion in activated B cells (Hatanaka et al, 2005)). Recent work in yeast, where HR during G1 still occurs with 5–10% efficiency, has indicated that the main impediment for HR during G1 is that DSB processing into ssDNA 3′-tails requires cyclin-dependent kinase (CDK) activity (Ira et al, 2004). However, generation of extended ssDNA regions by other pathways did not require CDK activity (Ira et al, 2004), indicating that HR as such should well be possible in G1 cells as long as ssDNA substrates are sufficiently long to form Rad51 filaments. A second question is why would the repair of MLH1-dependent ssDNA gaps even require HR rather than simple gap closure by repair polymerases. A possible explanation could be that closely spaced base lesions or abasic sites on opposite strands cause repair polymerases to stall. Direct recruitment of persistent ssDNA gaps into the HR pathway would avoid converting them into secondary DSBs, which in G1 cells are preferentially repaired by the error-prone NHEJ pathway, and which in large numbers would increase the risk of chromosome translocations.
MLH1 is best known for its role in the MMR pathway, which is formally defined as a postreplicative pathway for the repair of mismatches occurring during S phase. The role of MLH1 in our system is therefore unrelated to its postreplication repair function, as ASCIZ focus formation occurs efficiently in G1 cells and within a few hours of MMS treatment (Figures 1 and 2), in contrast to the ∼48 h delayed postreplicative formation of RPA-ATR foci in G2/M cells after low-dose MNNG treatment (Stojic et al, 2004). However, it is now clear that MLH1 and other MMR proteins can also have more immediate, non-postreplicative functions (Adamson et al, 2002; Wang and Qin, 2003), and the role of MLH1 in the ASCIZ pathway, which is unlikely to involve actual ‘mismatches', would fall into the latter category. We do not yet know if additional MMR proteins are involved in the ASCIZ pathway, but as outlined above, MLH1-dependent recruitment of EXO1 could represent a plausible mechanism for the generation of extended ssDNA gaps. A related mechanism where recombinational repair of activation-induced cytosine deaminase (AID)-dependent abasic sites involves MMR components has recently been proposed for immunoglobulin class-switch recombination (Rada et al, 2004). While class-switch recombination depends on NHEJ proteins (Li et al, 2004) and is therefore unlikely to involve Rad51 foci, it is tempting to speculate that AID-induced immunoglobulin variable chain gene conversion with upstream pseudogenes as another means of generating antibody diversity (Hatanaka et al, 2005) could employ the ASCIZ pathway.
In conclusion, we have identified ASCIZ as a human DNA damage response protein that plays a key role as a possible scaffold in a novel Rad51 focus formation pathway that is required to repair cytotoxic DNA structures resulting from processing of base lesions. Our data provide a basis for future studies to determine precise molecular mechanisms by which ASCIZ forms foci in a highly lesion-specific manner, how it regulates Rad51 recruitment into these foci and which other proteins are involved in this pathway. It will also be important to determine if ASCIZ foci are sites of Rad51 loading onto ssDNA, or whether they are ‘search engines' that recruit loaded Rad51 filaments in the hunt for homologous repair templates (nota bene (n.b.), in the absence of sister chromatids). ASCIZ was isolated as a CHK2 FHA domain-interacting protein and potential ATM/ATR kinase substrate (Supplementary Figure S1), but overexpression of dominant-negative kinase-defective CHK2 or inhibition of ATM/ATR-like kinase activity by caffeine had only a subtle or no effect on ASCIZ focus formation (Supplementary Figure S4). Another interesting question is therefore which functions of ASCIZ are regulated by checkpoint kinases and how these are related to Rad51 foci. Finally, our findings could also have clinical relevance: firstly, as ASCIZ is involved in the repair of base lesions similar to those generated by a range of chemotherapeutics (Liu et al, 2003), it could represent an interesting drug target; and secondly, as mutations in proteins required for Rad51 focus formation can lead to genome instability and cancer predisposition (D'Andrea and Grompe, 2003; West, 2003), it will be important to search for disease association of the human ASCIZ gene that is located close to the major fragile site FRA16D on chromosome 16q23.2, a region frequently mutated in diverse cancers (Bednarek et al, 2001).
Materials and methods
Cell culture and DNA damage treatment
U2OS cells were used for up to 15 passages. The complete ASCIZ open reading frame (667 amino-acid residues; GenBank accession numbers BC002701 and NM_015251) was cloned into pEGFP-C1 or pCDNA3 and transfected using Fugene 6 (Roche). CHK2 and kinase-defective CHK2-D347A were cloned into pCDNA4. Stable GFP-ASCIZ cell lines and chromosome-complemented HCT116 lines were selected using G418. For siRNA experiments (Elbashir et al, 2001), cells were transfected using Oligofectamine (Life Technologies) and 20–60 nM synthetic RNA duplexes (Dharmacon) corresponding to ASCIZ cDNA residues 579–597 (5′-CCC UGA UCC UCG GCC UAG AdTdT-3′ and 5′-UCU AGG CCG AGG AUC AGG GdTdT-3′; si579) or 226–244 (5′-CUG UGC ACA AAA CCA GAA GdTdT-3′ and 5′-CUU CUG GUU UUG UGC ACA GdTdT-3′; si226) relative to the translational initiation codon. Experiments on siRNA-treated cells were performed 48–72 h after transfection. DNA-damaging agents were added in antibiotic-free medium. Colonies were stained using crystal violet. For flow cytometry, cells were fixed in ethanol, stained with 20 μg/ml propidium iodide and analyzed using Becton-Dickinson flow cytometers and CellQuest software. To determine S-phase indices, 10 μg/ml BrdU in medium was added to cells grown on coverslips.
Antibodies
A recombinant ASCIZ fragment (residues 285–477) was cloned and purified essentially as described (Pike et al, 2003) for immunization of two rabbits according to standard procedures approved by the St Vincent's Hospital Melbourne Animal Ethics Committee. Antibodies were affinity-purified by glycine elution from an ASCIZ(285–477) column and used at 50–200 ng/ml for immunoblots, where they detect endogenous ASCIZ protein with an apparent electrophoretic mobility ∼115 kDa (Supplementary Figure S4), and are available from Chemicon. Other antibodies used were as follows: mouse anti-actin (MAB1501R; Chemicon), mouse anti-BrdU (#1170376; Roche), mouse anti-CHK2 (A12; Santa Cruz); mouse anti-γH2AX (05-636; Upstate), mouse anti-lamin B (101-B7; Oncogene), mouse anti-p53 (DO-1; Santa Cruz), rabbit anti-phospho-p53 (Ser15; Cell Signaling #9284) and rabbit anti-Rad51 (PC130; Oncogene). The TUNEL assay kit was from Roche. Secondary antibodies were from Amersham or Molecular Probes.
Microscopy
For fluorescence microscopy, cells were fixed in 3.7% paraformaldehyde or methanol/acetone, permeabilized in Tris-buffered saline containing 0.2% Triton X-100 for 10 min, and incubated with primary antibodies at 4°C overnight and secondary antibodies for 1 h at 37°C. In double-labeling with GFP fluorescence, primary antibodies were detected using Alexa-594-labeled secondary antibodies. For detection of ssDNA foci, cells were incubated for 30 h with 10 μg/ml BrdU (Sigma) prior to MMS treatment and anti-BrdU staining without DNA denaturation (Raderschall et al, 1999). For determination of S-phase indices of BrdU-labeled cells, coverslips were incubated with 80 U/ml DNase I for 1 h at 37°C after fixation and permeabilization. Photomicrographs were taken on Kodak 320T film using a Zeiss Axiovert-25 inverted fluorescence microscope at × 100–250 original magnification, scanned and assembled using Adobe Photodeluxe and Adobe Illustrator as described (Conlan et al, 2004).
Western and Northern blot analyses
The human multi-tissue Northern blot was from Clontech. Other Northern and Western blots were performed as described (Du et al, 2002; Conlan et al, 2004).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Acknowledgments
We thank Virginia Leopold for tissue culture maintenance; Patrick Humbert and Christine Latif for help with FACS and IR; Carleen Cullinane, David Thomas and Alexander Dobrovic for cell lines; and Patrick Humbert, Michael Lisby, Brietta Pike, Helena Richardson and Ana Traven for comments on the manuscript. This work was supported by a grant-in-aid from the Cancer Council Victoria (to JH), an Australian Postgraduate Award (to CJM) and a National Health and Medical Research Council of Australia Senior Research Fellowship (to JH).
References
- Adamson AW, Kim WJ, Shangary S, Baskaran R, Brown KD (2002) ATM is activated in response to N-methyl-N′-nitro-N-nitrosoguanidine-induced DNA alkylation. J Biol Chem 277: 38222–38229 [DOI] [PubMed] [Google Scholar]
- Barnes DE, Lindahl T (2004) Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet 38: 445–476 [DOI] [PubMed] [Google Scholar]
- Bednarek AK, Keck-Waggoner CL, Daniel RL, Laflin KJ, Bergsagel PL, Kiguchi K, Brenner AJ, Aldaz CM (2001) WWOX, the FRA16D gene, behaves as a suppressor of tumor growth. Cancer Res 61: 8068–8073 [PubMed] [Google Scholar]
- Bishop DK, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum RR, Shinohara A (1998) Xrcc3 is required for assembly of Rad51 complexes in vivo. J Biol Chem 273: 21482–21488 [DOI] [PubMed] [Google Scholar]
- Chen CF, Chen PL, Zhong Q, Sharp ZD, Lee WH (1999) Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J Biol Chem 274: 32931–32935 [DOI] [PubMed] [Google Scholar]
- Cline SD, Hanawalt PC (2003) Who's on first in the cellular response to DNA damage? Nat Rev Mol Cell Biol 4: 361–372 [DOI] [PubMed] [Google Scholar]
- Conlan LA, McNees CJ, Heierhorst J (2004) Proteasome-dependent dispersal of PML nuclear bodies in response to alkylating DNA damage. Oncogene 23: 307–310 [DOI] [PubMed] [Google Scholar]
- D'Andrea AD, Grompe M (2003) The Fanconi anaemia/BRCA pathway. Nat Rev Cancer 3: 23–34 [DOI] [PubMed] [Google Scholar]
- Du X-J, Cole TJ, Tenis N, Gao X-M, Köntgen F, Kemp BE, Heierhorst J (2002) Impaired cardiac contractility response to hemodynamic stress in S100A1-deficient mice. Mol Cell Biol 22: 2821–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D, Jackson SP (2002) The FHA domain. FEBS Lett 513: 58–66 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Glaab WE, Risinger JI, Umar A, Barrett JC, Kunkel TA, Tindall KR (1998) Cellular resistance and hypermutability in mismatch repair-deficient human cancer cells following treatment with methyl methanesulfonate. Mutat Res 398: 197–207 [DOI] [PubMed] [Google Scholar]
- Glaab WE, Tindall KR, Skopek TR (1999) Specificity of mutations induced by methyl methanesulfonate in mismatch repair-deficient human cancer cells. Mutation Res 427: 67–78 [DOI] [PubMed] [Google Scholar]
- Hammet A, Pike BL, McNees CJ, Conlan LA, Tenis N, Heierhorst J (2003) FHA domains as phospho-threonine binding modules in cell signaling. IUBMB Life 55: 23–27 [DOI] [PubMed] [Google Scholar]
- Hatanaka A, Yamazoe M, Sale JE, Takata M, Yamamoto K, Kitao H, Sonoda E, Kikuchi K, Yonetani Y, Takeda S (2005) Similar effects of Brca2 truncation and Rad51 paralog deficiency on immunoglubulin V gene diversification in DT40 cells support an early role for Rad51 paralogs in homologous recombination. Mol Cell Biol 25: 1124–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry-Mowatt J, Jackson D, Masson JY, Johnson PA, Clements PM, Benson FE, Thompson LH, Takeda S, West SC, Caldecott KW (2003) XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol Cell 11: 1109–1117 [DOI] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Nagase T, Nakajima D, Seki N, Ohira M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O (1997) Prediction of the coding sequences of unidentified human genes. VIII. 78 new cDNA clones from brain which code for large proteins in vitro. DNA Res 4: 307–313 [DOI] [PubMed] [Google Scholar]
- Jager AC, Rasmussen M, Bisgaard HC, Singh KK, Nielsen FC, Rasmussen LJ (2001) HNPCC mutations in the human DNA mismatch repair gene hMLH1 influence assembly of hMutLalpha and hMLH1-hEXO1 complexes. Oncogene 20: 3590–3595 [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J (2004) Cell-cycle checkpoints and cancer. Nature 432: 316–323 [DOI] [PubMed] [Google Scholar]
- Li MJ, Maizels N (1997) Nuclear Rad51 foci induced by DNA damage are distinct from Rad51 foci associated with B cell activation and recombination. Exp Cell Res 237: 93–100 [DOI] [PubMed] [Google Scholar]
- Li Z, Woo CJ, Iglesias-Ussel MD, Ronai D, Scharff MD (2004) The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev 18: 1–11 [DOI] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R (2003) Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol 5: 572–577 [DOI] [PubMed] [Google Scholar]
- Liu L, Yan L, Donze JR, Gerson SL (2003) Blockage of abasic site repair enhances antitumor efficacy of 1,3-bis-(2-chloroethyl)-1-nitrosourea in colon tumor xenografts. Mol Cancer Ther 2: 1061–1066 [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ (1998) Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282: 1893–1897 [DOI] [PubMed] [Google Scholar]
- Mirzoeva OK, Petrini JHJ (2001) DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol Cell Biol 21: 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu K, Kowalczykowski SC (2003) RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol Cell 11: 1337–1347 [DOI] [PubMed] [Google Scholar]
- O'Regan P, Wilson C, Townsend S, Thacker J (2001) XRCC2 is a nuclear RAD51-like protein required for damage-dependent RAD51 focus formation without the need for ATP binding. J Biol Chem 276: 22148–22153 [DOI] [PubMed] [Google Scholar]
- Paull TT, Rogaku EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM (2000) A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10: 886–895 [DOI] [PubMed] [Google Scholar]
- Pike BL, Yongkiettrakul S, Tsai MD, Heierhorst J (2003) Diverse but overlapping functions of the two forkhead-associated (FHA) domains in Rad53 checkpoint kinase activation. J Biol Chem 278: 30421–30424 [DOI] [PubMed] [Google Scholar]
- Pike BL, Yongkiettrakul S, Tsai MD, Heierhorst J (2004) Mdt1, a novel Rad53 FHA1 domain-interacting protein, modulates DNA damage tolerance and G2/M cell cycle progression in Saccharomyces cerevisiae. Mol Cell Biol 24: 2779–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C, Di Noia JM, Neuberger MS (2004) Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell 16: 163–171 [DOI] [PubMed] [Google Scholar]
- Raderschall E, Golub EI, Haaf T (1999) Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc Natl Acad Sci USA 96: 1921–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C, Jasin M (2000) Recombination between two chromosomes: implications for genomic integrity in mammalian cells. Cold Spring Harb Symp Quant Biol 65: 553–560 [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM (1997) Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 90: 425–435 [DOI] [PubMed] [Google Scholar]
- Stojic L, Mojas N, Cejka P, Di Pietro M, Ferrari S, Marra G, Jiricny J (2004) Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase. Genes Dev 18: 1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P, Krejci L, Van Komen S, Sehorn MG (2003) Rad51 recombinase and recombination mediators. J Biol Chem 278: 42729–42732 [DOI] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S (2001) Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol 21: 2858–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas M, Davies D, West SC (2003) BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene 22: 1115–1123 [DOI] [PubMed] [Google Scholar]
- Tindall KR, Glaab WE, Umar A, Risinger JI, Koi M, Barrett JC, Kunkel TA (1998) Complementation of mismatch repair gene defects by chromosome transfer. Mutat Res 402: 15–22 [DOI] [PubMed] [Google Scholar]
- Traven A, Heierhorst J (2005) SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. BioEssays 27: 397–407 [DOI] [PubMed] [Google Scholar]
- Wang Y, Qin J (2003) MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proc Natl Acad Sci USA 100: 15387–15392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SC (2003) Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol 4: 435–445 [DOI] [PubMed] [Google Scholar]
- Yu VP, Koehler M, Steinlein C, Schmid M, Hanakahi LA, van Gool AJ, West SC, Venkitaraman AR (2000) Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev 14: 1400–1406 [PMC free article] [PubMed] [Google Scholar]
- Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY (1999) BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res 59: 3547–3551 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6


