Abstract
We have identified two LIM domain proteins, LimF and ChLim, from Dictyostelium that interact with each other and with the small, Rab5-related, Rab21 GTPase to collectively regulate phagocytosis. To investigate in vivo functions, we generated cell lines that lack or overexpress LimF and ChLim and strains that express activating or inhibiting variants of Rab21. Overexpression of LimF, loss of ChLim, or expression of constitutively active Rab21 increases the rate of phagocytosis above that of wild type. Conversely, loss of LimF, overexpression of ChLim, or expression of a dominant-negative Rab21 inhibits phagocytosis. Our studies using cells carrying multiple mutations in these genes further indicate that ChLim antagonizes the activating function of Rab21-GTP during phagocytosis; in turn, LimF is required for Rab21-GTP function. Finally, we demonstrate that ChLim and LimF localize to the phagocytic cup and phago-lysosomal vesicles. We suggest that LimF, ChLim, and activated Rab21-GTP participate as a novel signaling complex that regulates phagocytic activity.
Keywords: Dictyostelium, GTPases, phagocytosis, Rab21, vesicle formation
Introduction
‘Professional' phagocytes (Rabinovitch, 1995), including macrophages and Dictyostelium, a free-living, soil amoebae, have an ability to rapidly and efficiently internalize a variety of organisms or particles, a function that is vital for host defenses toward foreign invasion, tissue remodeling, and nutrient uptake. Macrophages, myeloid dendritic cells, and neutrophils can phagocytose microbial pathogens, which potentiates innate and adaptive immunity. The elimination of apoptotic cells by phagocytes is also essential to control immune and inflammatory response, as well as to regulate tissue homeostasis during development and to prevent necrosis. In Dictyostelium, phagocytosis of bacteria and fungi is essential for nutrient capture in the wild.
The phagocytic process begins with particle recognition and cell surface binding (Rabinovitch, 1995; Duhon and Cardelli, 2002; Vieira et al, 2002; Henry et al, 2004). As the plasma membrane and cortical cytoskeleton become reorganized, a phagocytic cup is formed. The particle is then engulfed and internalized, forming the early phagosome, a unique organelle that is trafficked through a series of early and late endocytic membrane compartments that define the phago-lysosomal pathway. Ultimately, an ingested microbe is destroyed.
Phagosome biogenesis has recently been subject to superb proteomic approaches (Garin et al, 2001; Gotthardt et al, 2002). While, many new and unexpected components have been described that provide unique perspective, mechanistic linkages remain to be fully explored. Furthermore, although protection from microbial invasion is a primary immunity response, little is known about recognition specificity or the molecular mechanisms that regulate phagocytic uptake and phagosome trafficking (Vieira et al, 2002). In addition, the infectious nature of Mycobacterium, Salmonella, Legionella, and other pathogenic parasites, which are readily susceptible to cellular uptake but are able to subvert intracellular destruction and to propagate productively within phagocytes, is only partially understood at the host level.
The professional phagocyte Dictyostelium is a genetically and biochemically tractable organism (see Kreppel et al, 2004) for defining molecular mechanisms that regulate phagocytosis. Biochemical and genetic analyses indicate that Dictyostelium and mammalian macrophages/neutrophils have strong similarities in endocytosis (Duhon and Cardelli, 2002; Vieira et al, 2002), despite their separation by more than 500 000 000 years. The shared role of Rac1 during the phagocytosis of yeast in Dictyostelium (Faix, 2002) and of apoptotic cells in Caenorhabditis elegans and mammalian systems (Grimsley et al, 2004) underscores this relationship. In addition, Dictyostelium exhibit phagocytic rates that are several-fold in excess of those in macrophages and neutrophils and, like macrophages, Dictyostelium cells are subject to pathogenic infection by Mycobacterium and Legionella (Skriwan et al, 2002).
We have identified two novel LIM domain proteins, LimF and ChLim, from Dictyostelium that are binding partners and that coordinate with activated, GTP-bound Rab21 to regulate phagocytosis. The in vivo functions of LimF, ChLim, and Rab21 in Dictyostelium were studied in cell lines that carry specific gene deficiencies or that overexpress certain protein variants. Cells that lack ChLim, overexpress LimF, or express a constitutively active form of Rab21 exhibit significantly increased rates of phagocytosis as compared with wild type. To establish genetic and biochemical hierarchies in the pathway, we examined cells carrying mutations in multiple genes. We conclude that activated Rab21-GTP is required to promote high rates of phagocytosis, but that ChLim inhibits the function of Rab21-GTP. Furthermore, LimF is required for the activating function of Rab21-GTP during phagocytosis. Finally, we have confirmed a physical relationship during phagocytosis: GFP-ChLim and YFP-LimF are enriched at the phagocytic cup and associated with intracellular phago-lysosomal vesicles. We suggest that LimF, ChLim, and Rab21-GTP participate in a unique signaling complex that regulates phagocytic activity and discuss potentially related paths in other systems.
Results
A novel LIM domain protein and interacting partners of Dictyostelium
Recent observations have suggested that LIM domain-containing proteins can regulate cellular response through the reorganization of actin filaments at the cell cortex (Prassler et al, 1998; Chien et al, 2000; Khurana et al, 2002a, 2002b; Schneider et al, 2003). LIM domains are unique motifs comprised of two conserved, zinc-finger-like modules: CX2CX16–23HX2CX2CX2CX16–21CX2C. Structural stability of LIM domains depends on the coordinated tetrahedral binding of two zinc-atoms via cysteine and histidine residues. LIM domains act as adapters that mediate protein–protein interactions for a variety of cellular and developmental activities and LIM-regulated cytoskeletal reorganizations are required for cell motility, cell–cell interaction, cell–substratum attachment, and other dynamic processes (Khurana et al, 2002b).
In an effort to identify novel regulators of cytoskeletal function in Dictyostelium, we focused on a new LIM domain protein, LimF, identified in a bioinformatics domain search. PCR amplifications were used to generate full-length genomic and cDNA sequences of LimF. LimF has 197 amino-acid residues organized into three perfect LIM domain (CX2CX16–23HX2CX2CX2CX16–21CX2C) repeats, at residues 9–61, 79–136, and 143–196 (see Figure 1A and Supplementary Figures 1A and 2); no additional motifs are present, thus defining LimF as a LIM-only protein. LimF has ∼50% amino-acid identity within the LIM and spacer regions of the LIM-containing carboxyl-terminus of LimA of Dictyostelium (Supplementary Figure 1B), but we did not detect strong sequence similarity (at <e−15) to any other protein.
Figure 1.
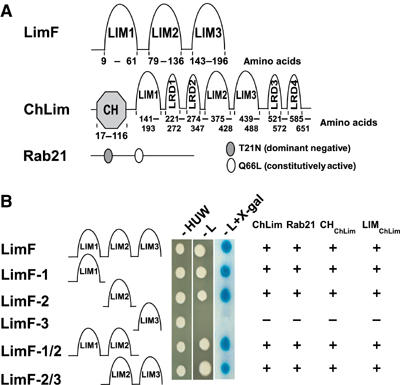
The interacting partners LimF, ChLim, and Rab21. (A) LimF has three LIM domains. ChLim has an N-terminal CH domain, three LIM domains, and four LRDs. Rab21 is a small GTPase; the dominant-negative (T21N) and constitutively activated (Q66L) mutations are indicated. (B) Interactions of LimF with ChLim and Rab21 were studied in a yeast two-hybrid system. All cells were able to grow in the absence of histidine (H), uracil (U), and tryptophan (W). Positive interaction (+) is indicated by growth in the absence of leucine (L) and by enhanced β-galactosidase activity using X-gal as substrate. ChLim is full-length ChLim, CHChLim is the CH domain of ChLim, and LIMChLim is the Lim domain of ChLim. Images of yeast are representative data for full-length ChLim.
To determine the biochemical function of LimF, we searched for interacting partners in a yeast two-hybrid screen. Eight putative LimF-interacting partners were identified in an initial screen. Of these, two were identified multiple times in several screens and continued to display high selectivity as assayed both by growth dependency and intensity of β-galactosidase expression during all subsequent rounds of analyses (see Figure 1B); both of these proteins were selected for additional study. Although neither cDNA fusion was full length, sequence analyses predicted that one of the proteins, designated ChLim, is novel with both calponin homology (CH) and LIM domains (Figure 1A and Supplementary Figures 2, 3 and 4). The other LimF-interacting protein is the small GTPase Rab21, which had been previously named RabB (Bush et al, 1993; see below). The mRNAs for LimF, ChLim, and Rab21 are expressed during growth and throughout all developmental stages examined (Bush et al, 1993; Van Driessche et al, 2002; http://dictybase.org/db/cgi-bin//gene_page.pl?gene_name=jc2v2_0_02067; data not shown), consistent with potential interactions in vivo.
Full-length genomic DNA and cDNAs for both ChLim and Rab21 were amplified using information derived from the completed genomic sequence of Dictyostelium (see Kreppel et al, 2004). ChLim encodes a protein of 686 amino-acid residues and contains a single 126 nt intron. Residues 17–116 form the CH domain with >40% identity (>60% similarity) to CH domains of calponin-like proteins of other species (see Figure 1A and Supplementary Figures 3 and 4). The remainder of the protein is comprised of three bona fide LIM domains and four elements that we have termed LIM-related domains (LRDs) (see Figure 1A and Supplementary Figures 2 and 3). Residues 141–193, 375–428, and 439–488 have consensus LIM domain sequences. Four LRDs are apparent at residues 221–272, 274–347, 521–572, and 585–651. Although each LRD diverges sufficiently from the classic LIM sequence within the central HX2CX2CX2C element to exclude their classification as true LIM domains (see Supplementary Figures 2 and 3), the differences only alter the spacing between conserved cysteine/histidine residues. Each LRD is, thus, still likely to coordinate the tetrahedral binding of two zinc atoms and to exhibit a three-dimensional structure that is similar to the true LIM domains. In addition, the four predicted LRD sequences also align a precisely positioned tyrosine that is often conserved among LIM domain family members (see Supplementary Figures 1 and 3).
Rab proteins form a subgroup of the Ras superfamily of small G proteins that individually appear to regulate the trafficking of specific membrane compartments (Pereira-Leal and Seabra, 2001; Segev, 2001; Colicelli, 2004). Dictyostelium Rab21 was originally named RabB (Bush et al, 1993), but has been reclassified as a Rab21 ortholog since it is 72% identical (85% similar) to the ∼200 amino acids that define the Rab domain of human Rab21, and is >60% identical in overall sequence (see Bush et al, 1993). Little is known of Rab21 function in any system (Pereira-Leal and Seabra, 2001; Colicelli, 2004). There are no known overt orthologs of ChLim or LimF. We have not identified a protein in any other system that has CH and LIM features organized exactly as ChLim, although several mammalian proteins that interact with Rab or the actin machinery possess CH and LIM domains. No LIM-only proteins have been found with the precise organization of LimF.
Functional interactions among LimF, ChLim, and Rab21
To map the interactions among LimF, ChLim, and Rab21, we generated a series of truncated fusion proteins for analyses in the yeast two-hybrid system (Figure 1B). The three LIM domains of LimF were tested individually against full-length Rab21, full-length ChLim, the CH domain (CHChLim; residues 1–116) of ChLim, and the entire LIM region (LIMChLim; residues 138–686) of ChLim (see Figure 1A). Fusion protein expressions were confirmed by Western blot assay. Strong interactions of LIM domains 1 and 2 of LimF were seen with all the Rab21 and ChLim constructs (Figure 1B). In contrast, LIM domain 3 of LimF did not interact with either ChLim or Rab21. This C-terminal LIM domain does not seem to function as a negative regulator for interaction since constructs with LIM domains 2+3 show identical strength of interaction with Rab21 and ChLim as do constructs with LIM domains 1+2 (Figure 1B). Finally, we have shown that residues 413–686 of ChLim (see Figure 1A), which encompass the carboxyl-terminal half of the LIMChLim region, are sufficient for strong interaction with full-length LimF (data not shown). We did not detect LimF–LimF homologous interactions in either a direct two-hybrid assay or a far Western blot overlay (data not shown), under conditions where LimF and ChLim do interact (data not shown).
We also examined the ability of LimF to interact with ChLim and Rab21 proteins using extracts from Dictyostelium. Glutathione S-transferase (GST) and GST-LimF proteins were expressed in Escherichia coli, purified, absorbed to glutathione-coated beads, and used for in vitro pull-down assays. Lysates were prepared from growing Dictyostelium that were engineered to express GFP-ChLim and incubated with the GST or GST-LimF beads. Immunoblot analyses using α-GFP antibody confirmed the specific interaction of GFP-ChLim with GST-LimF, whereas no GFP-ChLim binding was observed with GST control beads (Figure 2A). LimF was also able to interact specifically with endogenous Rab21 in lysates of the GFP-ChLim-expressing cells (Figure 2A).
Figure 2.
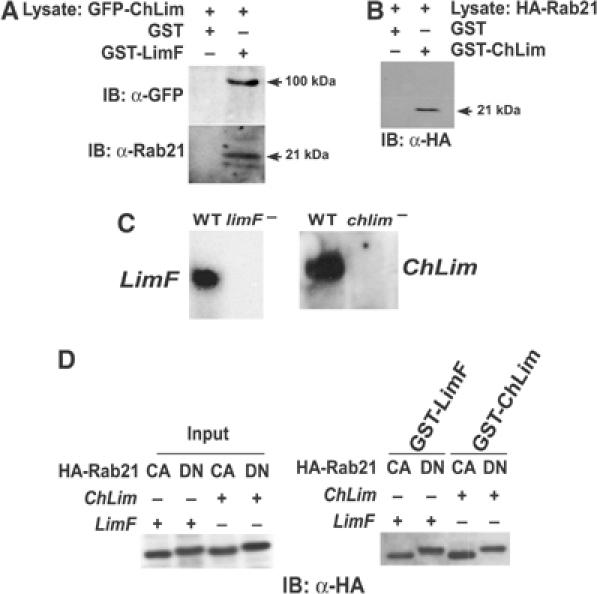
Interactions among LimF, ChLim, and Rab21. (A) Lysates of cells expressing GFP-ChLim were mixed with immobilized GST-LimF and GST control beads, and bound proteins analyzed by immunoblotting (IB) with α-GFP and α-Rab21. (B) Lysates of cells expressing HA-Rab21 were mixed with immobilized GST-ChLim and GST control beads, and bound proteins analyzed by immunoblotting (IB) with α-HA. (C) Total RNA from growing wild-type (WT), limF-null, and chlim-null cells was fractionated on formaldehyde–agarose gels and probed with full-length LimF or ChLim cDNA as indicated. (D) Lysates of limF-null and chlim-null cells expressing HA-Rab21Q66L (CA; constitutively active) or HA-Rab21T21N (DN; dominant-negative) were mixed with immobilized GST-LimF or GST-ChLim, and bound proteins analyzed by immunoblotting (IB) with α-HA. The left panel shows the relative input of equivalent lysate proportions for the indicated cell lines. Constitutively active, GTP-bound Rab21Q66L migrates with a slightly faster mobility than does the dominant-negative Rab21T21N form.
Using lysates from cells expressing HA-Rab21 and glutathione-coated beads in complex with GST-ChLim, we demonstrated that Rab21 is capable of interacting with ChLim, as well as with LimF (Figure 2B). These data further substantiate a functional relationship among the LimF, ChLim, and Rab21 proteins.
Rab proteins cycle between activated, GTP-bound and inactive, GDP-bound forms. Thus, it was critical to examine if the conformational state of Rab21 altered its ability to interact with either LimF or ChLim. In addition, we wished to determine if these interactions required the simultaneous presence of LimF and ChLim. First, using standard methods for targeted mutagenesis, we constructed cell lines specifically deficient in either LimF or ChLim (see Figure 2C). HA fusions of Rab21 were engineered to incorporate either a GTP-locked, activating (Q66L) or inactivating (T21N) mutation (see Figure 1A). The Rab21 fusions were expressed in wild-type, limF-null, and chlim-null cell lines and extracts were prepared for pull-down assays using GST, GST-LimF, and GST-ChLim beads. All forms of the Rab21 proteins were able to bind specifically and equivalently to GST-LimF and GST-ChLim, regardless of the cell extract (Figure 2D). We did not observe nonspecific interaction with GST controls (see Figure 2B). Since the relative inputs of Rab21WT (data not shown), Rab21Q66L, and Rab21T21N were comparable (Figure 2D), we further conclude that the different nucleotide-specific conformations of Rab21 are able to bind to LimF and to ChLim with largely similar efficiencies. These data suggest that neither LimF nor ChLim is an effector of activated Rab21-GTP. Rather, we suggest that both these proteins may regulate the ability of Rab21 to bind and activate downstream components. Finally, the data from the limF- and chlim-null studies indicate that specific interaction of Rab21 with LimF or with ChLim does not require a higher order complex dependent upon the presence of both LimF and ChLim (Figure 2D).
Cortical actin organization in cells carrying mutations in LimF, ChLim, and Rab21
LIM domain proteins in Dictyostelium have been implicated in a variety of actin-mediated processes, including cell polarization, phagocytosis, chemotaxis, and morphogenesis (Prassler et al, 1998; Chien et al, 2000; Khurana et al, 2002a, 2002b; Schneider et al, 2003). We, thus, examined the cytoskeletal organization of F-actin in wild-type and mutant cell lines, using the specific binding component rhodamine-conjugated phalloidin. In wild-type cells, F-actin is primarily observed as a cortical band at the cell periphery with only diffuse cytoplasmic staining (Figure 3A). limF- and chlim-nulls exhibited nearly indistinguishable distributions of F-actin. However, overexpression of LimF (LimFOE) or ChLim (ChLimOE) altered normal F-actin patterns. Both LimFOE and ChLimOE cells show a modest but consistent increase in F-actin-rich filopodia (Figure 3B).
Figure 3.
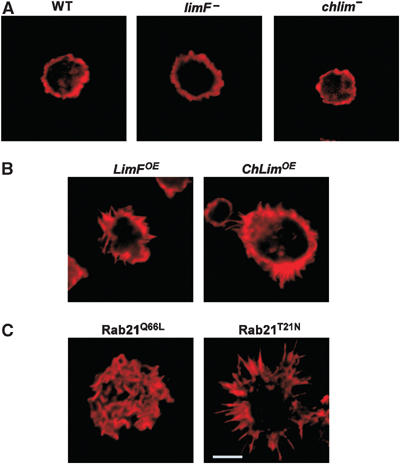
Actin organization in LimF, ChLim, and Rab21 mutant cells. Various strains of Dictyostelium were grown in shaking suspension, fixed with formaldehyde, stained with rhodamine-labeled phalloidin, and analyzed by confocal microscopy. (A) limF- and chlim-null cells display a cortical ring of F-actin similar to wild type (WT). (B) LimFOE and ChLimOE cells show a modest increase in F-actin-rich filopodia. (C) Cells expressing the constitutively active Rab21Q66L have actin-rich, ruffles over their entire surfaces. Cells expressing the dominant-negative Rab21T21N show enhanced actin microspikes; the bar inset represents 10 μm and is comparable for all panels.
Cells expressing the constitutively active Rab21Q66L showed enhanced ruffling of the cell surface, characterized by the prominent display of F-actin-rich blebs (Figure 3C). Expression of the dominant-negative Rab21T21N also alters the organization of cortical F-actin, but in a quite different manner. Rab21T21N-expressing cells have extremely pronounced actin-rich ‘microspikes' and filopodia-like structures.
Mutation of the Rab21, LimF, or ChLim genes did not seem to correlate with many cellular or developmental changes. Protein overexpression or deficiency did not alter patterns of random movement or directed motility to either folate or cAMP or of growth in liquid, axenic media (data not shown), although we did observe a mild cytokinesis defect for chlim-null cells; ∼5% of chlim-nulls had >4 nuclei during growth in shaking culture (data not shown). In addition, all cell lines have substantially normal patterns of development under submerged conditions or on solid matrices (data not shown). Cellular adhesive properties to a plastic substrate were similarly unchanged (Table I). On average, identical shear forces (±10%) were required to disrupt cell–substrate interactions among the different cell lines, although the ChLimOE cells may be slightly less adhesive. This contrasts the major adhesion defects in cells carrying deficiencies of other genes that regulate organization of the cell surface (see Table I). Rab21, LimF, or ChLim also does not appear to have a significant function for fluid-phase uptake (Figure 4). We did, however, notice differences in the ability of the various cell lines to utilize bacteria as a food source. In the most striking phenotype, ChLimOE cells consistently have smaller plaques on bacterial lawns than wild-type controls (data not shown), a phenotype that may be associated with a reduced ability to use bacteria as a nutrient source; reciprocally, chlim-null cells have expanded growth zones when grown on bacteria.
Table 1.
Substrate adhesion of LimF, ChLim, and Rab21 mutant cells
| Cell line | % cells attached (±5%) at 65 r.p.m. |
|---|---|
| WT | 50 |
| limF− | 50 |
| LimFOE | 50 |
| chlim− | 55 |
| ChLimOE | 35 |
| Rab21Q66L | 45 |
| Rab21T21N | 50 |
| sadA− | <5a |
| phg1− | <5a |
| phg2− | <10a |
| myoVII− | <5a |
|
talin− |
<5a |
| Log-phase cells were plated in plastic culture dishes and shaken at varying speeds (0–90 r.p.m.) for 60 min at room temperature. Unattached cells were counted at each speed and normalized to the input cell number. A total of 50% of wild-type (WT) cells remained attached after shaking at 65 r.p.m. For the other cell lines, numbers listed reflect the percentage of cells attached under identical (65 r.p.m.) shaking conditions. | |
| aData for the sadA−, phg1−, phg2−, myoVII−, and talin− cell lines are extrapolated from previously published studies (Niewohner et al, 1997; Cornillon et al, 2000; Tuxworth et al, 2001; Fey et al, 2002; Gebbie et al, 2004) in a similar comparison with wild type. | |
Figure 4.
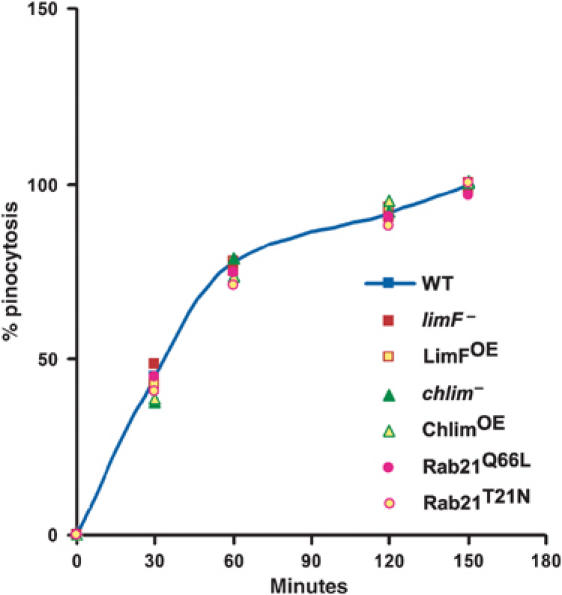
Pinocytosis in LimF, ChLim, and Rab21 mutant cells. Log-phase cells were incubated with TRITC-dextran at room temperature at 160 r.p.m. At the indicated times, samples were withdrawn, quenched with Trypan blue, and intracellular rhodamine fluorescence was measured for equivalent cell numbers. Arbitrary fluorescence units were normalized to the maximal value obtained for wild-type (WT) cells within the same experiment. All the experiments were performed 3–4 times.
LimF, ChLim, and Rab21-GTP cooperatively regulate phagocytosis through specific activating and inhibitory functions
The altered growth patterns on bacterial lawns suggested an altered ability of the various cell lines to utilize bacteria as a nutrient source. This prompted us to investigate directly differences in phagocytosis among these cell lines. Phagocytosis initiates as the particle associates with the cell surface. A phagocytic cup is formed around the particle by membrane progression and particles are drawn into the cell by actin-dependent contractile forces and trafficked into the phago-lysosomal pathway.
We quantitatively measured rates of phagocytosis in the various null and overexpressing cell lines. Equivalent numbers of cells of the different strains were mixed with a defined number of tetramethyl rhodamine isothiocyanate (TRITC)-labeled, heat-killed yeast. Samples were removed throughout a 120–150 min period, treated with Trypan blue to quench the fluorescence of non-internalized yeast, and phagocytosis quantitatively determined by measuring cell-associated fluorescence that persists after an extensive wash treatment.
The limF- and chlim-null cells exhibited completely opposite effects on phagocytosis in comparison to wild-type controls (Figure 5A and Table II). In the absence of ChLim, the rate of phagocytosis was significantly greater than that of wild-type levels. In contrast, loss of LimF reduced overall phagocytosis by ∼40%. These data are in fundamental agreement with growth phenotypes on bacteria, where chlim-nulls exhibit expanded growth (i.e. enhanced phagocytosis), while limF-nulls appear to be prematurely starved for nutrients. The differing effects on phagocytosis were replicated when cells overexpressing LimF (LimFOE cells) or ChLim (ChLimOE cells) were compared. LimFOE cells were more active for phagocytosis than wild type, whereas the ChLimOE cells were more compromised. Thus, LimF and ChLim function antagonistically during phagocytosis. Strikingly, mutations of ChLim were reproducibly more severe than those of LimF.
Figure 5.
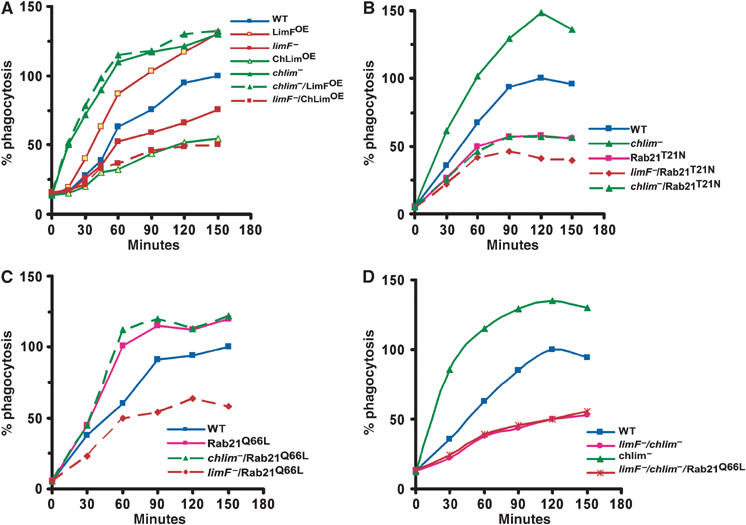
LimF, ChLim, and Rab21 regulation of phagocytosis. (A) Various strains of Dictyostelium were incubated with TRITC-labeled, heat-killed yeast particles at room temperature under shaking conditions. At the indicated time points, samples were collected and processed for fluorimetric analyses. Arbitrary fluorescence units were normalized to the maximal value obtained for wild-type (WT) cells within the same experiment. All the experiments were performed 3–4 times (see Table II). (B) Comparative analyses of Rab21T21N function for quantitative phagocytosis. (C) Comparative analyses of Rab21Q66L function for quantitative phagocytosis. (D) Comparative analyses for quantitative phagocytosis in limF−/chlim− cells.
Table 2.
LimF, ChLim, and Rab21 regulation of phagocytosis
| Cell line | % relative phagocytosis |
|---|---|
| WT | 100±5 |
| limF− | 66±7 |
| ChLimOE | 35±7 |
| limF−/ChLimOE | 40±7 |
| LimFOE | 165±12 |
| chlim− | 230±23 |
| chlim−/LimFOE | 240±30 |
| Rab21T21N | 61±5 |
| Rab21Q66L |
175±16 |
| Various strains of Dictyostelium were incubated with TRITC-labeled, heat-killed yeast particles for 60 min at room temperature under shaking conditions. Arbitrary fluorescence units were normalized to the value obtained for wild-type (WT) cells within the same experiment. All the experiments were performed 3–4 times. Statistical analyses using the Student's t-test indicate significant differences between the mutant strains, at P-values <0.05. 0 times were nearly identical for all cell lines (see Figure 5A). | |
Since LimF and ChLim interact physically, we determined their genetic relationship in chlim-null lines that overexpress LimF (chlim−/LimFOE) and in limF-null lines that overexpress ChLim (limF−/ChLimOE). The phagocytosis phenotype of each double-mutant cell line matched exactly that of the corresponding single ChLim mutation (Figure 5A and Table II). chlim−/LimFOE cells were as phagocytically activated as the parental chlim-nulls, whereas the limF−/ChLimOE cells exhibited a phenotype more similar to ChLimOE than to that of the limF-null (Figure 5A and Table II). These data indicate that ChLim has an inhibitory role during phagocytosis, whereas LimF must be activating. The data may suggest that, genetically, LimF may lie upstream of the inhibitory function of ChLim during phagocytosis.
We next focused on the role of Rab21 as part of a potential phagocytic regulatory complex. Expression of the dominant-negative Rab21T21N significantly inhibited phagocytosis compared to parental wild-type controls (Figure 5B). More significantly, expression of Rab21T21N reduced the normally elevated rate of phagocytosis of chlim-null cells to the low levels characteristic of Rab21T21N-expressing cells. These data indicate that Rab21 must lie in a path that is functionally downstream of the inhibitory action of ChLim. The additional loss of LimF further compromised phagocytosis. limF−/Rab21T21N cells were phagocytically less active than are wild-type cells expressing Rab21T21N (Figure 5B).
Consistent with Rab21 functioning to activate phagocytosis, Rab21Q66L-expressing cells were hyperactive for phagocytosis (Figure 5C). We also monitored phagocytosis in chlim−/Rab21Q66L and limF−/Rab21Q66L cells. Expression of the constitutively active Rab21Q66L is unable to rescue phagocytosis in cells deficient for LimF, while Rab21Q66L and chlim−/Rab21Q66L cells show similarly elevated rates of phagocytosis.
To examine further the inter-relationships among LimF, ChLim, and Rab21, we analyzed phagocytosis in cells deficient for both LimF and ChLim (limF−/chlim− cells) and in limF−/chlim− cells expressing the constitutively active form of Rab21 (limF−/chlim−/Rab21Q66L cells). If LimF functioned simply to antagonize the inhibitory role of ChLim, limF−/chlim− cells would be predicted to have elevated rates of phagocytosis. This was not observed (Figure 5D); expression of constitutively active Rab21 in limF−/chlim− cells was unable to restore phagocytosis.
Collectively, these data indicate a functional and highly complex interactive relationship among LimF, ChLim, and Rab21. Activated Rab21 directs phagocytosis; in contrast, ChLim serves to inhibit the activating function of Rab21, whereas LimF is required for the activating function of Rab21. The data do not predict a simple linear relationship among the proteins, but rather suggest that LimF/ChLim/Rab21 function as a regulatory unit to direct phagocytic uptake (see Figure 8).
Figure 8.
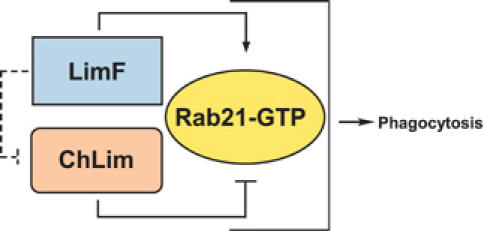
The antagonistic action of LimF, ChLim, and Rab21 during phagocytosis. We propose that phagocytosis in Dictyostelium is regulated by activating and inhibitory interactions of LimF, ChLim, and Rab21-GTP and suggest that similar regulatory pathways involving Rab21 are required in other professional phagocytes.
The CH domain of the ChLim inhibitor and LimF regulate association with the phagocytic cup
To study the subcellular localizations of the Rab/LIM phagocytic proteins in live cells, we separately expressed GFP-ChLim, YFP-LimF, and YFP-Rab21, and examined fluorescence in growing cells using confocal laser scanning microscopy. GFP-ChLim can be observed at the cortex of the cell, at the periphery of some intracellular vesicles/vacuoles, and additionally with a diffuse intracellular pattern (Figure 6A). To examine these localizations further, we separately expressed the CH and LIM regions (see Figure 1) of ChLim as fusions with GFP. GFP-CHChLim shows exclusive cortical localization, whereas GFP-LIMChLim is only detected in the cytosol. In general, GFP-LIMChLim exhibits a diffused cytosolic distribution, but minimal association with intracellular vacuoles, suggesting that this latter association of full-length ChLim may require both the CH domain and the LIM region (Figure 6A). The YFP patterns of LimF and Rab21 exhibited diffuse cytosolic fluorescence with enrichment around discrete intracellular vacuoles, which is also suggestive of association with intracellular phago-lysosomal vesicles (Figure 6A).
Figure 6.
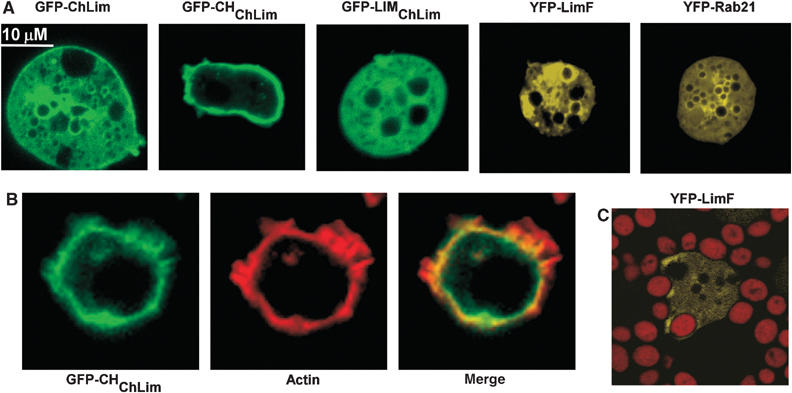
Cellular localizations of ChLim, LimF, and Rab21. (A) Live, wild-type cells expressing GFP-ChLim, GFP-CHChLim, GFP-LIMChLim, YFP-LimF, and YFP-Rab21 were resuspended in phosphate buffer, plated in chambered coverglasses, and analyzed by confocal microscopy. (B) Cells expressing GFP-CHChLim were fixed with formaldehyde, stained with rhodamine-phalloidin, and analyzed by confocal microscopy. (C) Cells expressing YFP-LimF were fed TRITC-labeled, heat-killed yeast particles. Individual cells were imaged simultaneously for YFP and rhodamine fluorescence by confocal microscopy.
The enrichment of CHChLim at the cell cortex is consistent with an association with F-actin. Therefore, we stained cells expressing GFP-CHChLim with rhodamine-labeled phalloidin and simultaneously imaged the cellular localization of both CHChLim and actin (Figure 6B). The images confirm the tight association of CHChLim at the cell cortex. There does appear to be some colocalization of CHChLim and actin at the intracellular face of the cytoskeletal cortex, but not at the actin-rich projections, a pattern similar to cornonin and cofilin.
The presence of ChLim near the cell surface and its clear involvement in phagocytosis (see Figure 5) suggests the possibility of ChLim engagement with the phagocytic cup. Thus, we examined dynamically the subcellular localization of GFP-ChLim in cells that were challenged with TRITC-labeled yeast cells (Figure 7A; see Supplementary Figure 5, video 1). GFP-ChLim rapidly accumulates at the site of attachment of yeast and remains in association throughout the engulfing process and as the yeast particles traffic within the cell. As with full-length ChLim, GFP-CHChLim is recruited rapidly to the phagocytic cup; however and in contrast with ChLim, GFP-CHChLim fluorescence is rapidly lost from the vesicles as the yeast-containing vesicles are fully engulfed (Figure 7C; see Supplementary Figure 5, video 2). In many respects, CHChLim association with the phagocytic cup resembles that of actin itself (Schreiner et al, 2002). To visualize actin localization, we expressed GFP-ABD, the actin binding domain (ABD) of ABP-120/filamin in fusion with GFP (Pang et al, 1998). In the presence of yeast, there is an initial enrichment of actin at the site of phagocytosis, but also rapid dissociation once the yeast particle becomes internalized (Figure 7D; see Supplementary Figure 5, video 3).
Figure 7.
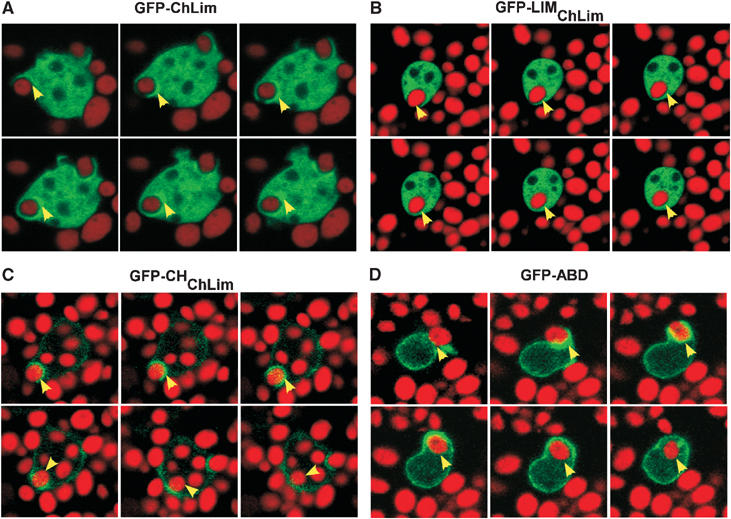
Dynamic distribution of ChLim during phagocytosis. Live wild-type cells expressing (A) GFP-ChLim, (B) GFP-CHChLim, (C) GFP-LIMChLim, and (D) GFP-ABD were fed TRITC-labeled, heat-killed yeast particles. Individual cells were imaged simultaneously for GFP and rhodamine fluorescence during a time course by confocal microscopy. See Supplementary Figure 5.
We were unable to detect consistent association of the LIMChLim region with internalized yeast (Figure 7B). Although LIMChLim may be too broadly distributed to permit localized imaging, ChLim association with phagosomes is clearly functionally initiated in a CH-dependent manner. The LIM region appears to be required to maintain association with the yeast particles as they transit from the cortex; however, it may not be capable of secondary association in the absence of the initiating function of the CH domain.
Finally, we have demonstrated that YFP-LimF is also enriched at the phagocytic cup in a manner similar to that of ChLim (Figure 6C), although we have been unable to observe unequivocal association of YFP-Rab21 with phagocytic vesicles.
Discussion
Phagocytosis is fundamental to the survival of a broad variety of organisms. Many unicellular species rely on microbial and particle engulfment for nutrient capture. Multicellular animals have extended this response for purposes of host defense against invading microbial pathogens and for the regulation of specific developmental pathways. We have described a novel complex comprised of a Rab21 ortholog and LIM proteins that regulates phagocytic activity in Dictyostelium and suggest that a functionally similar pathway for Rab21 regulation may mediate phagocytic control in other species.
Activated Rab21-GTP positively regulates phagocytosis (see Figure 8). In contrast, loss of ChLim increases the rate of phagocytosis indicating that it has an inhibitory role. Further, since loss of ChLim is unable to rescue the low phagocytic rates of cells expressing the dominant-negative Rab21T21N, ChLim must lie upstream in the regulatory pathway. Like Rab21-GTP, LimF is a phagocytic activator.
The Rab GTPases comprise a large family of proteins that target specific intracellular compartments and regulate distinct pathways of membrane trafficking (Somsel Rodman and Wandinger-Ness, 2000; Zerial and McBride, 2001; Henry et al, 2004). In the activated, GTP-bound state, Rab subtypes recruit discrete effectors to promote vesicle formation and movement and membrane fusion. Rab subtypes can be classified into broad groupings on the basis of their amino-acid sequence relationships. Group V (Pereira-Leal and Seabra, 2001) includes mammalian Rab subtypes 5, 22A, 21, and 31 (Pereira-Leal and Seabra, 2001; Colicelli, 2004). Rab5 is the most ancient and may be expressed ubiquitously. Rab21 is absent in yeast, but is expressed in Dictyostelium, nematodes, and Drosophila. Rab5 and Rab22A appear to exhibit unique involvement in phagosome formation and early endosome trafficking (Roberts et al, 1999; Clemens et al, 2000; Duclos et al, 2000, 2003; Kauppi et al, 2002; Perskvist et al, 2002; Vieira et al, 2003; Saito-Nakano et al, 2004). Little, however, has been published on the function of Rab21 in any system. Our data suggest that Rab21-GTP, like Rab5, plays a fundamental role in phagosome formation and early endocytic pathways. The colocalization of the Rab21 regulators ChLim and LimF with the phagocytic cup and the forming phagosome lend strong support to this model. The absence of Rab22A and Rab31 in nematodes, Drosophila, and Dictyostelium (see Pereira-Leal and Seabra, 2001; Colicelli, 2004) further emphasizes the likelihood for a conserved and critical role for Rab21 in host defense and development.
While Rab GTPases are fundamental for phagosome maturation and trafficking, actin remodeling is also critical. Actin is polymerized at the site of particle attachment and directs membrane extension during phagosome formation. In Dictyostelium, many actin-associated proteins are enriched at the phagocytic cup and play essential roles in phagocytosis. For example, cells that are deficient in the two F-actin crosslinking proteins α-actinin and ABP120 have reduced uptake and intracellular growth of Legionella (Fajardo et al, 2004). LIM proteins LimC and LimD are also involved in cytoskeletal organization and associate with the phagocytic cup (Khurana et al, 2002a, 2002b), and cells lacking both limC and limD have phagocytic properties that are similar to those of the α-actinin−/ABP120− cells (Fajardo et al, 2004). However, the actions of Rab21 on phagocytosis do not appear to function by simply altering actin remodeling. Rather, we suggest that Rab21 acts in parallel with Rab5 to regulate vesicle trafficking. None of the LimF, ChLim, or Rab21 mutant cell lines show adhesion abnormalities (see Table I), defects that are associated with changes in cytoskeletal structure and that are known to indirectly alter phagocytic rates (see Niewohner et al, 1997; Cornillon et al, 2000; Tuxworth et al, 2001; Fey et al, 2002; Gebbie et al, 2004). In addition, while we have observed some alterations in F-actin assembly, they do not correlate with phagocytic changes. Finally, no abnormalities were observed for any of the cell lines for pinocytosis or growth in liquid media (see Figure 4), thus separating LimF, ChLim, and Rab21 function from a mechanism that is common to both phagocytosis and pinocytosis (see Cardelli, 2001).
Central to our model for phagocytic regulation is activated Rab21-GTP. Since, inactive Rab21T21N and activated Rab21Q66L are able to interact equally with and independently of both LimF and ChLim, neither would appear to function as an effector protein for Rab21-GTP. We suggest that Rab21, LimF, and ChLim participate as a novel interactive regulatory complex, where ChLim possesses an inhibitory role within a complex (see Figure 8); the activating function of Rab21Q66L for phagocytosis is dependent on the presence of LimF. As mediators of protein–protein interactions, LimF and ChLim may have functional counterparts in other systems that are not immediately identified by simple bioinformatics. In this regard, LimF and ChLim have unique functions among the characterized LIM proteins of Dictyostelium, which primarily regulate cytoskeletal function and cell adhesion and motility (Khurana et al, 2002a, 2002b).
Materials and methods
Growth, development, and transformation of Dictyostelium
For growth on bacteria, Dictyostelium were plated on SM agar plates overlaid with Klebsiella aerogenes. Cells were also cultivated in shaking suspension or in submerged culture at 21°C in D3T medium (KD Medical). The mutant cell lines were maintained in the presence of 5 μg/ml blasticidin and/or 10–20 μg/ml of G418 and/or 25 μg/ml of hygromycin B, as appropriate. Null mutants of LimF and Chlim were generated by homologous recombination of targeting constructs disrupted by insertion of a blasticidin resistance cassette. For generation of limF−/chlim− double null mutant, gene replacement vector was constructed by insertion of G418 cassette in the coding region of ChLim. The limF− cells were transformed with the resulting vector and transformants were selected in the presence of blasticidin and G418. All the transformants were screened by PCR and further confirmed by RT–PCR and Northern blot hybridization.
For expression of GFP fusion proteins, full-length and deletion constructs of Chlim were fused to the C-terminus of GFP into the pDexRH expression vector (Westphal et al, 1997). YFP-tagged versions of LimF and Rab21 were generated by fusing the respective cDNAs at the C-terminus of YFP into pDXA-YFP expression vector (Knetsch et al, 2002). For overexpression, cells were electroporated with 5–10 μg of plasmid DNA and selected with 20 μg/ml of G418 (Invitrogen) or 25 μg/ml of Hygromycin B (Calbiochem).
Cell lines used were as follows: WT (wild type)—Ax3; limF−—limF-null, BLAST resistant; LimFOE—LimF overexpressor, G418 resistant; chlim−—chlim-null, BLAST resistant; ChLimFOE—ChLim overexpressor, G418 resistant; Rab21Q66L—Rab21 constitutively active, G418 resistant; Rab21T21N—Rab21 dominant negative, G418 resistant; limF−/ChLimOE—BLAST resistant+G418 resistant; chlim−/LimFOE—BLAST resistant+G418 resistant; limF−/Rab21Q66L—BLAST resistant+G418 resistant; limF−/Rab21T21N—BLAST resistant+G418 resistant; chlim−/Rab21Q66L—BLAST resistant+G418 resistant; chlim−/Rab21T21N—BLAST resistant+G418 resistant; limF−/chlim−—BLAST resistant+G418 resistant; limF−/chlim−/Rab21Q66L—BLAST resistant+G418 resistant+hygromycin resistant; GFP-ChLim—GFP fused to full-length ChLim, G418 resistant; GFP-CHChLim—GFP fused to CH domain of ChLim, G418 resistant; GFP-LIMChLim—GFP fused to Lim domain of ChLim, G418 resistant; YFP-Rab21—YFP fused to full-length Rab21, G418 resistant; YFP-LimF—YFP fused to full-length LimF, G418 resistant.
Rab21 mutagenesis
RabB (Bush et al, 1993) has been renamed Rab21 and we have used this new convention throughout. pAD-HARab21WT and pAD-HARab21QL were kindly provided by Dr James A Cardelli and used to express, respectively, wild-type Rab21, constitutively active Rab21Q66L. Dominant-negative Rab21T21N was generated by PCR-based mutagenesis using the primers GAAGGTTGTGTTGGTAAAAACTCAATAG TTTTTAGATATATAGAC and GTCTATATATCTAAAAACTATTGAGTTT TTACCAACACAACCTTC and QuickChange II (Stratagene). Mutation sites, in bold, change the threonine at position 21 to asparagine. All of the constructs were verified by sequence analyses.
Yeast two-hybrid interactions
For bait constructs, protein coding regions of LimF were cloned in-frame with the DNA binding domain (BD) of pLexA (Clontech Matchmaker). Yeast cultures were grown under standard conditions in liquid medium or on solid medium with minimal synthetic dropout medium. All yeast transformations were carried out using standard TE/lithium acetate. In brief, the reporter strain EGY48/p8op-lacZ was sequentially transformed with the bait plasmid as the pLexA-LimF fusion and a Dictyostelium cDNA library (8–16 h) cloned into pB42AD (kindly provided by Dr Richard A Firtel). Transformants were screened for leucine auxotrophic marker (Leu) and β-galactosidase (β-gal).
For characterization of individual positives, plasmids were isolated from yeast and transformed into E. coli KC8 cells (hsdR, leuB600, trpC9830, pyrF∷Tn5, hisB463, lacΔX74, strA, galU, galK) and selected on M9 plates supplemented with ampicillin and thiamine. The plasmids were isolated from bacterial cells and were sequenced using the prey plasmid-specific primers.
Domain-specific interactions were also tested. EGY48/p8op-lacZ yeast cells were simultaneously transformed with prey plasmids containing full-length ChLim or Rab21 or truncated versions of ChLim and with bait plasmids containing the different LIM domains of LimF. The selected transformants were tested on minimal plates lacking Leu in the presence of X-gal as above.
All the constructs were verified by sequence and Western blot analyses.
Protein purification and pull-down assays
The full-length cDNA coding regions of LimF and ChLim were cloned downstream of the GST sequences in pGEX vectors (Amersham-Pharmacia Biotech). GST alone and both fusion proteins were expressed in E. coli strain BL21 (DE3; Stratagene) under the control of the tac promoter.
For lysates, 5 × 107 Dictyostelium cells were incubated on ice for 30 min in 100 μl of lysis buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 0.5% Triton X-100 plus protease inhibitors). The lysate was cleared by centrifugation at 12 000 r.p.m. for 10 min at 4°C.
Gutathione-Sepharose beads (BD Biosciences Clontech) complexed with 20–30 μg of purified GST, GST-LimF, or GST-CH-LIM were incubated with 100 μl of lysate at 4°C for 3 h. Beads were harvested by centrifugation and washed at 4°C in lysis buffer. A 50 μl portion of 1 × SDS gel loading buffer was added to the pelleted beads and the mixture was boiled for 10 min. Proteins were fractionated by NuPage 4–12% acrylamide gels (Invitrogen) and blotted onto nitrocellulose membranes. Membranes were blocked in TBS-T (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20) containing 5% nonfat milk and probed with α-GFP (Covance), α-Rab21 (provided by James A Cardelli), or α-HA (Sigma-Aldrich) antibody.
Fluorescence microscopy
The cells expressing GFP- and YFP-tagged fusion proteins were harvested in log phase, washed, and resuspended in PBS. Cells were plated on either chambered coverglasses (Nalge Nunc International) or glass bottom dishes (MatTek Corporation), allowed to settle for 15 min, and observed using the Axiovert 100M inverted microscope (Carl Zeiss). In vivo phagocytosis, using TRITC-labeled yeast, was performed as described previously (Maniak et al, 1995).
For F-actin staining, cells were resuspended in PBS and layered on glass coverslips. After 15 min, PBS was replaced by 0.5 ml of fix solution (3.7% formaldehyde in PBS). After 5 min, the fix solution was removed and replaced with 0.5 ml of 0.5% NP-40 in PBS to permeabilize the cells. After 5 min, detergent solution was removed and the coverslips were immediately washed three times in PBS. F-actin was stained with TRITC-phalloidin (Sigma) in PBS for 30 min in a covered humid chamber at room temperature.
Fixed and live cells were observed using an inverted microscope (Axiovert 100M) equipped with a confocal laser system (LSM 510, Carl Zeiss) and an oil immersion × 63/1.4 objective lens. An argon laser (488 nm) was used for excitation of GFP and YFP and HeNe1 (535 nm) was used for rhodamine.
Quantitative phagocytosis
Quantitative phagocytosis using heat-killed yeast cell was performed as described (Maniak et al, 1995). Dictyostelium were washed, and resuspended in fresh medium at 2 × 106 cells/ml. After recovery for 15 min in shaking suspension at room temperature, TRITC-labeled heat-killed yeast cells were added to the cell suspension. Samples of 1 ml were withdrawn at indicated time points and added to Trypan blue solution (20 mg/ml dissolved in 20 mM sodium citrate containing 150 mM NaCl) to quench the fluorescence of noninternalized yeast cells. Cell pellets were resuspended in Phosphate buffer, and fluorescence was measured in a luminescence spectrometer LS50B (Perkin-Elmer) using 544 nm light for excitation and 574 nm for emission.
Quantitative pinocytosis
Log-phase cells were incubated with TRITC-dextran at room temperature at 160 r.p.m. (Hacker et al, 1997). At selected times, 1 ml samples were withdrawn and quenched with Trypan blue. The cell pellets were resuspended in phosphate buffer and intracellular rhodamine fluorescence was measured; excitation was at 544 nm and emission at 574 nm. Arbitrary fluorescence units were normalized to the maximal value obtained for wild-type cells within the same experiment. All the experiments were performed 3–4 times.
Cell adhesion assay
Adhesion of cells to substrata was performed as described (Fey et al, 2002). Briefly, log-phase cells were plated overnight in tissue culture dishes. The next day, the dishes were shaken for 60 min at varying speeds, from 0 to 90 r.p.m. Unattached cells were removed and counted using a hemocytometer.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Video 1
Supplementary Video 2
Supplementary Video 3
Acknowledgments
We are exceedingly grateful to Drs D Rosel, C Parent, J Hammer, B Khurana, and R Siegel for their many helpful and interesting discussions. Finally, we cannot express our gratitude sufficiently to Joshua Krupp during the early phase of these studies and to Drs J Cardelli and R Firtel for generously providing valuable reagents.
References
- Bush J, Franek K, Daniel J, Spiegelman GB, Weeks G, Cardelli J (1993) Cloning and characterization of five novel Dictyostelium discoideum rab-related genes. Gene 136: 55–60 [DOI] [PubMed] [Google Scholar]
- Cardelli J (2001) Phagocytosis and macropinocytosis in Dictyostelium: phosphoinositide-based processes, biochemically distinct. Traffic 2: 311–320 [DOI] [PubMed] [Google Scholar]
- Chien S, Chung CY, Sukumaran S, Osborne N, Lee S, Ellsworth C, McNally JG, Firtel RA (2000) The Dictyostelium LIM domain-containing protein LIM2 is essential for proper chemotaxis and morphogenesis. Mol Biol Cell 11: 1275–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, Horwitz MA (2000) Deviant expression of Rab5 on phagosomes containing the intracellular pathogens Mycobacterium tuberculosis and Legionella pneumophila is associated with altered phagosomal fate. Infect Immun 68: 2671–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colicelli J (2004) Human RAS superfamily proteins and related GTPases. Sci STKE 2004: RE13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillon S, Pech E, Benghezal M, Ravanel K, Gaynor E, Letourneur F, Bruckert F, Cosson P (2000) Phg1p is a nine-transmembrane protein superfamily member involved in Dictyostelium adhesion and phagocytosis. J Biol Chem 275: 34287–34292 [DOI] [PubMed] [Google Scholar]
- Duclos S, Corsini R, Desjardins M (2003) Remodeling of endosomes during lysosome biogenesis involves ‘kiss and run' fusion events regulated by rab5. J Cell Sci 116: 907–918 [DOI] [PubMed] [Google Scholar]
- Duclos S, Diez R, Garin J, Papadopoulou B, Descoteaux A, Stenmark H, Desjardins M (2000) Rab5 regulates the kiss and run fusion between phagosomes and endosomes and the acquisition of phagosome leishmanicidal properties in RAW 264.7 macrophages. J Cell Sci 113 (Part 19): 3531–3541 [DOI] [PubMed] [Google Scholar]
- Duhon D, Cardelli J (2002) The regulation of phagosome maturation in Dictyostelium. J Muscle Res Cell Motil 23: 803–808 [DOI] [PubMed] [Google Scholar]
- Faix J (2002) The actin-bundling protein cortexillin is the downstream target of a Rac1-signaling pathway required for cytokinesis. J Muscle Res Cell Motil 23: 765–772 [DOI] [PubMed] [Google Scholar]
- Fajardo M, Schleicher M, Noegel A, Bozzaro S, Killinger S, Heuner K, Hacker J, Steinert M (2004) Calnexin, calreticulin and cytoskeleton-associated proteins modulate uptake and growth of Legionella pneumophila in Dictyostelium discoideum. Microbiology 150: 2825–2835 [DOI] [PubMed] [Google Scholar]
- Fey P, Stephens S, Titus MA, Chisholm RL (2002) SadA, a novel adhesion receptor in Dictyostelium. J Cell Biol 159: 1109–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garin J, Diez R, Kieffer S, Dermine JF, Duclos S, Gagnon E, Sadoul R, Rondeau C, Desjardins M (2001) The phagosome proteome: insight into phagosome functions. J Cell Biol 152: 165–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebbie L, Benghezal M, Cornillon S, Froquet R, Cherix N, Malbouyres M, Lefkir Y, Grangeasse C, Fache S, Dalous J, Bruckert F, Letourneur F, Cosson P (2004) Phg2, a kinase involved in adhesion and focal site modeling in Dictyostelium. Mol Biol Cell 15: 3915–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt D, Warnatz HJ, Henschel O, Bruckert F, Schleicher M, Soldati T (2002) High-resolution dissection of phagosome maturation reveals distinct membrane trafficking phases. Mol Biol Cell 13: 3508–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsley CM, Kinchen JM, Tosello-Trampont AC, Brugnera E, Haney LB, Lu M, Chen Q, Klingele D, Hengartner MO, Ravichandran KS (2004) Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. J Biol Chem 279: 6087–6097 [DOI] [PubMed] [Google Scholar]
- Hacker U, Albrecht R, Maniak M (1997) Fluid-phase uptake by macropinocytosis in Dictyostelium. J Cell Sci 110 (Part 2): 105–112 [DOI] [PubMed] [Google Scholar]
- Henry RM, Hoppe AD, Joshi N, Swanson JA (2004) The uniformity of phagosome maturation in macrophages. J Cell Biol 164: 185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi M, Simonsen A, Bremnes B, Vieira A, Callaghan J, Stenmark H, Olkkonen VM (2002) The small GTPase Rab22 interacts with EEA1 and controls endosomal membrane trafficking. J Cell Sci 115: 899–911 [DOI] [PubMed] [Google Scholar]
- Khurana B, Khurana T, Khaire N, Noegel AA (2002a) Functions of LIM proteins in cell polarity and chemotactic motility. EMBO J 21: 5331–5342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana T, Khurana B, Noegel AA (2002b) LIM proteins: association with the actin cytoskeleton. Protoplasma 219: 1–12 [DOI] [PubMed] [Google Scholar]
- Knetsch ML, Tsiavaliaris G, Zimmermann S, Ruhl U, Manstein DJ (2002) Expression vectors for studying cytoskeletal proteins in Dictyostelium discoideum. J Muscle Res Cell Motil 23: 605–611 [DOI] [PubMed] [Google Scholar]
- Kreppel L, Fey P, Gaudet P, Just E, Kibbe WA, Chisholm RL, Kimmel AR (2004) dictyBase: a new Dictyostelium discoideum genome database. Nucleic Acids Res 32 (Database issue): D332–D333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniak M, Rauchenberger R, Albrecht R, Murphy J, Gerisch G (1995) Coronin involved in phagocytosis: dynamics of particle-induced relocalization visualized by a green fluorescent protein Tag. Cell 83: 915–924 [DOI] [PubMed] [Google Scholar]
- Niewohner J, Weber I, Maniak M, Muller-Taubenberger A, Gerisch G (1997) Talin-null cells of Dictyostelium are strongly defective in adhesion to particle and substrate surfaces and slightly impaired in cytokinesis. J Cell Biol 138: 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KM, Lee E, Knecht DA (1998) Use of a fusion protein between GFP and an actin-binding domain to visualize transient filamentous-actin structures. Curr Biol 8: 405–408 [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC (2001) Evolution of the Rab family of small GTP-binding proteins. J Mol Biol 313: 889–901 [DOI] [PubMed] [Google Scholar]
- Perskvist N, Roberg K, Kulyte A, Stendahl O (2002) Rab5a GTPase regulates fusion between pathogen-containing phagosomes and cytoplasmic organelles in human neutrophils. J Cell Sci 115: 1321–1330 [DOI] [PubMed] [Google Scholar]
- Prassler J, Murr A, Stocker S, Faix J, Murphy J, Marriott G (1998) DdLIM is a cytoskeleton-associated protein involved in the protrusion of lamellipodia in Dictyostelium. Mol Biol Cell 9: 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitch M (1995) Professional and non-professional phagocytes: an introduction. Trends Cell Biol 5: 85–87 [DOI] [PubMed] [Google Scholar]
- Roberts RL, Barbieri MA, Pryse KM, Chua M, Morisaki JH, Stahl PD (1999) Endosome fusion in living cells overexpressing GFP-rab5. J Cell Sci 112 (Part 21): 3667–3675 [DOI] [PubMed] [Google Scholar]
- Saito-Nakano Y, Yasuda T, Nakada-Tsukui K, Leippe M, Nozaki T (2004) Rab5-associated vacuoles play a unique role in phagocytosis of the enteric protozoan parasite Entamoeba histolytica. J Biol Chem 279: 49497–49507 [DOI] [PubMed] [Google Scholar]
- Schneider N, Weber I, Faix J, Prassler J, Muller-Taubenberger A, Kohler J, Burghardt E, Gerisch G, Marriott G (2003) A Lim protein involved in the progression of cytokinesis and regulation of the mitotic spindle. Cell Motil Cytoskeleton 56: 130–139 [DOI] [PubMed] [Google Scholar]
- Schreiner T, Mohrs MR, Blau-Wasser R, von Krempelhuber A, Steinert M, Schleicher M, Noegel AA (2002) Loss of the F-actin binding and vesicle-associated protein comitin leads to a phagocytosis defect. Eukaryot Cell 1: 906–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N (2001) Ypt/rab gtpases: regulators of protein trafficking. Sci STKE 2001: RE11. [DOI] [PubMed] [Google Scholar]
- Skriwan C, Fajardo M, Hagele S, Horn M, Wagner M, Michel R, Krohne G, Schleicher M, Hacker J, Steinert M (2002) Various bacterial pathogens and symbionts infect the amoeba Dictyostelium discoideum. Int J Med Microbiol 291: 615–624 [DOI] [PubMed] [Google Scholar]
- Somsel Rodman J, Wandinger-Ness A (2000) Rab GTPases coordinate endocytosis. J Cell Sci 113 (Part 2): 183–192 [DOI] [PubMed] [Google Scholar]
- Tuxworth RI, Weber I, Wessels D, Addicks GC, Soll DR, Gerisch G, Titus MA (2001) A role for myosin VII in dynamic cell adhesion. Curr Biol 11: 318–329 [DOI] [PubMed] [Google Scholar]
- Van Driessche N, Shaw C, Katoh M, Morio T, Sucgang R, Ibarra M, Kuwayama H, Saito T, Urushihara H, Maeda M, Takeuchi I, Ochiai H, Eaton W, Tollett J, Halter J, Kuspa A, Tanaka Y, Shaulsky G (2002) A transcriptional profile of multicellular development in Dictyostelium discoideum. Development 129: 1543–1552 [DOI] [PubMed] [Google Scholar]
- Vieira OV, Botelho RJ, Grinstein S (2002) Phagosome maturation: aging gracefully. Biochem J 366: 689–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira OV, Bucci C, Harrison RE, Trimble WS, Lanzetti L, Gruenberg J, Schreiber AD, Stahl PD, Grinstein S (2003) Modulation of Rab5 and Rab7 recruitment to phagosomes by phosphatidylinositol 3-kinase. Mol Cell Biol 23: 2501–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal M, Jungbluth A, Heidecker M, Muhlbauer B, Heizer C, Schwartz JM, Marriott G, Gerisch G (1997) Microfilament dynamics during cell movement and chemotaxis monitored using a GFP-actin fusion protein. Curr Biol 7: 176–183 [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H (2001) Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2: 107–117 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Video 1
Supplementary Video 2
Supplementary Video 3


