Abstract
γ-Tubulin functions as a multiprotein complex, called the γ-tubulin complex (γ-TuC), and composes the microtubule organizing center (MTOC). Fission yeast Alp4 and Alp6 are homologues of two conserved γ-TuC proteins, hGCP2 and hGCP3, respectively. We isolated a novel gene, alp16+, as a multicopy suppressor of temperature-sensitive alp6-719 mutants. alp16+ encodes a 759-amino-acid protein with two conserved regions found in all other members of γ-TuC components. In addition, Alp16 contains an additional motif, which shows homology to hGCP6/Xgrip210. Gene disruption shows that alp16+ is not essential for cell viability. However, alp16 deletion displays abnormally long cytoplasmic microtubules, which curve around the cell tip. Furthermore, alp16-deleted mutants are hypersensitive to microtubule-depolymerizing drugs and synthetically lethal with either temperature-sensitive alp4-225, alp4-1891, or alp6-719 mutants. Overproduction of Alp16 is lethal, with defective phenotypes very similar to loss of Alp4 or Alp6. Alp16 localizes to the spindle pole body throughout the cell cycle and to the equatorial MTOC at postanaphase. Alp16 coimmunoprecipitates with γ-tubulin and cosediments with the γ-TuC in a large complex (>20 S). Alp16 is, however, not required for the formation of this large complex. We discuss evolutional conservation and divergence of structure and function of the γ-TuC between yeast and higher eukaryotes.
INTRODUCTION
Microtubules, consisting of polymers of α- and β-tubulin heterodimers, possess intrinsic polarity, which stems from the fast-growing plus end and the slow-growing minus end (Nogales, 2000). In most eukaryotes, the minus end of microtubules is embedded in specialized structures, including the centrosome in animal cells, the basal body in Ciliata, and the spindle pole body (SBP) in yeast. These structures, although different in morphology, are functionally referred to as the microtubule organizing centers (MTOCs) (Pickett-Heaps, 1969), because they play a role in organizing microtubules in vivo. γ-Tubulin is a conserved member of tubulin superfamily (Oakley and Oakley, 1989; Joshi, 1993; Dutcher, 2001; McKean et al., 2001), and instead of being incorporated into microtubule filaments, it localizes to the MTOC; in particular, in animal cells, it is enriched in the pericentriolar material around the centrosome as well as in the cytosol (Moritz et al., 1995). A number of experiments performed in various organisms and systems have illuminated an important role of γ-tubulin in nucleation of microtubules and the formation of mitotic bipolar spindles (Oakley et al., 1990; Horio et al., 1991; Stearns et al., 1991; Zheng et al., 1991; Joshi et al., 1992; Oakley, 1992; Joshi, 1993; Felix et al., 1994; Stearns and Kirschner, 1994).
One of the most significant characteristics of γ-tubulin, in contrast to α- and β-tubulin, is its existence as a multiprotein complex (called the γ-tubulin complex, γ-TuC) with nontubulin proteins. In animal cells, the γ-TuC comprises an open-ring structure of 25-nm diameter, and because of this shape, it is often called the γTubulin Ring Complex (γTuRC) (Moritz et al., 1995, 1998; Zheng et al., 1995). Genetic analysis performed in budding yeast was instrumental in terms of the characterization of nontubulin components, because it allowed identification, for the first time, of two other components of the γ-TuC. These are Spc97 and Spc98, which, together with γ-tubulin, form an ∼6-S or 300-kDa complex in this organism (Geissler et al., 1996; Knop et al., 1997; Knop and Schiebel, 1997). The size suggests that the budding yeast γ-TuC comprises two molecules of γ-tubulin and one molecule of Spc97 and Spc98 per complex.
Biochemical analysis showed that the size of the γ-TuC of higher eukaryotes is much larger (25–30 S, >2000 kDa), suggesting that in higher eukaryotes the composition of the γ-TuC might differ from that of budding yeast (Stearns and Kirschner, 1994; Zheng et al., 1995). Subsequent purification of the γ-TuC and identification of nontubulin components have shown that this is indeed the case (Martin et al., 1998; Murphy et al., 1998; Tassin et al., 1998). In terms of evolutional conservation, it turns out that Spc97 and Spc98 are conserved ubiquitously through evolution, such that mammals contain each homologue; human homologues are called γ-TuC protein (GCP) 2 and GCP3, respectively (Murphy et al., 1998), while frog homologues are designated gamma ring protein (GRIP) Xgrip110 and Xgrip109 (Martin et al., 1998). What is surprising and of interest is the revelation that Spc97/GCP2 and Spc98/GCP3 might have been derived from a common ancestor, because these vertebrate homologues share common motifs, which were not apparent from amino acid comparison between Spc97 and Spc98.
Further purification and homology searches for other components in vertebrates have highlighted evolutional divergence between budding yeast and animals. In addition to Spc97/GCP2 and Spc98/GCP3, animals contain additional nontubulin components. In humans, frogs, and flies, three other components (GCP4, -5, and -6; Xgrip75, 133, and 210; and Dgrip75, 128, and 163, respectively) have been identified (Fava et al., 1999; Gunawardane et al., 2000; Zhang et al., 2000; Murphy et al., 2001). Interestingly, it transpires that all three of these homologues also share structural motifs similar to GCP2 and GCP3, further supporting the notion that all the nontubulin components of the γ-TuC stem from a common origin and have then deviated.
We have shown previously that fission yeast contains Alp4 and Alp6, which are structural and functional homologues of GCP2/Spc97 and GCP3/Spc98, respectively (Vardy and Toda, 2000). Temperature-sensitive (ts) mutants of these corresponding genes were originally identified as those defective in growth polarity (Radcliffe et al., 1998) and display lethal mitotic defects, including monopolar spindles, chromosome segregation defects, and “cut” phenotypes (Vardy and Toda, 2000; Vardy et al., 2002). Size analysis of the fission yeast γ-TuC indicates that in fission yeast, unlike budding yeast, the γ-TuC exists as a large complex (>20 S), comparable to the size of the vertebrate γ-TuC. This leads to the prediction that fission yeast, like higher eukaryotes, may contain γ-TuC proteins other than Alp4 and Alp6.
In this study, we present the identification of a novel γ-TuC protein Alp16. The alp16+ gene, originally isolated as multicopy plasmids that suppress ts alp6-719 mutants, encodes a protein that contains conserved motifs found in all other members of γ-TuC components and is most homologous to GCP6 and Xgrip210. We show here genetic and biochemical properties of Alp16 and discuss a role of nontubulin components in γ-tubulin function.
MATERIALS AND METHODS
Strains, Media, and Genetic Methods
Strains used in this study are listed in Table 1. YPD (2% dextrose, 2% polypeptone and 1% yeast extract) and YE5S were used as rich media, and modified synthetic EMM2 was used as minimal medium. Standard methods were followed as described (Moreno et al., 1991). For color assay to examine synthetic lethal interaction, EMM2 or YE5S medium containing a lower amount of adenine (5 μg/ml) was used (Paluh et al., 2000).
Table 1.
Strain list
| Strains | Genotypes | Derivations |
|---|---|---|
| HM123 | h−leu1 | Our stock |
| 513 | h−leu1 ura4 | Our stock |
| DH225 | h−leu1 alp4-225 | Our stock |
| DH719 | h−leu1 alp6-719 | Our stock |
| DH1891 | h−leu1 alp4-1891 | Our stock |
| LV1 | h−leu1 ura4 alp4-1891 | Our stock |
| LV3 | h−leu1 ura4 alp6-719 | Our stock |
| LV8 | h−leu1 ura4 alp16∷ura4+ | This study |
| LV13 | h−leu1 alp16+-3HA-kanr | This study |
| LV15 | h−leu1 alp4+-3HA-kanr | This study |
| LV16 | h−leu1 alp6+-3HA-kanr | This study |
| LV66 | h−/h+leu1/leu1 ura4/ura4 his7/his7 ade6- M210/ade6-M216alp6+∷ura4+/+ | This study |
| LV223 | h−leu1 nmt1-alp16+-kanr | This study |
| AF1 | h−leu1 ura4 ade1-D1 ade6-M210 alp4-1891 | This study |
| AF2 | h−leu1 ura4 ade1-D1 ade6-M210 alp4-1891 containing pAF-alp4 | This study |
| AF3 | h−leu1 ura4 ade1-D1 ade6-M210 alp4-1891 alp16∷kanrcontaining pAF-alp4 | This study |
| AF4 | h−leu1 ura4 alp16∷kanr | This study |
| AF6 | h−leu1 alp16+-13myc-kanr | This study |
| AF7 | h−leu1 alp4+-13myc-kanr | This study |
| AF8 | h−leu1 alp6+-13myc-kanr | This study |
Mutant alleles used for leu1 and ura4 are leu1-32 and ura4-D18.
Cloning of the alp16+ Gene
A Schizosaccharomyces genomic library in pUR19 (Barbet et al., 1992) was used for the isolation of genes which complemented ts alp4-225, alp4-1891, and alp6-719 mutants (LV1, LV2, and LV3, respectively; Table 1). In total, 30 (alp4) and 7 (alp6) plasmids were independently isolated. Restriction enzyme mapping showed that they were classified into two (pLV4-1 and -2) and 4 different plasmids (pLV6-1, -2, -3, and -4), respectively. pLV4-1 and -2 and pLV6-1 plasmids contained alp4+, whereas pLV6-2 and -3 contained alp6+. The remaining pLV6-4 contains a novel gene, which we designate alp16+.
Nucleic Acids Preparation and Manipulation
Enzymes were used as recommended by the suppliers (New England Biolabs, Beverly, MA). Nucleotide sequence data reported in this article are in the DDBJ/EMBL/GenBank databases under accession number AB074978 (alp16+).
Gene Disruption, C-Terminus Tagging, and Overexpression
The method based on polymerase chain reaction-generated fragments (Bähler et al., 1998) was used for complete gene disruption, epitope tagging (green fluorescent protein [GFP], 3 hemagglutinin (3HA), or 13myc) in the C terminus under the endogenous promoter, and overexpression using a thiamine-repressible nmt1 promoter (Maundrell, 1990). The alp6+ or alp16+ gene was deleted in diploid. Dissection of asci from heterozygous diploid cells showed that the alp16+ gene is not essential for cell viability, because four viable spores were obtained and uracil auxotroph was segregated 2:2 from 20 tetrads. In contrast, gene disruption of alp6+ shows that it is essential.
Gel Filtration Chromatography
Soluble protein extracts were prepared in buffer A (20 mM Tris-HCl, pH 7.5, 20% glycerol, 0.1 mM EDTA, 1 mM mercaptoethanol, and 5 mM ATP, plus a cocktail of inhibitors; Sigma, St. Louis, MO). Gel filtration chromatography was performed on a Superose-6 column by fast protein liquid chromatography (Amersham Pharmacia Biotech, Piscataway, NJ). The column was equilibrated with 2 column volumes of buffer A containing 100 mM NaCl. To determine molecular weight, a parallel column was run with standards consisting of dextran (2000 kDa), thyroglobulin (669 kDa), and α-amylase (232 kDa). Fractions (50 μl each) were separated by SDS-PAGE on 7.5 or 10% gels, and fractionated proteins were detected with individual antibodies.
Immunochemical Assays
Affinity-purified rabbit polyclonal anti-GFP antibody was provided by Dr. Ken Sawin (Institute of Cell & Molecular Biology, University at Edinburgh, UK). Mouse monoclonal anti–α-tubulin antibody (TAT-1) was obtained from Dr. Keith Gull (University of Manchester, UK). Mouse monoclonal anti-HA (16B12) and anti-myc antibodies (9E10) were purchased from BAbCO (Richmond, CA), and anti–γ-tubulin antibody (T6557) was from Sigma. Horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin (IgG), goat anti-mouse IgG (Bio-Rad Laboratories, Hercules, CA), and a chemiluminescence system (ECL, Amersham, Arlington Heights, IL) were used to detect bound antibody. Fission yeast whole cell extracts were prepared using glass beads to disrupt cells as described previously (Vardy and Toda, 2000). For immunoprecipitation, 1 mg of total protein extracts was used.
Indirect Immunofluorescence Microscopy
Cells were fixed with methanol, and primary antibody (affinity-purified anti-GFP, 1/200, or TAT-1, 1/50) was applied, followed by Cy3-conjugated goat anti-rabbit or anti-mouse IgG (Sigma). Immunofluorescence images were viewed with a Zeiss Axioplan 2 (Zeiss, Oberkochen, Germany) equipped with a chilled video CCD camera (C4742-95) and a PC computer containing AQM software (Kinetic Imaging, Merseyside, UK) or with a Zeiss LSM510 laser scanning confocal microscope. Images were then processed by use of Adobe Photoshop (version 5.5).
Synthetic Lethality Analysis
The procedures developed by Drs. Michael J. Moser and Trisha Davis (University of Washington, Seattle, WA) were followed according to Paluh et al. (2000). A strain (AF1, Table 1) that contains alp4-1891, ade1-D25, ade6-M210, ura4-D18, and leu1–32 was first constructed. A multicopy plasmid (pAF-alp4) that carries the alp4+, ade1+, and ura4+ genes was made from pLV6-4 by inserting a 4-kb polymerase chain reaction-amplified fragment containing ade1+ with its native promoter (300 base pairs upstream sequence). AF1 was transformed with pAF-alp4 (designated AF2). Usually, pAF-alp4 is unstable and can be lost mitotically, which results in the appearance of subpopulations displaying white (or red-sectored), Ade−, Ura−, and resistance to 5-FOA. If AF1 is combined with another mutation (such as Δalp16) that shows synthetic lethal interaction with alp4-1891, pAF-alp4 becomes stable, and the resulting transformant colonies display only red, Ade−, Ura+, and 5-FOA sensitivity.
RESULTS
Isolation of the alp16+ Gene as a Multicopy Plasmid That Suppresses Mutations in the γ-TuC
To identify novel genes that are involved in γ-TuC function, a fission yeast genomic library was used to isolate plasmids that complemented the mutations in the γ-TuC. Fission yeast γ-TuC comprises at least three components: Gtb1/Tug1 (γ-tubulin), Alp4 (the GCP2/Spc97 homologue), and Alp6 (the GCP3/Spc98 homologue) (Horio et al., 1991; Stearns et al., 1991; Vardy and Toda, 2000). Mutant strains used were ts alp4-225, alp4-1891, and alp6-719 (Radcliffe et al., 1998). From a number of plasmids that suppressed ts phenotypes of these strains, one plasmid (pLV6-4), isolated from screening of alp6-719, contained inserts distinct from those carrying either alp4+ or alp6+ gene. Subcloning and subsequent nucleotide sequencing of pLV6-4 showed that a gene that rescued the mutation, which we designate alp16+, is novel.
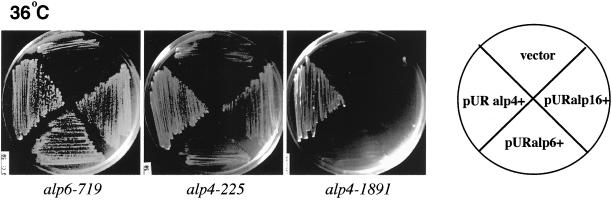
Next, multicopy plasmids containing alp4+, alp6+, alp16+, or gtb1+ were used to examine high-dosage suppression of ts alp4 and alp6 mutants. Plasmids containing alp16+ were capable of complementing both alp6-719 and alp4-225, but not alp4-1891 (Figure 1). As reported previously, multicopy plasmids containing alp4+ were capable of rescuing alp6-719 (Vardy and Toda, 2000). In contrast, plasmids containing gtb1+ could not suppress any ts mutations examined.
Figure 1.
Isolation of alp16+ as a multicopy suppressor of alp6-719. The ts alp4 or alp6 mutants indicated were transformed with an empty vector or multicopy plasmids containing alp4+, alp6+, or alp16+ (pLV4-1, pLV6-1, or pLV6-4, respectively), and transformants were streaked on rich YE5S plates and incubated at 36°C for 3 d.
The Alp16 Protein Contains Structural Motifs Common to All γ-TuC Proteins
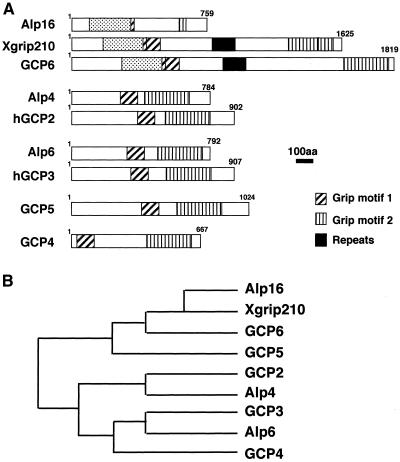
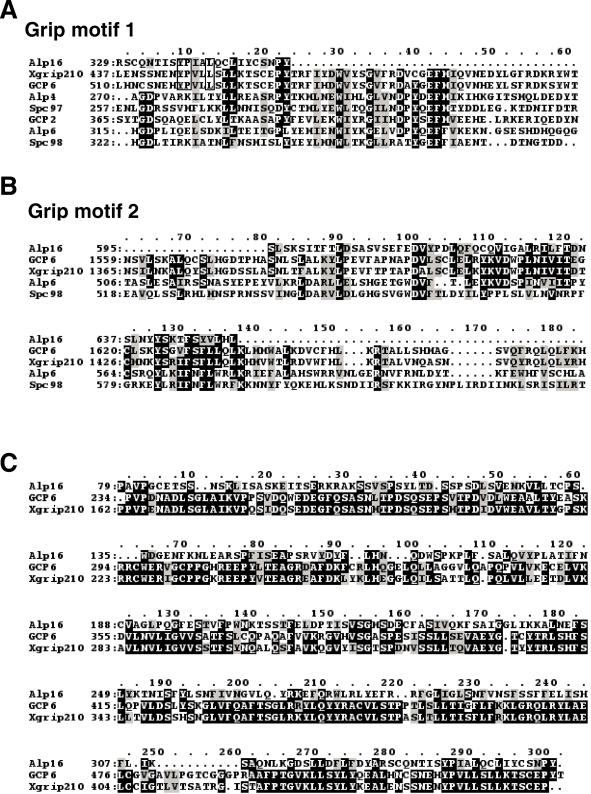
Nucleotide sequencing of an entire open reading frame encoding alp16+ showed that Alp16 consists of 759 amino acid residues. A homology search against the budding yeast database did not show any homologues in the entire genome of this organism. In contrast, careful comparison between Alp16 and proteins from vertebrate species revealed that Alp16 is a distant member of GCPs (Murphy et al., 1998) or GRIPS (Oegema et al., 1999). As shown in Figure 2, Alp16 contains two domains (Grip1 and Grip2), albeit only partly, which exist in all γ-TuC proteins. Comparison of individual amino acid residues in Grip1 and Grip2 is shown in Figure 3, A and B. This analysis suggests that Alp16 is a novel member of γ-TuC proteins.
Figure 2.
Structural comparison between Alp16 and vertebrate γ-TuC proteins. (A) Schematic diagram showing conservation of structural motifs between fission yeast Alp4, Alp6, Alp16, and the vertebrate GCPs. Hatched and vertically striped boxes show Grip1 and Grip2 motifs, respectively (Gunawardane et al., 2000), and stippled boxes show a domain common between Alp16 and GCP6 (and Xgrip210). Solid boxes show tandem repeats of a 27-amino-acid sequence, which is found only in GCP6 and Xgrip210 (Zhang et al., 2000; Murphy et al., 2001). (B) A phylogenetic tree of the GCPs and fission yeast Alp4, Alp6, and Alp16.
Figure 3.
Amino acid comparison between Alp16 and other GCPs. (A–C) Alignment of amino acid sequence of Grip motif 1 (A) or Grip motif 2 (B) between Alp16 and various GCPs and budding yeast Spc98. (C) Amino acid sequence in the N-terminal domain (shown in stippled boxes in Figure 2A) is also compared between Alp16 and GCP6 (and Xgrip210). Identical amino acid residues are marked with black boxes with white letters (A–C) and black letters with open boxes (A), and conserved amino acid residues are shown by gray boxes with black letters.
In addition to the Grip domains, Alp16 contains, in the N-terminally proximal region, ∼300 amino acid residues that show sequence similarity to the corresponding region of human GCP6 and Xenopus Xgrip210 (26% identity and 43% similarity, emphasized with stippled boxes in Figure 2A). As mentioned above, this region does not exist in other GCPs. A phylogenetic tree also showed that Alp16 is closest to these two proteins (Figure 2B; amino acid comparison is shown in Figure 3C) (Zhang et al., 2000; Murphy et al., 2001). Comparison of GCP homologues in various organisms is shown in Table 2.
Table 2.
Comparison of γ-tubulin complex proteins in different species
| H. sapiens | X. laebis | D. melanogastera | S. pombe | S. cerevisiae |
|---|---|---|---|---|
| GCP1 | γ-tubulin | γTub23C, γTub37CD | Gtb1/Tug1 | Tub4 |
| GCP2 | Xgrip110 | Dgrip84 | Alp4 | Spc97 |
| GCP3 | Xgrip109 | dd4/Dgrip91 | Alp6 | Spc98 |
| GCP4 | Xgrip75b | Dgrip75 | ||
| GCP5 | XGRIP133b | Dgrip128 | ||
| GCP6 | XGRIP210 | Dgrip163 | Alp16 |
Drosophila genome sequencing predicts the existence of two other ORFs, which could encode the GCPs (AAF44968 and AAF50536) (Murphy et al., 2001).
Sequence information has not been available, and the value of molecular weight was adapted from (Martin et al., 1998; Zhang et al., 2000).
Alp16 Is a γ-TuC Protein
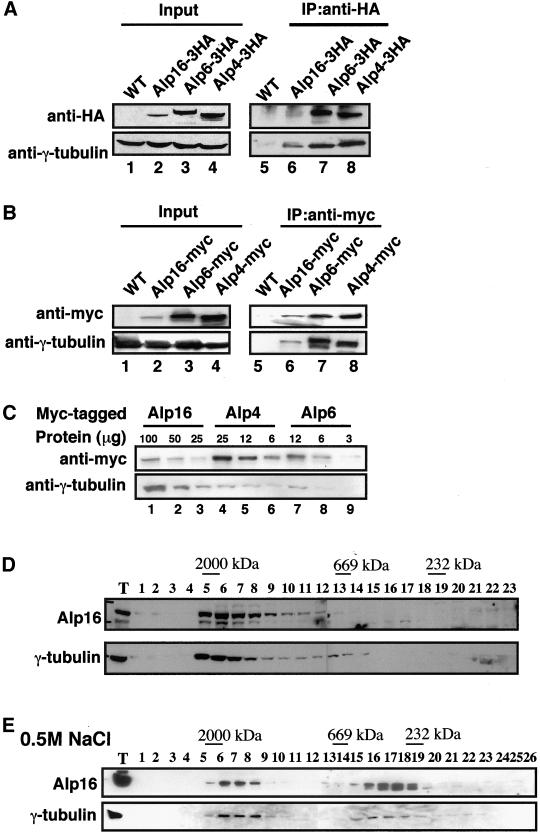
To examine whether Alp16 interacts physically with γ-tubulin in the cell, immunoprecipitation was performed. For this purpose, Alp16 was tagged under its endogenous promoter with the HA epitope in its C terminus (three repeats of HA, Alp16-3HA). We previously constructed similar tagged strains in which chromosomal alp4+ and alp6+ are fused with HA (Vardy and Toda, 2000). In addition, these three genes were also tagged with the myc epitope (13myc, Alp4-myc, Alp6-myc, and Alp16-myc). Any C-terminal tagging does not interfere with protein function, because growth properties of tagged strains, including generation time and sensitivity to microtubule-destabilizing drugs, are indistinguishable from those of nontagged parental strains. Immunoblotting using anti-HA or anti-myc antibody identified Alp16-3HA or Alp16-myc on an SDS-PAGE gel (lane 2 in Figure 4, A and B, respectively). Parallel immunoblotting using strains containing Alp4-3HA, Alp4-myc, Alp6-3HA, or Alp6-myc showed that Alp16 is not as abundant as these two proteins in the cell (lanes 2–4). Further analysis of serially diluted protein samples and densitometric calibration allowed us to estimate a stoichiometric ratio of Alp4, Alp6, and Alp16, which is ∼10:8:1 (Figure 4C).
Figure 4.
Physical interactions between Alp16 and the γ-TuC. (A, B) Identification of the alp16+ gene product and coimmunoprecipitation between Alp16 and γ-tubulin. Genomic copies of alp4+, alp6+, and alp16+ were tagged under their endogenous promoters at the C terminus with 3HA or 13myc. Immunoblotting was performed with anti-HA (A) or anti-myc antibody (B). Protein samples were prepared from appropriate tagged strains containing Alp16-3HA or Alp16-myc (lane 2), Alp6-3HA or Alp6-myc (lane 3), or Alp4-3HA or Alp4-myc (lane 4). Extracts from untagged wild-type strain were also prepared (lane 1). Anti–γ-tubulin was used as a loading control. Immunoprecipitation was performed with anti-HA (A) or anti-myc antibody (B) (lanes 5–8). Forty micrograms of extracts (lanes 1–4) and 1 mg equivalent of total proteins (lanes 5–8) were run. Precipitated proteins were detected with anti-HA (A), anti-myc (B), or anti–γ-tubulin antibody. (C) Stoichiometric analysis of Alp16. Indicated amount of extracts from strains containing Alp16-myc (lanes 1–3), Alp4-myc (lanes 4–6), or Alp6-myc (lanes 7–9) were run in each lane. Immunoblotting was performed with anti-myc and anti–γ-tubulin antibodies. (D) Gel filtration chromatography. Soluble cell extracts were prepared from an Alp16-3HA strain and separated by gel filtration. Each fraction (1–23), together with total extract (T, 10 μg), was analyzed by immunoblotting with anti-HA (top) or anti–γ-tubulin antibody (bottom). Positions of size markers (2000, 669, and 232 kDa) are shown at the top. (E) The size of the γ-TuC in the presence of high salt concentrations. Protein extracts were prepared in the presence of high salt concentrations (0.5 M NaCl) and separated through a Superose-6 column. In this case, a strain containing Alp16-myc was used to enhance the detection of the Alp16 protein.
Immunoprecipitation experiments showed that both Alp16-3HA and Alp16-myc coprecipitated with γ-tubulin (lane 6 in Figure 4, A and B). This result indicated that Alp16 interacts with γ-tubulin in the cell and substantiated the possibility that Alp16 is a novel component of the γ-TuC.
Next, the native size of the Alp16-containing protein complex was examined by gel filtration chromatography. As shown in Figure 4D, Alp16 sedimented in a large fraction (∼2000 kDa), and γ-tubulin also fractionated around this size. In Drosophila and vertebrates, it is known that the γ-TuC consists of two populations in terms of its size: a large complex (>25 S) and a smaller complex (9.8 S or 280 kDa) (Moritz et al., 1998; Oegema et al., 1999). This smaller complex is composed only of γ-tubulin, GCP2, and GCP3. This particular form of the complex becomes apparent when the γ-TuC is treated with high concentrations of salt (Oegema et al., 1999). We have reported previously that the fission yeast γ-TuC also apparently forms large as well as small complexes (Vardy and Toda, 2000). We therefore examined the size of the fission yeast γ-TuC in the presence of a high concentration of salt (0.5 M NaCl). As shown in Figure 4E, under this condition, the γ-TuC still fractionated as a large complex, but the position of the peak fractions appeared to shift toward a smaller size (from fractions 5–7 in Figure 4D to fractions 6–8 in Figure 4E). Furthermore, more than 50% of the complex was present in smaller fractions (fractions 16–19). It is important to point out that Alp16 is also present in both peaks. Taking these results together, we conclude that Alp16 is the fourth component of the fission yeast γ-TuC and that Alp16 is capable of interacting with the γ-tubulin in both large and small forms of the complex.
The Cellular Localization of Alp16 to the MTOC
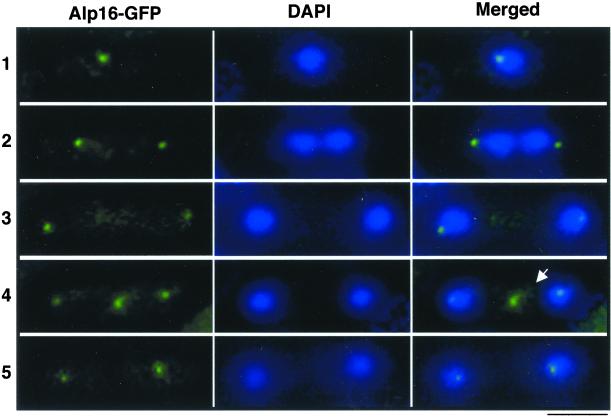
Next, the cellular localization of Alp16 was addressed. To this end, the GFP gene was fused to the C terminus of the chromosomal alp16+ gene. We have shown previously that a similar GFP tagging of alp4+ or alp6+ is sufficient to visualize the localization of these two proteins at the MTOCs (Hagan, 1998; Heitz et al., 2001) under fluorescence microscope (Vardy and Toda, 2000). In contrast, in the case of Alp16-GFP, probably because of lower protein abundance (see Figure 4, A and B), GFP signals were too weak to detect Alp16 localization. However, Alp16-GFP could be visualized by use of a polyclonal anti-GFP antibody (provided by Dr. Ken Sawin). As shown in Figure 5 (left), in exponentially growing cells, signals were seen as either a single spot (row 1) or double spots (rows 2, 3, and 5) around the nuclear periphery, most likely corresponding to the SPBs. Furthermore, at postanaphase (row 4), in addition to two nuclear dots, the medial dot located between the two SPBs was evident (arrow in row 4). This localization pattern is identical to that reported for Alp4 and Alp6 (Vardy and Toda, 2000) and Gtb1 (Horio et al., 1991), and the medial dot displays the equatorial MTOC (Heitz et al., 2001). Taken together, these results confirmed that Alp16 is a γ-TuC protein and localizes to the MTOC through the cell cycle.
Figure 5.
The cellular localization of Alp16. A C-terminally tagged strain with GFP under the endogenous promoter (Alp16-GFP) was fixed with methanol and processed for immunofluorescence microscopy. Affinity-purified rabbit polyclonal anti-GFP antibody (left) was used as a primary antibody. DAPI was also used for nuclear staining (middle). Merged images are shown at right. Representative images from interphase (1 and 5), anaphase (2 and 3), and postanaphase (4) are shown. In row 5, cytokinesis has been completed. Alp16 at the equatorial MTOC is marked with arrow in row 4. Bar, 10 μm.
Alp16 Is Dispensable under Normal Conditions but Required for Intact Cytoplasmic Microtubules
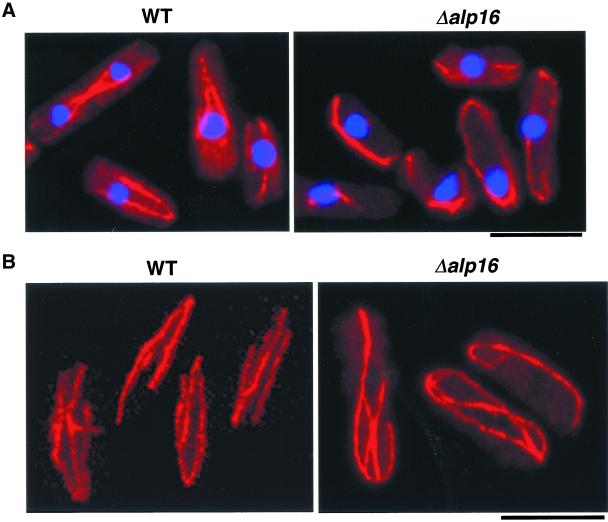
To examine phenotypes in the absence of Alp16, gene disruption was performed, in which the complete open reading frame of one of the alp16+ genes in a diploid cell was deleted. Tetrad dissection of this heterozygous diploid showed that the alp16+ gene is not essential for cell viability at any temperature tested (20, 30, and 36°C). Immunofluorescence microscopy using anti-tubulin antibody showed that in alp16 mutants, cytoplasmic microtubules are defective; instead of normal arrays that terminate near the cell tips (Hagan, 1998), microtubules are longer and often curve around the cell tips (Figure 6A). Curved microtubules are also observed under confocal microscopy, as shown in Figure 6B. The appearance of these long interphase microtubules is more evident when cultures are incubated at 36°C. These defective microtubules are very similar to those seen in ts alp4 or alp6 mutants at restrictive temperature (Vardy and Toda, 2000). In contrast to alp4 or alp6 mutants, which display mitotic defects, alp16 deletion did not show any noticeable mitotic phenotypes. This result indicates that Alp16 is nonessential for normal growth but is required for the formation of array-like cytoplasmic microtubules.
Figure 6.
Abnormally long interphase microtubules in alp16-deleted mutants. (A, B) An exponentially growing wild-type (WT, left) or alp16-deleted strain (right) was fixed with methanol and processed for immunofluorescence microscopy with anti-tubulin antibody (TAT-1). Merged images of microtubules (red) and DAPI (blue) are shown (A). Images from conventional fluorescence microscopy (A) or confocal microscopy (B) are presented. The culture was incubated at 36°C for 6 h, because microtubule defects are more evident at this temperature. Bar, 10 μm.
Alp16 Is Essential When γ-TuC Function Is Compromised
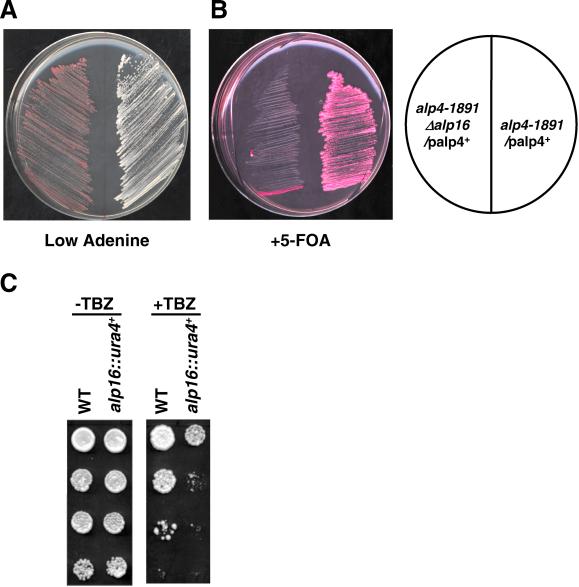
Despite apparent dispensability, we found that the alp16 deletion is synthetically lethal with a ts alp4-1891 mutant at permissive temperature. As shown in Figure 7, A and B, use of a standard assay system by which synthetic lethality could be examined by colony colors (Figure 7A) and resistance to 5-FOA (Figure 7B) (Paluh et al., 2000) (see MATERIALS AND METHODS for detail), clearly shows that the Δalp16 alp4-1891 double mutant containing multicopy plasmids carrying the alp4+ gene is unable to lose these plasmids at the permissive temperature of alp4-1891. This is shown by red colonies and sensitivity to 5-FOA. In contrast, an alp4-1891 single-mutant strain carrying the same plasmid was capable of losing plasmids, which resulted in white colonies and resistance to 5-FOA.
Figure 7.
Synthetic lethal interaction and hypersensitivity of alp16-deleted mutants to antimicrotubular drugs. (A, B) Two strains (AF2, left, and AF3, right, Table 1; see MATERIALS AND METHODS for strain construction) were streaked onto plates containing lower concentration of adenine (A) or 5-FOA and phloxine B (red dye) (B) and incubated at 26°C for 3 d. (C) Hypersensitivity of alp16-deletion mutants to TBZ. Wild-type and alp16-deletion mutants were spotted onto plate (−TBZ, left) or plate containing 20 μg/ml TBZ (+TBZ, right) as serial dilutions (106 cells in the top row and then diluted 10-fold in each subsequent spot below) and incubated at 30°C for 3 d.
Tetrad analysis between Δalp16 and alp4-1891 mutants confirmed synthetic lethality between these two mutants (Table 3). Furthermore, we found that Δalp16 strains are also inviable when combined with alp4-225 or alp6-719. It was also found that double mutants between alp4 (-225 or -1891) and alp6-719 are lethal. Microscopic observation of inviable Δalp16 alp4-225 or Δalp16 alp6-719 mutants showed that spores germinated, divided several times, and then ceased division with bent morphology (A.F. and T.T., unpublished observation). This terminal phenotype is very similar to that of alp6-deleted cells (Vardy and Toda, 2000). Synthetic lethal interaction of Δalp16 appears to be specific to compromised γ-tubulin function, but not as a result of defects in microtubule function in general, because double mutants between Δalp16 and mutations in α- or β-tubulin (nda2-52, Δatb2, or nda-311, defective in α1-, α2-, or β-tubulin, respectively) (Hiraoka et al., 1984; Toda et al., 1984) are all viable (Table 3).
Table 3.
Synthetic lethal interaction between the alp16 deletion and mutations in alp4+ and alp6+
| alp4-225 | alp4-1891 | alp6-719 | nda2-52 | Δatb2 | nda3-311 | |
|---|---|---|---|---|---|---|
| Δalp16 | Lethal | Lethal | Lethal | Viable | Viable | Viable |
| alp4-225 | Lethal | |||||
| alp4-1891 | Lethal |
nda2+, atb2+, and nda3+ encode α1-, α2-, and β-tubulin respectively, and nda2-52 and nda3-311 mutants are cold-sensitive (cs) for growth (Hiraoka et al., 1984; Toda et al., 1984). Appropriate strains were crossed, and tetrad analysis was performed. Spores were germinated at permissive temperature (26°C for ts alp4 and alp6 mutants, and 32°C for cs nda2 and nda3 mutants), whilst spores of Δalp16 Δatb2 double mutants were germinated at 30°C.
We showed previously that ts alp4 and alp6 mutants are hypersensitive to the microtubule-depolymerizing drug thiabendazole (TBZ) at permissive temperature (Vardy and Toda, 2000). To examine whether Alp16 is also involved in the resistance to this drug, sensitivity to TBZ was examined in Δalp16 mutants. As shown in Figure 7C, Δalp16 cells were hypersensitive to this drug. Taken together, these results showed that Alp16 is dispensable under normal growth conditions; however, it becomes essential when γ-TuC function is compromised by either mutations or microtubule damages.
Fission Yeast γ-Tubulin Is Capable of Forming a Large Complex without Alp16
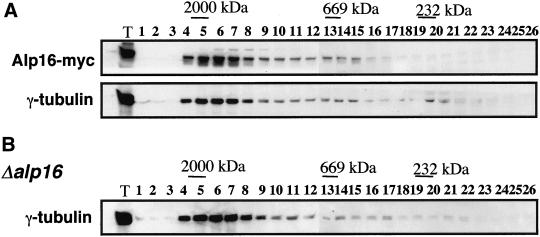
The size of the γ-TuC in fission yeast is large (>20 S), comparable to that of animal cells (Vardy and Toda, 2000; see also Figure 4D). We sought to clarify the role of Alp16 in this complex formation. For this purpose, gel filtration analysis was performed in wild-type cells containing Alp16-myc and alp16-deletion mutants. As shown in Figure 8, the size of the complex is indistinguishable between wild-type and Δalp16, judged from a pattern of γ-tubulin that cofractionated with Alp16-myc in wild-type cells. This result showed that fission yeast γ-tubulin is capable of forming a large complex, consisting of Gtb1, Alp4, and Alp6, without the fourth component, Alp16.
Figure 8.
The size of the γ-TuC in the absence of Alp16. Gel filtration was performed in wild-type cells containing Alp16-myc (top two panels) or alp16 deletion mutants (bottom). Soluble cell extracts were prepared as in Figure 4D. Immunoblotting was performed with anti-myc and anti–γ-tubulin antibodies.
Overexpression of alp16+ Results in Mitotic Spindle Defects, Similar to Loss of γ-TuC Function
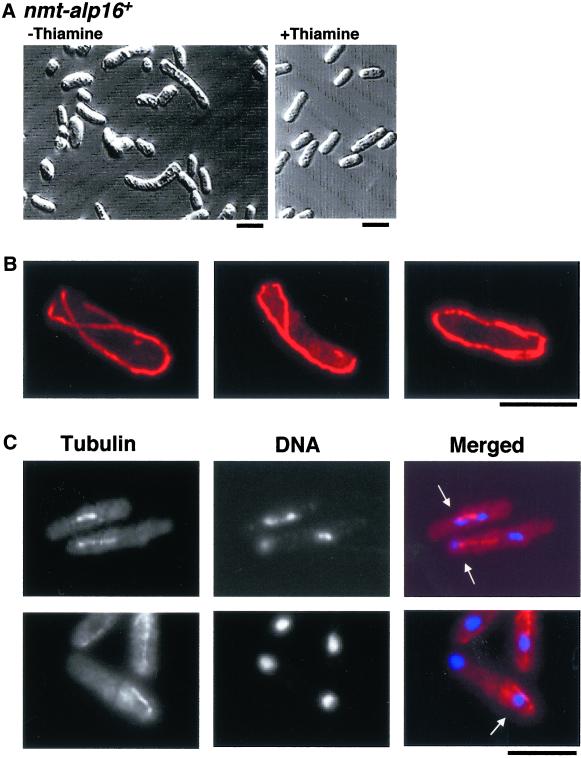
To examine the phenotypic consequences that arise from overproduction of Alp16, the strong thiamine-repressible nmt1 promoter (Maundrell, 1993) was integrated into the chromosomal locus just before the initiation codon of the alp16+ gene. It was found that overexpression of alp16+ is toxic and inhibits colony formation on plates in the absence of thiamine. Microscopic observation of these cells on thiamine-free plates indicated that many cells showed bent or branched morphology (Figure 9A), which is reminiscent of microtubule dysfunction in fission yeast.
Figure 9.
Lethal overexpression of the alp16+ gene. (A) Cell morphology. Cells containing the integrated nmt1-alp16+ gene were grown on minimal medium in the absence (left) or presence (right) of thiamine and incubated at 30°C for 2 d. (B) Long cytoplasmic microtubules. The same strain was grown in the liquid culture in the absence of thiamine for 20 h, processed for immunofluorescence microscopy using anti-tubulin antibody, and observed under confocal microscopy. (C) Mitotic defects with monopolar spindles. Cells that display mitotic phenotypes are shown (the same culture condition as in B). Conventional fluorescence microscopy was used for anti-tubulin staining (left) and DNA staining with DAPI (middle), and merged images are shown at right. Cells displaying monopolar mitotic spindles are marked with arrows (C). Bar, 10 μm.
Defective phenotypes of alp16+-overexpressing cells were further examined in the liquid culture. Cells were collected for immunofluorescence microscopy at 14, 16, 18, 20, and 24 h after induction at 30°C. Cells were stained with anti-tubulin antibody and DAPI. Two characteristic defects of microtubules were evident, one in interphase cells and the other in mitotic cells. In interphase, as shown in Figure 9B, cytoplasmic microtubules were often longer, like Δalp16 cells (see Figure 6), than those seen in wild-type cells, and curved around the ends of the cell. In contrast, in mitotic cells, spindle defects were evident, in particular the formation of monopolar spindles (Figure 9C). These two defects were in fact very similar to those observed in ts alp4-1891 and alp6-719 mutants at restrictive temperature (Vardy and Toda, 2000). It therefore appears that massive overproduction of Alp16 absorbs other structural components of the γ-TuC, which results in loss of γ-TuC function.
DISCUSSION
In this study, we describe the identification and characterization of a novel component (Alp16) of the fission yeast γ-TuC. The γ-TuC in this organism comprises three nontubulin components (Alp4, Alp6, and Alp16). Compared with animals, in which the complex consists of at least five nontubulin components (GCP2–6), the fission yeast complex is simpler despite its comparable size. In clear contrast, the budding yeast γ-TuC consists of only two nontubulin components (Spc97 and Spc98) with a smaller complex size. The fission yeast could therefore be regarded evolutionarily as an intermediate between animals and budding yeast.
Role of Nontubulin γ-TuC Proteins
The most surprising result arising from this study is the nonessentiality of Alp16 in fission yeast. In contrast, we and others have shown that γ-tubulin, Alp4, and Alp6 are absolutely required for cell viability and essential for various aspects of microtubule function. These include growth polarity, morphogenesis of intact cytoplasmic microtubules, and the formation of mitotic bipolar spindles (Horio et al., 1991; Paluh et al., 2000; Vardy and Toda, 2000; Hendrickson et al., 2001; Vardy et al., 2002). Furthermore, in virtually all eukaryotic cells, γ-tubulin, GCP2, and GCP3 play an indispensable role. This is based on both biochemical and genetic characterization in various organisms (Oakley, 1992; Barbosa et al., 2000; Schiebel, 2000; Sampaio et al., 2001). Analysis of the GCPs other than the three proteins described above have not been fully addressed, at least genetically; thus, it is premature at present to argue whether GCP4–6 are required for γ-TuC function. However, biochemical analysis such as protein depletion using specific antibodies has suggested that, like GCP2 and GCP3, they are essential (Fava et al., 1999; Zhang et al., 2000). Although a definite proof has to await the results from genetically amenable systems such as Drosophila, the requirement of GCP4–6 may become more crucial in animals than in fission yeast.
Despite being dispensable, Alp16 plays an important accessory role in microtubule function. It is required for the formation of normal interphase microtubules. Without Alp16, cytoplasmic microtubules fail to terminate at the cell tips; instead, they further elongate and curve. This phenotype is identical to that seen in ts alp4 or alp6 mutants, suggesting that Alp16 is essential for γ-TuC function, at least during interphase. The curved ends of long interphase microtubules correspond to their plus end (Drummond and Cross, 2000; Tran et al., 2001), whereas the γ-TuC localizes to the SPB, in which their minus end is embedded. Thus, it is likely that in vivo microtubule dynamics, such as the rate of growth and shrinkage and the frequency of catastrophe and rescue, might be somehow regulated not only at the plus end but also at the minus end in a γ-TuC–dependent manner.
The mitotic role of Alp16 becomes apparent and indispensable under conditions in which γ-TuC function is partially compromised, including the permissive temperature of ts alp4 or alp6 mutants and the presence of microtubule-depolymerizing drugs. In terms of stoichiometry, compared with other components of the γ-TuC, such as Alp4 and Alp6, the cellular amount of Alp16 needs to be low (1/8 to 1/10); otherwise, too much Alp16 results in the loss of γ-TuC function, probably because of the absorption of other essential components. It should be pointed out that, at least in fission yeast, γ-tubulin, Alp4, and Alp6 appear to be sufficient to form a large complex consisting of multiple copies of these three proteins (see below). Thus, it appears that Alp16, unlike Alp4 and Alp6, is a peripheral subunit of the γ-TuC, as suggested in GCP6/Xgrip210 (Keating and Borisy, 2000; Moritz et al., 2000; Wiese and Zheng, 2000; Zhang et al., 2000).
Divergence and Conservation of γ-TuC Proteins
We have performed a careful homology search against the predicted open reading frames in the fission yeast genome sequence (Wood et al., 2002) using GCPs or Grip motifs as queries. The only significant hits have been Alp4, Alp6, and Alp16. Therefore, it appears that nontubulin GCPs are composed of these three proteins in fission yeast, although it is formally possible that other GCPs, which do not contain Grip motifs, might exist. We have been attempting to purify the γ-TuC biochemically; however, probably because of a small amount of nontubulin components, we have not succeeded in biochemical identification of γ-TuC proteins (i.e., <400 molecules of Alp16 per cell; A.F. and T.T., unpublished results). We have searched for potential Alp16 homologues, other than GCP6, in organisms evolutionarily closer to fission yeast. These include Neurospora crassa and Aspergillus fumigatus, in which we have not detected the homologues. Because the genome sequencing of these organisms remains to be completed, we could not yet argue that Alp16 is conserved in other lower eukaryotes.
Recent molecular analysis of nontubulin γ-TuC components in vertebrates has revealed interesting structural features of these proteins. It appears that compared with GCP2 and GCP3, which are conserved in fungi, plants, and animals, the other three members (GCP4, GCP5, and GCP6) are more divergent (Murphy et al., 2001). GCP6 and Xgrip210 are unique, because they contain nine tandem repeats of a 27-amino-acid sequence, which is not found in other GCPs (Zhang et al., 2000; Murphy et al., 2001). Sequence analysis of Alp16 suggests that its closest GCP is GCP6. Despite this, Alp16 does not contain these tandem repeats, nor does the fission yeast entire genome contain any open reading frames, which show homology to these repeats. At the moment, the molecular function of these repeats remains to be identified.
Minimal Requirement of γ-TuC Function
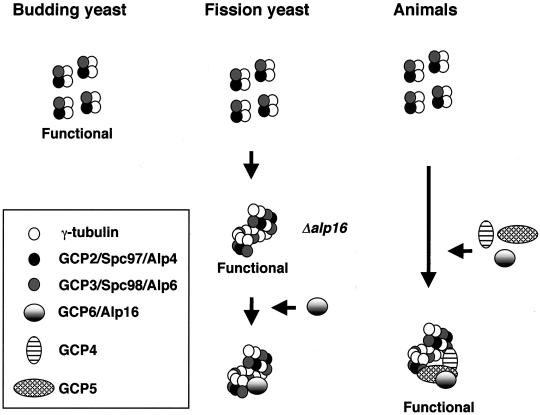
On the basis of the current results, we speculate the following evolutionary divergence in the γ-TuC (Figure 10). In budding yeast, γ-tubulin, Spc97, and Spc98, which form only a small complex, are sufficient to execute its role. In fission yeast, conversely, the three homologues (Gtb1/Tug1, Alp4, and Alp6) are capable of forming a large complex by self-multimerization, and this large γ-TuC without Alp16 is functional at least for its mitotic role. This suggests that, unlike in budding yeast, in fission yeast a small form of the γ-TuC is not functional as the MTOC in vivo. Finally, in animals, like budding yeast, the three proteins (γ-tubulin, GCP2, and GCP3) are capable of forming only a small complex, but unlike budding yeast and like fission yeast, this small complex is nonfunctional or at least severely impaired. This difference in the properties of the core γ-TuC among species might result in the development of an apparently more crucial role of GCP4, -5, and -6 in microtubule organization in animal cells.
Figure 10.
Assembly and function of the γ-TuC in yeast and animals. In budding yeast, γ-tubulin, Spc97 (GCP2), and Spc98 (GCP3) form a small complex (300 kDa; ratio 2:1:1, respectively), and this complex is sufficient for all aspects of γ-TuC function in this organism. Conversely, in fission yeast, the γ-TuC contains the fourth component, Alp16 (GCP6), and exists as a large complex (∼2000 kDa). In the absence of Alp16, however, the remaining three components are still capable of forming an analogous large complex, probably via multimerization of a small complex, and execute its role, at least under normal physiological conditions. Only when γ-TuC function is compromised does Alp16 become essential. Finally, in animal cells, for proper function of the γ-TuC, multimeric assembly or the recruitment to the centrosome of the other three GCPs (4–6), in addition to GCP2 and GCP3, is necessary.
We suggest two possible molecular functions of Alp16. The first is that Alp16 plays a role in the formation of the stable γ-TuC. In this case, Alp16 acts as structural glue. The second possibility is that Alp16 acts as an anchor, which helps the γ-TuC localize to the SPBs. Whichever is the case, our work has established that Alp16 plays an accessory but important role in fission yeast γ-TuC, which is required for formation of both interphase microtubules and mitotic bipolar spindles.
ACKNOWLEDGMENTS
We thank Drs. Ken Sawin for affinity-purified anti-GFP antibody and Janet Paluh, Michael J. Moser, and Trisha Davis for generous gifts of a strain (MP18) and plasmids (pKS+/ADE1-FS, pZA-25, and pNPT/ADE1-3) for the study of synthetic lethality. We thank Dr. Jacqueline Hayles for her critical reading of the manuscript and useful suggestions. M.A.G. was supported by an EMBO long-term fellowship. The research was supported by Cancer Research UK and by a research grant from the Human Frontier Science Program. The Cancer Research UK London Research Institute comprises the Lincoln's Inn Fields and Clare Hall Laboratories of the former Imperial Cancer Research Fund after the merger of the ICRF with the Cancer Research Campaign in February 2002.
Abbreviations used:
- γ-TuC
γ-tubulin complex
- GFP
green fluorescent protein
- MTOC
microtubule organizing center
- SPB
spindle pole body
- ts
temperature-sensitive
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.02–01–0603. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.02–01–0603.
REFERENCES
- Bähler J, Wu J, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Barbet N, Muriel WJ, Carr AM. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene. 1992;114:59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- Barbosa V, Yamamoto RR, Henderson DS, Glover DM. Mutation of a Drosophila gamma tubulin ring complex subunit encoded by discs degenerate-4 differentially disrupts centrosomal protein localization. Genes Dev. 2000;14:3126–3139. doi: 10.1101/gad.182800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DR, Cross RA. Dynamics of interphase microtubules in Schizosaccharomyces pombe. Curr Biol. 2000;10:766–775. doi: 10.1016/s0960-9822(00)00570-4. [DOI] [PubMed] [Google Scholar]
- Dutcher SK. The tubulin fraternity: alpha to eta. Curr Opin Cell Biol. 2001;13:49–54. doi: 10.1016/s0955-0674(00)00173-3. [DOI] [PubMed] [Google Scholar]
- Fava F, Raynaud-Messina B, Leung-Tack J, Mazzolini L, Li M, Guillemot JC, Cachot D, Tollon Y, Ferrara P, Wright M. Human 76p: a new member of the γ-tubulin–associated protein family. J Cell Biol. 1999;147:857–868. doi: 10.1083/jcb.147.4.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix MA, Antony C, Wright M, Maro B. Centrosome assembly in vitro: role of γ-tubulin recruitment in Xenopus sperm aster formation. J Cell Biol. 1994;124:19–31. doi: 10.1083/jcb.124.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Souès S, Kilmartin J, Schiebel E. The spindle pole body component Spc98p interacts with the γ-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Gunawardane RN, Martin OC, Cao K, Zhang L, Dej K, Iwamatsu A, Zheng Y. Characterization and reconstitution of Drosophila γ-tubulin ring complex subunits. J Cell Biol. 2000;151:1513–1524. doi: 10.1083/jcb.151.7.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan IM. The fission yeast microtubule cytoskeleton. J Cell Sci. 1998;111:1603–1612. doi: 10.1242/jcs.111.12.1603. [DOI] [PubMed] [Google Scholar]
- Heitz MJ, Petersen J, Valovin S, Hagan IM. MTOC formation during mitotic exit in fission yeast. J Cell Sci. 2001;114:4521–4532. doi: 10.1242/jcs.114.24.4521. [DOI] [PubMed] [Google Scholar]
- Hendrickson TW, Yao J, Bhadury S, Corbett AH, Joshi HC. Conditional mutations in γ-tubulin reveal its involvement in chromosome segregation and cytokinesis. Mol Biol Cell. 2001;12:2469–2481. doi: 10.1091/mbc.12.8.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes β-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Horio T, Uzawa S, Jung MK, Oakley BR, Tanaka K, Yanagida M. The fission yeast γ-tubulin is essential for mitosis and is localized at microtubule organizing centers. J Cell Sci. 1991;99:693–700. doi: 10.1242/jcs.99.4.693. [DOI] [PubMed] [Google Scholar]
- Joshi HC. γ-Tubulin: the hub of cellular microtubule assemblies. Bioessays. 1993;15:637–643. doi: 10.1002/bies.950151002. [DOI] [PubMed] [Google Scholar]
- Joshi HC, Palacios MJ, McNamara L, Cleveland DW. Gamma-tubulin is a centrosomal protein required for cell cycle-dependent microtubule nucleation. Nature. 1992;356:80–83. doi: 10.1038/356080a0. [DOI] [PubMed] [Google Scholar]
- Keating TJ, Borisy GG. Immunostructural evidence for the template mechanism of microtubule nucleation. Nat Cell Biol. 2000;2:352–357. doi: 10.1038/35014045. [DOI] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the γ-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Spc98p and Spc97p of the yeast γ-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin OC, Gunawardane RN, Iwamatsu A, Zheng Y. Xgrip109: a γ tubulin-associated protein with an essential role in γ tubulin ring complex (γTuRC) assembly and centrosome function. J Cell Biol. 1998;141:675–687. doi: 10.1083/jcb.141.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- Maundrell K. Thiamine repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- McKean PG, Vaughan S, Gull K. The extended tubulin family. J Cell Sci. 2001;114:2723–2733. doi: 10.1242/jcs.114.15.2723. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analyses of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:773–782. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Dedat JW, Alberts B, Agard DA. Microtubule nucleation by γ-tubulin-containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Guénebaut V, Heuser J, Agard DA. Structure of the γ-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol. 2000;2:365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- Moritz M, Zheng Y, Alberts BM, Oegema K. Recruitment of the γ-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J Cell Biol. 1998;142:775–786. doi: 10.1083/jcb.142.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SM, Preble AM, Patel UK, O'Connell KL, Dias DP, Moritz M, Agard D, Stults JT, Stearns T. GCP5 and GCP6: two new members of the human γ-tubulin complex. Mol Biol Cell. 2001;12:3340–3352. doi: 10.1091/mbc.12.11.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SM, Urbani L, Stearns T. The mammalian γ-tubulin complex contains homologues of the yeast spindle pole body components Spc97p and Spc98p. J Cell Biol. 1998;141:663–674. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E. Structural insights into microtubule function. Annu Rev Biochem. 2000;69:277–302. doi: 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- Oakley BR. Gamma-tubulin: the microtubule organizer? Trends Cell Biol. 1992;2:1–5. doi: 10.1016/0962-8924(92)90125-7. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Yoon Y, Jung MK. γ-Tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- Oakley CE, Oakley BR. Identification of γ-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergilus nidulans. Nature. 1989;338:662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- Oegema K, Wiese C, Martin OC, Milligan RA, Iwamatsu A, Mitchison TJ, Zheng Y. Characterization of two related Drosophila γ-tubulin complexes that differ in their ability to nucleate microtubules. J Cell Biol. 1999;144:721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluh JL, Nogales E, Oakley BR, McDonald K, Pidoux AL, Cande WZ. A mutation in γ-tubulin alters microtubule dynamics and organization and is synthetically lethal with the kinesin-like protein Pkl1p. Mol Biol Cell. 2000;11:1225–1239. doi: 10.1091/mbc.11.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett-Heaps JD. The evolution of the mitotic apparatus: an attempt at comparative ultrastructural cytology in dividing plant cells. Cytobios. 1969;3:257–280. [Google Scholar]
- Radcliffe P, Hirata D, Childs D, Vardy L, Toda T. Identification of novel temperature-sensitive lethal alleles in essential β-tubulin and nonessential α2-tubulin genes as fission yeast polarity mutants. Mol Biol Cell. 1998;9:1757–1771. doi: 10.1091/mbc.9.7.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio P, Rebollo E, Varmark H, Sunkel CE, González C. Organized microtubule arrays in γ-tubulin-depleted Drosophila spermatocytes. Curr Biol. 2001;11:1788–1793. doi: 10.1016/s0960-9822(01)00561-9. [DOI] [PubMed] [Google Scholar]
- Schiebel E. γ-Tubulin complexes: binding to the centrosome, regulation and microtubule nucleation. Curr Opin Cell Biol. 2000;12:113–118. doi: 10.1016/s0955-0674(99)00064-2. [DOI] [PubMed] [Google Scholar]
- Stearns T, Evans L, Kirschner M. γ-Tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- Stearns T, Kirschner M. In vitro reconstitution of centrosome assembly and function: the central role of γ-tubulin. Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- Tassin A-M, Celati C, Moudjou M, Bornens M. Characterization of the human homologue of the yeast Spc98p and its association with γ-tubulin. J Cell Biol. 1998;141:689–701. doi: 10.1083/jcb.141.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Adachi Y, Hiraoka Y, Yanagida M. Identification of the pleiotropic cell cycle gene NDA2 as one of two different α-tubulin genes in Schizosaccharomyces pombe. Cell. 1984;37:233–242. doi: 10.1016/0092-8674(84)90319-2. [DOI] [PubMed] [Google Scholar]
- Tran PT, Marsh L, Doye V, Inoue S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–412. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy L, Fujita A, Toda T. The γ-tubulin complex protein Alp4 provides a link between the metaphase checkpoint and cytokinesis in fission yeast. Genes Cells. 2002;7:365–373. doi: 10.1046/j.1365-2443.2002.00530.x. [DOI] [PubMed] [Google Scholar]
- Vardy L, Toda T. The fission yeast γ-tubulin complex is required in G1 phase and is a component of the spindle-assembly checkpoint. EMBO J. 2000;19:6098–6111. doi: 10.1093/emboj/19.22.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Zheng Y. A new function for the γ-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol. 2000;2:358–364. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- Wood V, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Zhang L, Keating TJ, Wilde A, Borisy GG, Zheng Y. The role of Xgrip210 in γ-tubulin ring complex assembly and centrosome recruitment. J Cell Biol. 2000;151:1525–1536. doi: 10.1083/jcb.151.7.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Jung MK, Oakley BR. γ-Tubulin is present in Drosophila melanogaster and Homo sapiens and is associated with the centrosome. Cell. 1991;65:817–823. doi: 10.1016/0092-8674(91)90389-g. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]