Abstract
The minimum structure of the Raf-1 serine/threonine kinase that recognizes active Ras was used to create a green fluorescent fusion protein (GFP) for monitoring Ras activation in live cells. In spite of its ability to bind activated Ras in vitro, the Ras binding domain (RBD) of Raf-1 (Raf-1[51-131]GFP) failed to detect Ras in Ras-transformed NIH 3T3 fibroblasts and required the addition of the cysteine-rich domain (CRD) (Raf-1[51-220]GFP) to show clear localization to plasma membrane ruffles. In normal NIH 3T3 cells, (Raf-1[51-220]GFP) showed minimal membrane localization that was enhanced after stimulation with platelet-derived growth factor or phorbol-12-myristate-13-acetate. Mutations within either the RBD (R89L) or CRD (C168S) disrupted the membrane localization of (Raf-1[51-220]GFP), suggesting that both domains contribute to the recruitment of the fusion protein to Ras at the plasma membrane. The abilities of the various constructs to localize to the plasma membrane closely correlated with their inhibitory effects on mitogen-activated protein kinase kinase1 and mitogen-activated protein kinase activation. Membrane localization of full-length Raf-1-GFP was less prominent than that of (Raf-1[51-220]GFP) in spite of its strong binding to RasV12 and potent activation of mitogen-activated protein kinase. These finding indicate that both RBD and CRD are necessary to recruit Raf-1 to active Ras at the plasma membrane, and that these domains are not fully exposed in the Raf-1 molecule. Visualization of activated Ras in live cells will help to better understand the dynamics of Ras activation under various physiological and pathological conditions.
INTRODUCTION
Compartmentalization of cellular signals has long been recognized as an effective means to ensure signaling specificity. Conventional techniques, whether biochemical or morphological, have been able to detect changes in redistribution of important signaling molecules only when the underlying process is strong enough to survive cell fractionation or fixation procedures. However, it is more difficult to follow labile molecular interactions with those techniques, even though those are equally important in cell regulation. Several recent studies explored the possibility of whether biochemical processes can be monitored in single living cells by fluorescent probes that specifically recognize the activated form(s) of signaling proteins or their enzymatic products (Teruel and Meyer, 2000). The present study was designed to investigate whether the minimum molecular determinants of Ras recognition by the Raf-1 serine/threonine kinase, the best-known downstream targets of the small GTP binding protein Ras (Avruch et al., 1994), could be used to visualize Ras activation in live cells by following the distribution of such domain fused to the green fluorescent protein (GFP).
Activation of Raf-1 by the GTP-bound form of Ras requires its recruitment to the plasma membrane followed by a chain of events that involve inter- and intramolecular rearrangements as well as multiple phosphorylations (Morrison and Cutler, 1997). Membrane recruitment of Raf-1 is believed to be primarily regulated by the GDP/GTP exchange on Ras proteins (Stokoe et al., 1994), although this is probably not the sole determinant of Raf-1 membrane translocation. Recently, a phosphatidic acid binding motif that affects Raf-1 distribution was identified within the Raf-1 molecule (Rizzo et al., 1999). The sequence responsible for the interaction of Raf-1 with the GTP-bound form of Ras (Ras binding domain, RBD) has been narrowed to residues 51–131 within the N-terminal regulatory domain of the Raf-1 molecule (Vojtek et al., 1993; Nassar et al., 1995). This domain has been successfully used to “pull down” activated Ras from cell lysates of various cell types (see Gorman et al., 1996 for a detailed discussion). However, several lines of evidence suggest that the adjacent cysteine-rich domain (CRD) of Raf-1 (residues 139–184) is also important for Ras-Raf interaction and creates an additional Ras binding site (Brtva et al., 1995; Hu et al., 1995a; Drugan et al., 1996). It is not yet certain whether binding of the CRD motif to Ras is regulated by GDP/GTP exchange on Ras, or through other interactions either with proteins such as the 14-3-3 proteins (Freed et al., 1994; McPherson et al., 1999) or with acidic phospholipids (Mott et al., 1996; McPherson et al., 1999). It is also not clear whether the CRD is important for the membrane localization or for the activation of Raf-1. Recently, the CRD motif of Raf-1 was shown to interact only with the lipid-modified form of H-Ras independent of the nucleotide-bound state of the latter (Williams et al., 2000). Most reports addressing these questions have assessed molecular interactions in cell-free systems using recombinant proteins or have investigated the functional properties of expressed mutant Ras or Raf-1 proteins to gain insight into their functionally important motifs. Although these studies have provided invaluable information, they could not identify the cellular compartments where such interactions take place and whether certain motifs participate in membrane recruitment, activation, or both under the conditions that exist at the intact cell membrane.
In the present study, we combine a biochemical and imaging approach to gain information about the structural features of Ras-Raf interaction using live cells. We demonstrate that the RBD of Raf-1 alone is not sufficient for plasma membrane localization of a GFP fusion construct by activated Ras, and only together with the CRD does it provide the binding strength for efficient membrane recruitment. We also show that the Ras recognition motifs are not fully exposed in the Raf-1 molecule and that factors other than Ras may contribute to the conformational change that allows its Ras-mediated activation.
MATERIALS AND METHODS
Reagents
Recombinant platelet-derived growth factor (PDGF) AB was purchased from Life Technologies (Grand Island, NY), and phorbol-12-myristate-13-acetate (PMA) was purchased from Sigma (St. Louis, MO). Ionomycin and 1,2-bis(o-aminophenoxy)ethane-N;N;N;N-tetraacetic acid were obtained from Calbiochem (Cambridge, MA). All other reagents were of high-performance liquid chromatography or analytical grade.
Cell Culture and Transfections
COS-7 cells and NIH 3T3 cells were cultured in DMEM/high glucose supplemented with l-Glutamine (Life Technologies) in the presence of 10% fetal bovine serum. For biochemical analysis, cells were cultured in 35-mm culture dishes and were transfected at ∼70% confluence with the LipofectAmine 2000 reagent (Life Technologies), according to the manufacturer's instructions. For confocal microscopy, cells were cultured and transfected on 25 mm diameter glass coverslips.
DNA Constructs
All DNA constructs were made by polymerase chain reaction amplification using the human Raf-1 sequence (Ferrier et al., 1997; kindly provided by Drs. Zoltan Olah and Wayne B. Anderson, National Cancer Institute) as template, and the Pfu-turbo DNA polymerase (Promega, Madison, WI) and primers that contained appropriate restriction sites for cloning into the pEGFP-N1 plasmid (CLONTECH, Palo Alto, CA). Some of the constructs (Raf[51-131]-GFP and Raf[51-200]) were also created in the pEGFP-C1 plasmid because proteolytic cleavage of the C-terminally GFP-tagged versions yielded some free GFP when the constructs were expressed in NIH 3T3 cells. Mutations were made with the QuikChange Mutagenesis kit from Stratagene (La Jolla, CA) and were verified with dideoxy sequencing. The same constructs were also created as GST fusion proteins for bacterial expression using the pGEX-6P plasmid system and purification on gluthathione-Sepharose columns (Amersham Pharmacia Biotech, Piscataway, NJ). Bacterial expression of the EGFP-fused proteins was achieved by inserting the GFP fusion proteins (created in the pEGFP-C1 plasmid) into the pET19b plasmid (Novagen, Madison, WI) and using the His6 tag for purification. For mammalian expression of GST-fused RBDs, a GST-C1 plasmid was created by inserting the GST sequence in place of that of EGFP between the NheI and PstI sites in the pEGFP-C1 plasmid. This plasmid was then used to create the constructs as described above for the GFP fusion proteins.
Confocal Microscopy
Twenty-four hours after transfection, cells were serum-deprived for 8–12 h and washed twice with a modified Krebs-Ringer buffer, containing (in millimoles): NaCl 120, KCl 4.7, CaCl2 1.2, MgSO4 0.7, glucose 10, and Na-HEPES 10, pH 7.4, before analysis. The coverslips containing the cells were placed into a chamber that was mounted on a heated stage with the medium temperature kept at 33°C. Cells were examined in an inverted microscope under a ×40 oil-immersion objective (Nikon, Melville, NY) and a laser confocal microscope system (MRC-1024) with the Lasersharp acquisition software (Bio-Rad, Hercules, CA) as previously described (Várnai and Balla, 1998). When mitochondria were also imaged, cells were preincubated with 250 nM MitoTracker (Molecular Probes, Eugene, OR) and were simultaneously excited with 488 and 568 nm laser lines.
Extracellular Signal-Regulated Kinase (ERK) 2 Activity Assay
COS-7 cells were transiently cotransfected with the appropriate Raf-1-GFP construct and hemagglutinin (HA)-tagged mitogen-activated protein kinase (MAPK; Erk2; Bondeva et al., 1998). Twenty-four hours post-transfection, cells were serum deprived for 6 h and then lysed on ice in a buffer containing 20 mM HEPES, pH 7.5, 10 mM EGTA, 2.5 mM MgCl2, 1% NP-40, 1 mM Na3VO4, 40 mM β-glycerophosphate, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 20 μg/ml aprotinin, and 20 μg/ml leupeptin. Lysates were cleared by centrifugation at 20,000 × g for 25 min at 4°C. The supernatant was incubated with 1.5 μg of anti-HA monoclonal anitbody (BabCo, Richmond, CA) for 2 h. The immunocomplexes were recovered using 20 μl of protein G-plus Sepharose beads (Calbiochem) followed by an additional incubation of 2 h at 4°C. Immunoprecipitates were washed twice with phosphate-buffered saline, pH 7.5, containing 0.1% NP-40 and 1 mM Na3VO4, once with 100 mM Tris-HCl, pH 7.5, and 0.1 M LiCl, and once with kinase reaction buffer, which consisted of 12.5 mM 3-(N-morpholino)propanesulfonic acid, pH 7.5, 12.5 mM β-glycerophosphate, 7.5 mM MgCl2, 0.5 mM EGTA, 0.5 mM NaF, and 1 mM Na3VO4. Finally, beads were resuspended in 30 μl of kinase reaction buffer containing 1 μCi [γ-32P]ATP, 20 μM ATP, 3.3 μM dithiothreitol, and 1.5 mg/ml myelin basic protein (MBP), and incubated at 30°C for 30 min. Reactions were terminated by the addition of 10 μl 5× SDS sample buffer. After denaturation, samples were separated on 8–16% gradient Tris-glycine SDS gels. The phosphorylated MBP was detected and quantitated after exposure of the dried gels on a PhosphorImager screen (Molecular Dynamics, Sunnyvale, CA).
Mitogen-Activated Protein Kinase Kinase (MEK1)-Coupled Assay
Transfection and immunoprecipitation was performed as described above for the MAPK assay except that HA-tagged MEK1 (Bondeva et al., 1998) was used for transfection instead of the HA-ERK2. MEK1 activity was assayed using the same assay conditions described above, but the reaction buffer was complemented with 50 ng of recombinant MAPK (Calbiochem). In the coupled assay, recombinant MAPK phosphorylated by the immunoprecipitated MEK1 was able to phosphorylate MBP as a substrate.
Ras/Raf Interactions
COS-7 cells were cotransfected with RasV12 (Rodriguez-Viciana et al., 1997) and the different Raf-1-GFP constructs. Cells were lysed after 6 h of serum deprivation in a lysis buffer (see above), but containing 5 mM MgCl2. Ras was immunoprecipitated from the lysates using an anti-pan-Ras monoclonal antibody (Oncogene, Cambridge, MA), and the immunocomplexes were washed as described above. Proteins were separated by SDS-PAGE and were transferred onto polyvinylidene difluoride membranes (Millipore, Bedford, MA) using a semidry transfer system (Bio-Rad) as described elsewhere (Bondeva et al., 1998). Membranes were probed with an affinity-purified anti-GFP polyclonal antibody (CLONTECH) for detection of the presence of the GFP-tagged Raf-1 fusion proteins or the anti-GST polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) when using the GST-fused forms.
The ability of recombinant GST- and GFP-fused RBDs of Raf-1 to interact with Ras in vitro was determined by incubating ∼5 μg of the purified bacterially expressed proteins (still attached to Sepharose beads) with cell lysates (equivalent of one-fifth of cells 80% confluent on a 10-cm culture dish) prepared from serum-deprived COS-7 cells that were either unstimulated or stimulated with epidermal growth factor (EGF; 100 ng/ml for 10 min) or transfected with RasV12. After incubation at 4°C for 90 min, the beads were separated from the rest of the lysate by centrifugation through a mixture of dimethyl-phtalate and bis(3,5,5-trimethyl-hexyl)phtalate (1:4, density, 1.0148 g/ml, 400 μl) into 40 μl of Laemmli buffer layered under the oil. This procedure allowed the capture of the complex at equilibrium without loosing proteins due to rapid dissociation upon washing (Gorman et al., 1996).
RESULTS
Localization of the RBD-GFP Fusion Proteins in NIH 3T3 cells
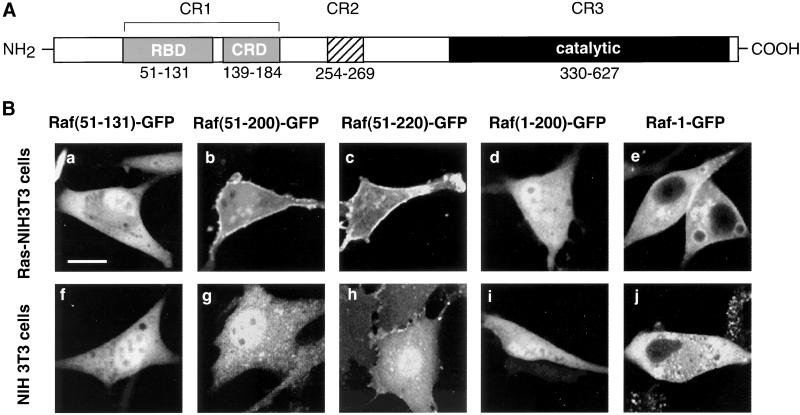
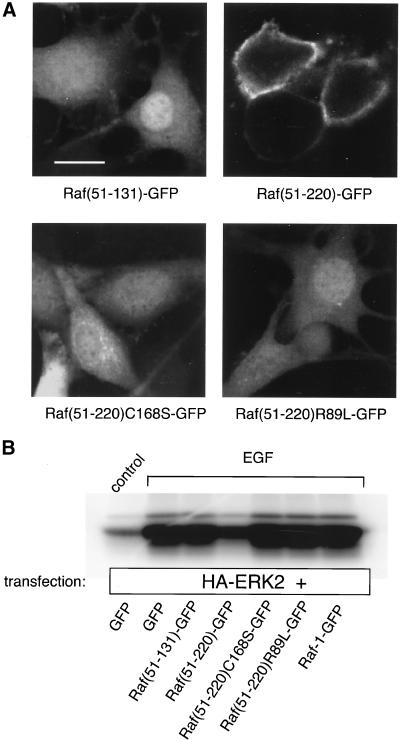
Although the interaction of Ras with the regulatory domain of Raf-1 has been studied in great detail, little is known about how the different elements that determine Ras-Raf-1 interactions contribute to the membrane recruitment of the latter within intact cells. Therefore, we created fusion proteins from the RBD of Raf-1 (Figure 1A) fused to GFP and compared the distribution of the expressed proteins in normal and Ras-transformed NIH 3T3 cells. First, we fused residues 51–131 of Raf-1 to GFP (Raf[51-131]GFP) and expressed the chimeric protein in H-Ras-transformed NIH 3T3 cells for analysis of the cellular fluorescence by confocal microscopy. Expression of the Raf(51-131)GFP construct yielded mostly cytosolic fluorescence with only a slight hint of membrane localization in some of the cells with low expression levels (Figure 1B, a). This finding was unexpected because this domain is believed to play a critical role in the membrane recruitment of Raf-1 by active Ras (Morrison and Cutler, 1997). Only when the construct was extended so that it also contained the CRD domain (Raf[51-200]GFP) was the fusion protein detected in the plasma membrane of Ras-transformed (but not in normal quiescent) NIH 3T3 cells (Figure 1B, b and g). Importantly, localization was observed only in cells that expressed the fusion protein at low levels, not in those showing moderate to high fluorescence, suggesting that the limited Ras binding sites become saturated even at moderate expression levels of the probes.
Figure 1.
Domain-structure of the Raf-1 serine/threonine kinase (A) and cellular distribution of various Raf-1-GFP fusion proteins in Ras-transformed and normal NIH 3T3 fibroblasts (B). (A) The N-terminal conserved region (CR1) contains two domains, the RBD and the CRD, both of which have been implicated in the interaction and activation of Raf-1 by the small GTP binding protein, Ras. CR2 is a serine/threonine-rich region that contains several (but not all) regulatory phosphorylation sites. CR3 is the catalytic domain of the kinase. (B) Confocal images show the distribution of various Raf-1 constructs containing the Ras-interaction domains of Raf-1 fused to GFP as expressed in Ras-transformed (a-e) or normal (f-j) NIH 3T3 cells.
Further extension of the construct toward the C terminus (Raf[51-220]) appeared to improve membrane localization in the Ras-transformed fibroblast. However, this improvement was not dramatic and was difficult to measure in quantitative terms. This fusion protein again showed a difference in its membrane localization between the Ras-transformed vs. normal NIH 3T3 cells (Figure 1B, d and i). In contrast, addition of residues 1–51 of Raf-1 to the GFP construct (Raf[1–200]) greatly reduced the number of cells that showed expression, and the fusion protein showed no membrane localization (Figure 1B, c and h). This was consistent with the reported inhibitory nature of the N-terminal part of the Raf-1 protein on Ras-Raf interaction and the apparent instability of such construct (Gorman et al., 1996). It is also possible that cells do not tolerate the expression of this protein and have been mostly eliminated.
When the full-length Raf-1 protein was fused to GFP, it was completely excluded from the nucleus, unlike the above-mentioned smaller fusion proteins. However, less prominent membrane localization was observed with this full-length protein in the Ras-transformed fibroblasts. First, fewer cells showed localization, and most of these had a weaker plasma membrane labeling than those expressing the shorter constructs (Figure 1B, e and j). This latter finding suggested that the RBD and CRD is not fully exposed in the full Raf protein and probably require intramolecular rearrangements (and dissociation of some interactive proteins) for full access to active Ras. This aspect of Raf-1 regulation was not further pursued in detail in the present study.
GFP-Fused Raf-1 Constructs Recognize Active Ras In Vitro
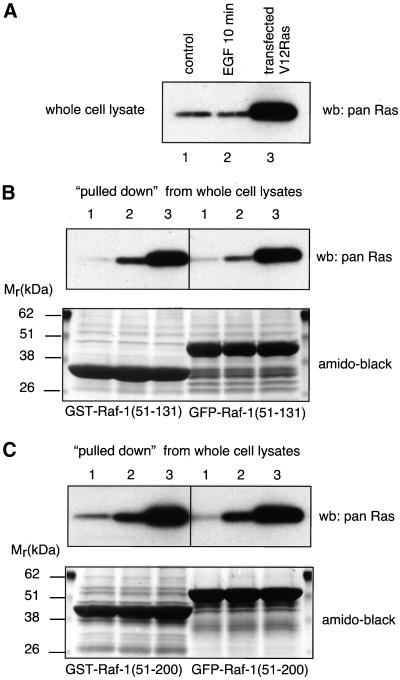
The inability of the RBD to localize to activated Ras in Ras-transformed NIH 3T3 cells was in contrast to the well-documented ability of this motif to recognize GTP-bound Ras in vitro. Therefore, the same GFP fusion constructs have been created for bacterial expression to compare their in vitro Ras-binding activity with their widely used GST-fused counterparts. The GFP-fused proteins were purified via a His6 tag placed at the N terminus of GFP. As shown in Figure 2, GFP-fused Raf(51-131) was clearly able to “pull down” active Ras from COS-7 cell lysates, but Raf(51-200) was more efficient in this regard. There was no major difference between the abilities of the domains to recognize Ras whether fused to GFP or GST. These data indicate that the lack of localization of the GFP-fused RBD to active Ras within the intact NIH 3T3 cell is not due to the inability of this fusion protein to recognize Ras.
Figure 2.
Interaction of recombinant GFP- and GST-fused RBDs of Raf-1 with activated Ras in vitro. Bacterially expressed and purified GFP- and GST-fused Raf(51-131) and Raf(51-200) domains were incubated with cell lysates prepared from COS-7 cells that were rendered quiescent, stimulated with EGF (100 ng/ml 10 min), or were transfected with RasV12. Ras associated with the recombinant domains was analyzed with Western blotting using an anti-pan-Ras antibody (see “Materials and Methods” for details). (A) The amount of Ras in the cell lysates. (B and C) Ras associated with the Raf(51-131) and Raf(51-200) constructs, respectively. In each case, the amounts of recombinant proteins present on the blots are shown after amido-black staining.
Interaction of Raf-1-GFP Constructs with RasV12 in COS-7 Cells
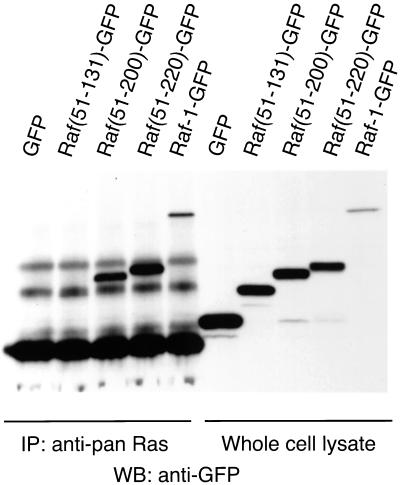
To substantiate that the membrane localization of the various Raf-1-GFP fusion proteins was due to their interaction with activated Ras and not with other membrane components, we investigated the ability of these proteins to form stable complex with RasV12. For this, we used COS-7 cells that were transfected with H-RasV12 together with the respective Raf-1-GFP fusion construct, and we immunoprecipitated Ras from the cell lysates to detect the associated GFP fusion protein. As shown in Figure 3, a clear interaction of RasV12 was observed with Raf(51-200)GFP, Raf(51-220)GFP, and the full-length Raf-1-GFP molecule. The interaction of RasV12 with Raf(51-131)GFP was poorly detectable with this procedure, partially because of the close migration of Raf(51-131)GFP with one of the nonspecific bands stained by the GFP antibody. Nevertheless, an interaction similarly strong as observed with Raf(51-200)GFP would still be detectable with this method and, therefore, we concluded that the binding of Raf(51-131) to RasV12 is significantly weaker than that of Raf(51-200). To confirm that this was not due to the presence of GFP in the fusion protein, similar experiments were performed with GST-fused proteins expressed in COS-7 cells. These experiments also confirmed that association of GST-Raf(51-131) with immunoprecipitated RasV12, although detectable, was very weak compared with GST-Raf(51-200) (our unpublished observations). However, it should be emphasized that these data are not inconsistent with the reported ability of RBD to bind Ras-GTP (see also above). A less stable interaction with rapid dissociation, such as that of RBD with Ras (Gorman et al., 1996), would not be detected with the washing procedure during the immunoprecipitation experiment.
Figure 3.
Interaction of various Raf-1-GFP fusion proteins with RasV12 coexpressed in COS-7 cells. COS-7 cells were transfected with plasmid DNAs encoding RasV12 and the respective Raf-1-GFP fusion constructs or GFP for 24 h. After 6 h of serum deprivation, cells were lysed and Ras was immunoprecipitated with an anti-pan-Ras antibody, followed by Western blotting for detection of the presence of GFP. The right panel shows the presence of the expressed proteins in the whole-cell lysates, and the left panel shows proteins that were associated with Ras. Representative data are shown from three similar observations.
Expressed Raf-1-GFP Fusion Proteins Inhibit Ras/Raf-Mediated ERK Activation
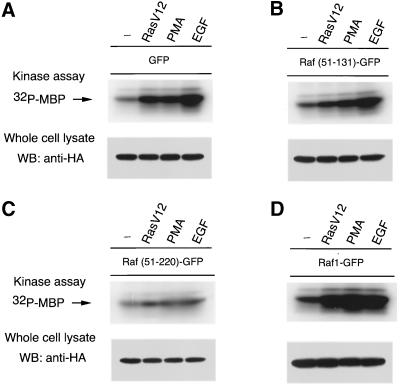
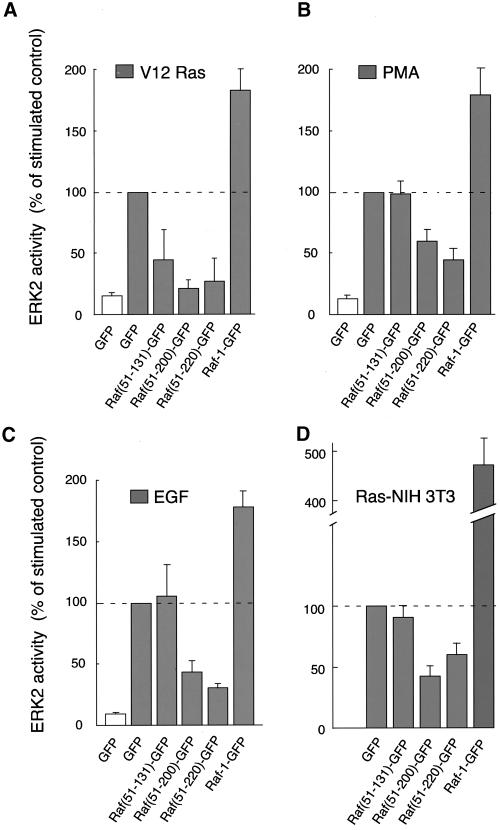
Interaction of the constructs with activated Ras at the plasma membrane is expected to exert an inhibitory effect on Ras-Raf-1-mediated signaling processes. To investigate such dominant-negative effects of the constructs, COS-7 cells were cotransfected with HA-ERK2 and GFP (Figure 4A) or the corresponding Raf-1-GFP fusion constructs (Figure 4, B-D), and they were stimulated with PMA, EGF, or RasV12 overexpression or were left untreated. This way, the effects of the various stimuli on ERK2 activity were compared for each expressed GFP construct separately. The results clearly demonstrated that the stronger the binding of a construct to active Ras, the more effective is its inhibitory effect on ERK2. For example, expression of Raf(51-220)GFP or Raf(51-200)GFP (our unpublished observations) showed significant inhibitory effects on ERK2 activation regardless of the stimuli applied (Figure 4C), but Raf(51-131)GFP expression failed to inhibit the ERK2 response to either EGF or PMA. However, even this latter construct had a 50% inhibitory effect on ERK2 activity when RasV12 was used as a stimulus (Figure 4, A and B). The expression levels of the Raf-1-GFP fusion proteins were comparable in these experiments, except for the full-length Raf-1-GFP, which was expressed at a reduced level (see Figure 3 as an example). Full-length Raf-1-GFP strongly stimulated ERK2 activation, indicating that the GFP-tagged Raf-1 was active in spite of the presence of the GFP molecule on its C terminus (Figure 4D). Similar results were obtained when MEK1 activity was assayed in a MEK1-ERK-coupled assay as described in “Materials and Methods” (our unpublished observations). Moreover, expression of the constructs had clearly detectable inhibitory effects on the activation of endogenous ERK2 in COS-7 cells as assessed by using a phospho-ERK2 antibody on total cell lysates (our unpublished observations).
Figure 4.
Stimulation of ERK2 activity by RasV12, PMA, or EGF in COS-7 cells expressing various Raf-1-GFP fusion proteins. COS-7 cells were transfected with plasmid DNAs encoding HA-epitope-tagged ERK2 and GFP only (A), Raf(51-131)GFP (B), Raf(51-220)GFP (C), or Raf-1-GFP (D) with or without RasV12. One day post-transfection and after 6 h of serum deprivation, cells were stimulated with PMA (200 nM) for 15 min or with EGF (100 ng/ml) for 5 min. HA-ERK2 was then immunoprecipitated from the cell lysates and its activity was measured using [γ-32P]ATP and MBP as substrate. Similar observations were obtained in two additional experiments.
For a better comparison, in a second set of experiments, ERK2 activation was tested for each of the stimuli in cells expressing the different GFP constructs. For example, the effect of RasV12 overexpression on ERK2 activity was different in the presence of the various GFP fusion proteins and could be plotted as percentage of the control response observed in the presence of GFP alone (Figure 5A). The effects of expression of the various constructs were then plotted similarly for the ERK2 responses to PMA and EGF treatment (Figure 5, B and C) or to the activated endogenous Ras in Ras-transformed NIH 3T3 cells (Figure 5D). When the results were presented in this way, the inhibitory effects of the Raf(51-200)GFP and Raf1(51-220)GFP constructs were even more obvious on the ERK2 responses regardless of the stimuli used. In contrast, Raf(51-131)GFP was inhibitory (∼50% inhibition) in experiments where RasV12 was overexpressed in COS-7 cells, but had no effects in PMA- or EGF-treated cells or in Ras-transformed fibroblasts (Figure 5). These results raised the possibility that when active Ras is acutely overexpressed, it may use mechanism(s) other than those that function in Ras-transformed fibroblasts for ERK2 activation. Interestingly, expression of full-length Raf-1-GFP was significantly more potent in activating ERK2 in Ras-transformed NIH 3T3 cells than any of the stimuli including expressed RasV12 in COS-7 cells (Figure 5), suggesting that Ras-transformed NIH 3T3 cells perhaps possess additional factors in their membranes that sensitize Raf-1 activation by Ras.
Figure 5.
Differential ability of Raf-1-GFP fusion proteins to inhibit ERK2 activation in response to different stimuli in COS-7 cells and Ras-transformed NIH 3T3 cells. In this series of experiments, the ERK2 responses to the various stimuli (expressed as 100%) were compared between cells transfected with GFP or the various Raf-1-GFP protein constructs. For experimental details see the legend to Figure 3. Means ± SEM of four to six experiments are shown.
Together, these results indicate that the signaling function of endogenous Ras can be effectively inhibited by some of the GFP-Raf-1 constructs, but only by those that are recruited to the plasma membrane, supporting the conclusion that they visualize active Ras in the membranes.
RBD and CRD Are Equally Important in Membrane Localization
To determine the relative importance of RBD and CRD in the membrane localization of Raf(51-220)GFP, we created mutants within this construct that are known to inhibit the activation of Raf-1 by Ras. It has been reported that the R89L mutation within the RBD and the C168S substitution within the CRD completely eliminate Raf-1 activation (Fabian et al., 1993; Luo et al., 1997). When the cellular distribution of such mutant Raf(51-220)GFP proteins was examined, both mutations eliminated the plasma membrane localization of the fusion protein in Ras-transformed NIH 3T3 cells (Figure 6A). Similarly, both mutants failed to inhibit EGF-stimulated or RasV12-stimulated (our unpublished observations) ERK2 activation when used as a dominant-negative inhibitor in COS-7 cells (Figure 6B).
Figure 6.
Mutations either within the RBD or CRD eliminate binding of Raf(51-220)GFP to Ras in the plasma membrane. Key residues within the RBD and CRD of Raf-1 known to affect its interaction with Ras were mutated within Raf(51-220)GFP, and the constructs were expressed in COS-7 or Ras-transformed NIH 3T3 cells. Both mutations (R89L and C168S) eliminate plasma membrane localization of the fusion protein in Ras-transformed NIH 3T3 cells (A) as well as its inhibitory effect on ERK2 activation in COS-7 cells (B).
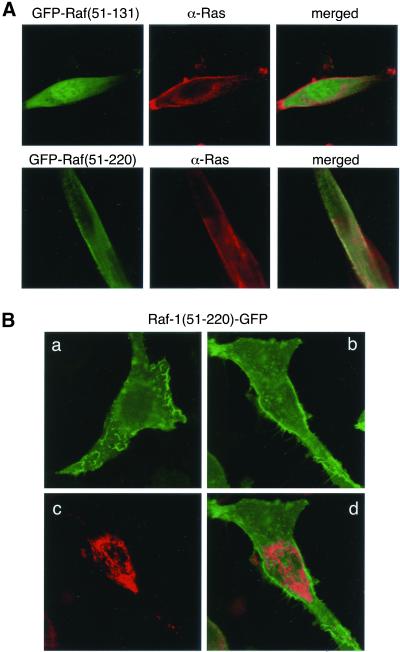
Localization of Active Ras in Membrane Ruffles
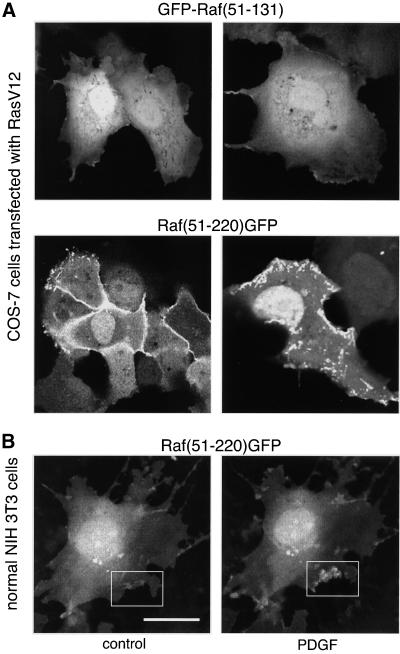
As mentioned above, the distribution of Raf(51-220)GFP in cells expressing the protein at low levels allowed analysis of the cellular sites where active Ras is found. In Ras-transformed NIH 3T3 cells, most of the Raf(51-220)GFP fluorescence (but not that of Raf[51-131]GFP) was associated with membranes in colocalization with Ras (Figure 7A), and was especially enriched in membrane ruffles (Figure 7B, a). However, some fluorescence was also associated with intracellular structures (Figure 7B, b). Because Raf-1 has been shown to associate with mitochondria (Wang et al., 1996), we examined whether those intracellular structures represent the mitochondria. However, the mitochondrial marker MitoTracker clearly indicated that the Raf(51-220)GFP protein did not localize to the mitochondria (Figure 7, B and C). Another question of interest was whether the fluorescent probes would detect unprocessed Ras located in intracellular membranes. It has been recently shown that during its synthesis and processing, Ras is present in intracellular membranes, most prominently in the Golgi, when overexpressed in COS-7 cells (Choy et al., 1999). Therefore, we also examined the distribution of the fluorescent probes in COS-7 cells overexpressing RasV12. As shown in Figure 8A, GFP-Raf(51-131) showed no localization to any membranes in these live cells, and Raf(51-220)GFP again localized to membrane ruffles, but was not detected over the Golgi (Figure 8A)1.
Figure 7.
Cellular distribution of Raf(51-131)GFP and Raf(51-220)GFP in Ras-transformed NIH 3T3 cells. (A) The distribution of Raf(51-131)GFP (upper) and Raf(51-220)GFP (lower) in fixed Ras-transformed NIH 3T3 cells immunostained with anti-Ras (red) antibody. (B) Panel a shows a cell in which the surface of the cell was imaged to demonstrate the high enrichment of the membrane associated fluorescence in membrane ruffles. This distribution was especially prominent in cells stimulated with PDGF. Panel b shows that fluorescence is also associated with small vesicular structures in the cytoplasm. These structures do not correspond to mitochondria as assessed by simultaneous imaging of the MitoTracker dye (c) and merging the two images (d).
Figure 8.
Cellular distribution of GFP-Raf(51-131) and Raf(51-220)GFP in COS-7 cells expressing RasV12 (A) and the effect of PDGF stimulation on the distribution of Raf(51-220)GFP in normal NIH 3T3 fibroblasts (B). COS-7 cells were cotransfected with RasV12 and the respective GFP fusion construct, and normal NIH cells were transfected with Raf(51-220)GFP for 24 h. Live cells were examined under a confocal microscope. NIH 3T3 cells were serum-deprived (control) for 6 h before stimulation with PDGF (50 ng/ml, 2.5 min). Note the decrease in cytoplasmic intensity and the increased localization of the protein in membrane ruffles in B.
Monitoring Ras Activation in Normal NIH 3T3 Cells
To follow Ras activation in normal NIH 3T3 cells, we used cells that were transfected with the Raf(51-220)GFP construct and rendered quiescent by serum-free incubation for 5–12 h. Because the amount of Ras in these cells is expected to be less than in Ras-transformed fibroblasts, we choose cells that expressed low levels of the GFP fusion protein so that the redistribution of a small amount of fluorescent protein could still be observed. In most cells, a variable amount of signal was already present at the plasma membrane. After stimulation with PDGF (50 ng/ml), the intensity of membrane-associated fluorescence was increased (Figure 8A), indicating the activation of Ras in the membrane. Similar changes were observed after stimulation with PMA (200 nM, not shown). These changes in the amounts of membrane-associated fluorescence were relatively small, and many cells (∼50%) showed no detectable response to stimulation. Also, the translocated Raf(51-220)GFP protein was mostly present on membrane ruffles that did not always present in the optical Z-section that was recorded before stimulation. A good index of translocation was often only the decrease in the cytosolic fluorescence (Figure 8B). These results suggest that the amount of activated Ras in the membrane of normal NIH 3T3 cells is relatively small (compared, for example, with the amounts of 3-phosphorylated inositides, which is easier to detect with a similar approach; Varnai et al., 1999).
As shown in Figure 1, full-length Raf-1-GFP showed only weak membrane localization even in Ras-transformed NIH 3T3 cells, indicating limited exposure of its Ras-binding sequences. This is in good agreement with the complex regulation of Raf-1 through multiple mechanisms that probably affects the conformation and access of its interactive domains. Several manipulations were tested for their ability to increase the interaction of full-length Raf-1 with the membrane in Ras-NIH 3T3 cells (including addition of cell permeable ceramide derivatives, exogenous phosphatidic acid, or stimulation with various agonists). Among these, only stimulation with phorbol esters or lysophosphatidic acid was found to cause a slight enhancement in the membrane localization of the full-length protein in some but not all of the cells (our unpublished observations).
DISCUSSION
The present study was designed to explore whether the minimum protein sequence of the Raf-1 protein kinase that is responsible for the recruitment of Raf-1 by active Ras to the plasma membrane can be used to detect Ras activation in living cells. Contrary to our expectations, our data indicate that the RBD alone is not sufficient to support a stable interaction with Ras in living cells in spite of its known ability to recognize active Ras in vitro. The present data show that the addition of the CRD to the RBD is necessary to obtain a functional probe for the detection of Ras in live cells. Mutation of critical single residues within either the RBD or CRD were found to prevent membrane localization of the GFP fusion protein in Ras-transformed NIH 3T3 cells, indicating that both motifs are equally important for membrane recruitment. Expression of the GFP-RBD was able to exert a moderate inhibition of ERK2 activation in COS-7 cells when expression of RasV12 was used as a “stimulus,” indicating that even this probe can interact with RasV12 to some extent. However, when endogenous Ras was activated by more physiological means (such as by PMA or EGF stimulation), only the constructs that also contained the CRD and showed membrane localization were found to inhibit MEK1 and ERK2 activation. These results suggest that the additional interaction provided by the CRD is required for the RBD to remain bound to active Ras and form a more stable complex in the plasma membrane. This additional binding can be achieved by alternative means as demonstrated elegantly in a recent report where a fusion protein was created by linking the RBD of Raf-1 to Ras and placing the construct between the cyan and yellow fluorescent proteins (Mochizuki et al., 2001). When targeting this construct to membranes by a CAAX motif, binding of RBD to Ras upon Ras activation has been demonstrated by detecting an increase in fluorescence energy transfer caused by the proximation of the two fluorophores. Those data clearly demonstrated that once tethered to Ras and targeted to membranes, the RBD alone is clearly able to interact with activated Ras without the need for the additional stabilizing force added by the CRD.
The requirement for the CRD for effective Raf-Ras interaction is consistent with the known importance of this domain for Raf-1 activation, especially in the case of lipid-modified, membrane-bound Ras (Hu et al., 1995b; Luo et al., 1997; Williams et al., 2000). That only lipid-modified Ras is recognized by the Raf(51-200)GFP construct is also supported by the finding that Ras was recognized only in the plasma membrane even in COS-7 cells expressing RasV12 that are known to have significant amounts of Ras in their internal membranes (Choy et al., 1999). This restriction is very likely be added by the CRD. The similarity between the solution structures of the CRD (Mott et al., 1996) and other proteins that interact with membrane lipids, such as the CRD of protein kinase Cδ (Zhang et al., 1995), Rabphillin 3A (Ostermeyer and Brünger, 1999), or the FYVE domains (Misra and Hurley, 1999), raises the possibility that the CRD may also confer lipid regulation to Ras-Raf interaction. Several lines of evidence suggest that lipids play an important role in Raf-1 activation, phosphatidylserine (McPherson et al., 1999), ceramide (Huwiler et al., 1996), and the product of phosphatidylcholine-specific phospholipase C (Cai et al., 1993) having been implicated. However, the exact nature of the elusive lipid regulator has yet to be uncovered. Interestingly, the CRD domain of Raf-1 is also a site where βγ-subunits of heterotrimeric G-proteins can interact with the protein (Pumiglia et al., 1995), a feature shared by the pleckstrin homology domains of βARK (Koch et al., 1993), and the Bruton's tyrosine kinase (Tsukada et al., 1994), both of which are sites for regulation by inositol phospholipids. Recently, the importance of localization to glycolipid-rich membrane microdomains that contain caveolin, cholesterol, and inositol phospholipids have been shown for H-Ras but not for K-Ras activation of Raf-1 (Roy et al., 1999). It is possible that the CRD contributes to sequestering H-Ras into those membrane domains, although the lipid modification of Ras itself is certainly more important in this regard (Rizzo et al., 2001).
The saturation of the Raf(51-220)GFP binding sites on the membrane already at moderate expression levels even in Ras-transformed NIH 3T3 cells indicates that active Ras is present in relatively low concentrations and only a small signal can be detected in normal NIH 3T3 cells after stimulation with either PMA or PDGF. This is in agreement with a recent report that found that Ras-Raf-1 interaction is not able to move bulk amounts of Raf-1-GFP to the plasma membrane of Rat-1 fibroblasts (Rizzo et al., 2000). However, our data clearly demonstrate the functional importance of these quantitatively small, yet functionally relevant interactions in Ras-mediated Raf-1 activation. It is important to note that all of the important regulatory phosphorylation sites of Raf-1 (Morrison and Cutler, 1997) are outside of the region used to create the GFP fusion proteins (Figure 1A). Accordingly, no electrophoretic mobility shift was observed with any of the constructs (other than Raf-1-GFP) after stimulation with PMA (T. Bondeva and T. Balla, unpublished observations). Therefore, it is not likely that the translocation responses seen after stimulation result from modifications of the GFP fusion protein.
The membrane localization of GFP-tagged full-length Raf-1 was significantly weaker than that of Raf(51-220)GFP, even in Ras-transformed fibroblasts. Because Raf-1-GFP was found to interact strongly with RasV12 and to activate MEK1 or ERK2, it appears functionally intact even with the C-terminal GFP tag attached. The fact that the best localization was seen with constructs that were also found inhibitory raises the possibility that an active mechanism is present in the full-length Raf-1 molecule (but not in the truncated constructs) that ensures its release from Ras, making it available for another activation cycle. This way the steady-state amount of Raf-1-GFP at the membrane may not have to be high and change noticeably even during more active cycling during stimulation. On the other hand, the availability of the Ras-binding motifs within the Raf-1 molecule is also likely to be regulated and could be just as important in limiting the amounts of active Raf in the membrane as the amount of RasGTP itself. In a recent study, phosphorylation of residue Ser259 of Raf-1 was shown to inhibit the interaction of Raf-1 with Ras in NIH 3T3 cells (Dhillon et al., 2002). Given the numerous proteins known to interact with Raf-1 and participate in its activation (e.g., Morrison and Cutler, 1997), active Ras is probably only one of several factors that determine the membrane recruitment of Raf-1. Such multiplicity of interaction of Raf-1 with Ras and the plasma membrane could account for the ability of full-length Raf-1 to interact with Ras in the membrane even when two critical cysteines (C165S and C168S) are mutated within the CRD (Roy et al., 1997). Recently, phosphatidic acid, through interaction with a sequence motif close to the catalytic site, was found to be more important than the Ras-binding motif for the regulation of membrane recruitment and internalization of Raf-1 in insulin-stimulated Rat-1 and HIRcB fibroblasts (Rizzo et al., 1999, 2000). No internalization of Raf-1 or any of the GFP-fused fragments was observed in the present study with any of the stimuli tested, including insulin, in the NIH 3T3. Whether this reflects cell type-specific regulation remains to be determined.
In summary, we created fluorescent fusion proteins from the RBD and CRD of Raf-1 and GFP for the detection of activated Ras in live cells. These probes recognized active Ras only in the plasma membrane of Ras-transformed NIH 3T3 cells and showed stimulus-induced recruitment to the membrane after stimulation of nontransformed NIH 3T3 cells with PDGF or PMA. Overexpression of the fusion proteins inhibited MEK1 and ERK2 activation, consistent with their binding to activated Ras. These novel research tools should facilitate our understanding of the spatial and temporal aspects of Ras-Raf signaling with the possibility of following these signaling events in single living cells.
ACKNOWLEDGMENTS
The authors thank Drs. Wayne B. Anderson and Zoltán Oláh (National Cancer Institute, National Institutes of Health, Bethesda, MD) for the full-length Raf-1 construct and the normal and Ras-transformed NIH 3T3 cells used in this study.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–01–0019. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–01–0019.
In contrast to live cells, localization of both GFP-Raf(51-131) and Raf(51-220)GFP to the plasma membrane and to Golgi-like intracellular membranes in apparent colocalization with Ras is observed in fixed COS-7 cells immunostained with an anti-Ras antibody. In Ras-transformed NIH 3T3 cells, the GFP constructs showed identical distribution in live cells and in fixed and immunostained cells. The reason for this discrepancy between live vs. fixed COS-7 cells is currently under investigation.
REFERENCES
- Avruch J, Zhang X, Kyriakis JM. Raf meets Ras: completing the framework of a signal transduction pathway. Trends Biochem Sci. 1994;19:279–283. doi: 10.1016/0968-0004(94)90005-1. [DOI] [PubMed] [Google Scholar]
- Bondeva T, Pirola L, Bulgarelli-Leva G, Rubio I, Wetzker R, Wymann MP. Bifurcation of lipid and protein kinase signals of PI3Kγ to the protein kinases PKB and MAPK. Science. 1998;282:293–296. doi: 10.1126/science.282.5387.293. [DOI] [PubMed] [Google Scholar]
- Brtva TR, Drugan JK, Ghosh S, Terrell RS, Campbell-Burk S, Bell RM, Der CJ. Two distinct Raf domains mediate interaction with Ras. J Biol Chem. 1995;270:9809–9812. doi: 10.1074/jbc.270.17.9809. [DOI] [PubMed] [Google Scholar]
- Cai H, Erhardt P, Troppmair J, Diaz-Meco MT, Sithanandam G, Rapp UR, Moscat J, Cooper GM. Hydrolysis of phosphatidylcholine couples Ras activation of Raf protein kinase during mitogenic signal transduction. Mol Cell Biol. 1993;13:7645–7651. doi: 10.1128/mcb.13.12.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy E, Chiu VK, Siletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, Philips MR. Endomembrane trafficking of Ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- Dhillon AS, Meikle S, Yazici Z, Eulitz M, Kolch W. Regulation of Raf-1 activation and signaling by dephosphorylation. EMBO J. 2002;21:64–71. doi: 10.1093/emboj/21.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drugan JK, Khosravi-Far R, White MA, Der CJ, Sung Y, Hwang Y, Campbell S. Ras interaction with two distinct binding domains in Raf-1 may be required for Ras transformation. J Biol Chem. 1996;271:233–237. doi: 10.1074/jbc.271.1.233. [DOI] [PubMed] [Google Scholar]
- Fabian JR, Daar IO, Morrison DK. Critical tyrosine residues regulate the enzymatic and biological activity of of Raf-1 kinase. Mol Cell Biol. 1993;13:7170–7179. doi: 10.1128/mcb.13.11.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrier AF, Lee M, Anderson WB, Benvenuto G, Morrison DK, Lowy DR, DeClue JE. Sequential modification of serines 621 and 624 in the Raf-1 carboxyl terminus produces alterations in its electrophoretic mobility. J Biol Chem. 1997;272:2136–2142. doi: 10.1074/jbc.272.4.2136. [DOI] [PubMed] [Google Scholar]
- Freed E, Symons M, Macdonald SG, McCormick F, Ruggieri R. Binding of 14-3-3 proteins to the protein kinase Raf and effects on its activation. Science. 1994;265:1713–1716. doi: 10.1126/science.8085158. [DOI] [PubMed] [Google Scholar]
- Gorman C, Skinner RH, Skelly JV, Neidle S, Lowe PN. Equilibrium and kinetic measurements reveal rapidly reversible binding of Ras to Raf. J Biol Chem. 1996;271:6713–6719. doi: 10.1074/jbc.271.12.6713. [DOI] [PubMed] [Google Scholar]
- Hu C, Kariya K, Tamada M, Akasaka K, Shirouzu M, Yokoyama S, Kataoka T. Cysteine-rich region of Raf-1 interacts with activator domain of post-translationally modified Ha-Ras. J Biol Chem. 1995a;270:30274–30277. doi: 10.1074/jbc.270.51.30274. [DOI] [PubMed] [Google Scholar]
- Hu C-D, Kariya K, Tamada M, Akasaka K, Shirouzu M, Yokoyama S, Kataoka T. Cysteine-rich region of Raf-1 interacts with activator domain of post-translationally modified Ha-Ras. J Biol Chem. 1995b;270:30274–30277. doi: 10.1074/jbc.270.51.30274. [DOI] [PubMed] [Google Scholar]
- Huwiler A, Brunner J, Hummel R, Vervoordeldonk M, Stabel S, Van Den Bosch H, Pfeilschifter J. Ceramide-binding and activation defines protein kinase c-Raf as a ceramide-activated protein kinase. Proc Natl Acad Sci USA. 1996;93:6959–6963. doi: 10.1073/pnas.93.14.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch WJ, Inglese J, Stone WC, Lefkowitz RJ. The binding site for the βγ subunits of heterotrimeric G proteins on the β-adrenergic receptor kinase. J Biol Chem. 1993;268:8256–8260. [PubMed] [Google Scholar]
- Luo Z, Diaz B, Marshall MS, Avruch J. An intact Raf zinc finger is required for optimal binding to processed Ras and for ras-dependent Raf activation in situ. Mol Cell Biol. 1997;17:46–53. doi: 10.1128/mcb.17.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson RA, Harding A, Lane RS, Hancock JF. Interaction of c-Raf-1 with phosphatidylserine and 14-3-3. Oncogene. 1999;18:3862–3869. doi: 10.1038/sj.onc.1202730. [DOI] [PubMed] [Google Scholar]
- Misra S, Hurley JH. Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell. 1999;97:657–666. doi: 10.1016/s0092-8674(00)80776-x. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Yamashita S, Kurokawa K, Ohba Y, Nagal T, Miyawaki A, Matsuda M. Spatio-temporal images of growth factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- Morrison DK, Cutler RE., Jr The complexity of Raf-1 regulation. Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- Mott HR, Carpenter JW, Zhong S, Ghosh S, Bell RM, Campbell SL. The solution structure of the Raf-1 cysteine-rich domain: a novel Ras and phospholipid binding site. Proc Natl Acad Sci USA. 1996;93:8312–8317. doi: 10.1073/pnas.93.16.8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar N, Horn G, Herrmann C, Scherer A, McCormick F, Wittinghofer A. The 2.2A crystal structure of the Ras-binding domain of the serine/threonine kinase cRaf1 in complex with Rap1A and a GTP analogue. Nature. 1995;375:554–560. doi: 10.1038/375554a0. [DOI] [PubMed] [Google Scholar]
- Ostermeyer C, Brünger AT. Structural basis of Rab effector specificity: crystal structure of the small GTP protein Rab3A complexed with the effector domain of rabphilin-3A. Cell. 1999;96:363–374. doi: 10.1016/s0092-8674(00)80549-8. [DOI] [PubMed] [Google Scholar]
- Pumiglia KM, LeVine H, Haske T, Habib T, Jove R, Decker SJ. A direct interaction between G-protein β γ subunits and the Raf-1 protein kinase. J Biol Chem. 1995;270:14251–14254. doi: 10.1074/jbc.270.24.14251. [DOI] [PubMed] [Google Scholar]
- Rizzo MA, Shome K, Vasudevan C, Stolz DB, Sung T-C, Frohman MA, Watkins SC, Romero G. Phospholipase D and its product, phosphatidic acid mediate agonist-dependent Raf-1 translocation to the plasma membrane and the activation of the mitogen-activated protein kinase pathway. J Biol Chem. 1999;274:1131–1139. doi: 10.1074/jbc.274.2.1131. [DOI] [PubMed] [Google Scholar]
- Rizzo MA, Kraft CA, Watkins SC, Levitan ES, Romero G. Agonist-dependent traffic of raft-associated Ras and Raf-1 is required for activation of the mitogen-activated protein kinase cascade. J Biol Chem. 2001;276:34928–34933. doi: 10.1074/jbc.M105918200. [DOI] [PubMed] [Google Scholar]
- Rizzo MA, Shome K, Watkins SC, Romero G. The recruitment of Raf-1 to membranes is mediated by direct interaction with phosphatidic acid and is independent of association with Ras. J Biol Chem. 2000;275:23911–23918. doi: 10.1074/jbc.M001553200. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Cell, Warne PH, Khwaja A, Marte BM, Pappin D, Das P, Waterfield MD, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- Roy S, Lane A, Yan J, McPherson R, Hancock JF. Activity of plasma membrane-recruited Raf-1 is regulated by Ras via the Raf zinc finger. J Biol Chem. 1997;272:20139–20145. doi: 10.1074/jbc.272.32.20139. [DOI] [PubMed] [Google Scholar]
- Roy S, Luetterforst R, Harding A, Apolloni A, Etheridge M, Stang E, Rolls B, Hancock JF, Parton RG. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat Cell Biol. 1999;1:98–105. doi: 10.1038/10067. [DOI] [PubMed] [Google Scholar]
- Stokoe D, Macdonald SG, Cadwallader K, Symons M, Hancock JF. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;26:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- Teruel MN, Meyer T. Translocation and reversible localization of signaling proteins: a dynamic future for signal transduction. Cell. 2000;103:181–184. doi: 10.1016/s0092-8674(00)00109-4. [DOI] [PubMed] [Google Scholar]
- Tsukada S, Simon M, Witte O, Katz A. Binding of the βγ subunits of heterotrimeric G-proteins to the PH domain of Bruton's tyrosine kinase. Proc Natl Acad Sci USA. 1994;91:11256–11260. doi: 10.1073/pnas.91.23.11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium-and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P, Rother KI, Balla T. Phosphatidylinositol 3-kinase-dependent membrane association of the Bruton's tyrosine kinase pleckstrin homology domain visualized in single living cells. J Biol Chem. 1999;274:10983–10989. doi: 10.1074/jbc.274.16.10983. [DOI] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Wang HG, Rapp UR, Reed JC. Bcl-2 targets the protein kinase Raf-1 to mitochondria. Cell. 1996;87:629–638. doi: 10.1016/s0092-8674(00)81383-5. [DOI] [PubMed] [Google Scholar]
- Williams JG, Drugan JK, Yi G-S, Clark GJ, Channing JD, Campbell SL. Elucidation of binding determinants and functional consequences of Ras/Raf-CRD interactions. J Biol Chem. 2000;275:22172–22179. doi: 10.1074/jbc.M000397200. [DOI] [PubMed] [Google Scholar]
- Zhang G, Kazainetz MG, Blumberg PM, Hurley JH. Crystal structure of the cys2 activator-binding domain of protein kinase Cδ in complex with phorbol ester. Cell. 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]