Abstract
The Listeria model system has been essential for the identification and characterization of key regulators of the actin cytoskeleton such as the Arp2/3 complex and Ena/vasodilator-stimulated phosphoprotein (VASP) proteins. Although the role of Ena/VASP proteins in Listeria motility has been extensively studied, little is known about the contributions of their domains and phosphorylation state to bacterial motility. To address these issues, we have generated a panel of Ena/VASP mutants and, upon expression in Ena/VASP-deficient cells, evaluated their contribution to Ena/VASP function in Listeria motility. The proline-rich region, the putative G-actin binding site, and the Ser/Thr phosphorylation of Ena/VASP proteins are all required for efficient Listeria motility. Surprisingly, the interaction of Ena/VASP proteins with F-actin and their potential ability to form multimers are both dispensable for their involvement in this process. Our data suggest that Ena/VASP proteins contribute to Listeria motility by regulating both the nucleation and elongation of actin filaments at the bacterial surface.
INTRODUCTION
A crucial step in the life cycle of intracellular pathogens such as the Gram positive bacterium Listeria monocytogenes is their ability to recruit components from the host actin cytoskeleton to their surface. These components are then rearranged into phase-dense actin comet tails that are required for the intracellular motility of these parasites and confer on them the ability to directly invade neighboring cells. Because these pathogens use key cytoskeletal components that are essential for actin-based processes such as cell motility, they have inadvertently provided us with a powerful model system to study the molecular mechanisms that control the dynamics of the actin cytoskeleton (Cossart and Bierne, 2001; Frischknecht and Way, 2001).
L. monocytogenes (simply referred to as Listeria in the following sections of the text) subvert the host cell actin cytoskeleton through the expression of a single virulence factor, the ActA protein (Domann et al., 1992; Kocks et al., 1992). ActA harbors three major regions that are required for its interaction with cytoskeletal proteins: an amino-terminal actin monomer-binding site, an adjacent positively charged motif, and a central proline-rich region. The positively charged motif binds to, and activates the Arp2/3 complex, a cytoskeletal component that can nucleate actin filaments and is essential for bacterial motility (May et al., 1999; Pistor et al., 2000; Skoble et al., 2000, 2001; Zalevsky et al., 2001). The G-actin–binding site is not required for intracellular bacterial motility, although it plays a role in the Arp2/3-mediated actin filament nucleation in vitro (Pistor et al., 2000; Skoble et al., 2000, 2001). The central proline-rich domain, which includes three to four copies of the E/DFPPPPXD/E motif (Pistor et al., 1995; Smith et al., 1996; Niebuhr et al., 1997), binds to proteins of the Ena/VASP family (Pistor et al., 1995; Smith et al., 1996; Niebuhr et al., 1997; Machner et al., 2001), which includes the mammalian proteins vasodilator-stimulated phosphoprotein (VASP), mammalian Enabled (Mena), Ena-VASP-like (EVL), and the Drosophila protein Ena (Gertler et al., 1990, 1996; Halbrugge et al., 1990).
Several lines of evidence support the notion that Ena/VASP proteins are key regulators of the dynamics of the actin cytoskeleton. They associate with the surface of motile Listeria in an asymmetric manner (Chakraborty et al., 1995; Gertler et al., 1996) and are necessary for efficient Listeria motility in both infected cells and cell-free extracts (Smith et al., 1996; Niebuhr et al., 1997; Laurent et al., 1999; Loisel et al., 1999). Ena/VASP proteins localize at subcellular regions where remodeling of the actin cytoskeleton takes place, such as the front of spreading lamellipodia in motile cells (Rottner et al., 1999), tips of growth cone filopodia, focal adhesions, and epithelial cell-cell junctions (Reinhard et al., 1992; Gertler et al., 1996; Lanier et al., 1999; Vasioukhin et al., 2000). In hematopoietic systems, Ena/VASP proteins localize at the immunological synapse in Jurkat T cells and at phagocytic cups during Fcγ receptor-mediated phagocytosis (Krause et al., 2000; Castellano et al., 2001; Coppolino et al., 2001), where they colocalize with the Ena/VASP-binding protein Fyb/SLAP (Krause et al., 2000; Coppolino et al., 2001). In both systems, the displacement of Ena/VASP proteins from these sites inhibits the remodeling of the actin cytoskeleton that accompanies both the formation of immunological synapses and phagocytic cups, suggesting that they are essential for these processes (Krause et al., 2000; Coppolino et al., 2001). Finally, experiments with Ena/VASP-deficient fibroblasts and Rat2 cells in which Ena/VASP proteins were neutralized indicate that these proteins negatively influence random cell motility (Bear et al., 2000). In addition, in vitro they stimulate actin polymerization by shortening the lag phase of actin filament formation (Laurent et al., 1999; Harbeck et al. 2000; Lambrechts et al., 2000).
Ena/VASP proteins are characterized by a common tripartite structure. Their N-terminal region, the EVH1 domain, interacts with the motif E/DFPPPPXD/E, which is present in ActA and in the cytoskeletal proteins vinculin, zyxin, palladin, and Fyb/SLAP (Brindle et al., 1996; Niebuhr et al., 1997; Carl et al., 1999; Drees et al., 2000; Krause et al., 2000; Mykkanen et al., 2001). The central domain of the Ena/VASP proteins harbors a proline-rich region that binds to profilin and, in addition, to SH3 and WW domains (Reinhard et al., 1995; Gertler et al., 1996; Ermekova et al., 1997). The C terminus of Ena/VASP proteins binds to F-actin in vitro and is thought to mediate the multimerization of these proteins (Bachmann et al., 1999).
The EVH1 domain is required for targeting Ena/VASP proteins to Listeria surface as well as to focal adhesions (Gertler et al., 1996; Niebuhr et al., 1997; Carl et al., 1999). In contrast, little is known about the functions of the other domains and whether the phosphorylation state of Ena/VASP proteins plays a role in bacterial motility. To address these points, we have generated several Mena and VASP mutants that either lack one of these domains or carry mutated phosphorylation sites. We expressed these mutants in Ena/VASP-deficient cells and analyzed their contribution to Listeria motility. In a parallel study, the ability of Mena mutants to rescue normal motile properties of this cell line was also evaluated (Loureiro et al., 2002). Our results clearly indicate that the interaction of Ena/VASP proteins with F-actin and their potential ability to form multimers are both dispensable for their function in actin-based Listeria movement, whereas the proline-rich region, the putative G-actin binding site, and the Ser/Thr phosphorylation of Ena/VASP proteins are required for efficient Listeria motility.
MATERIALS AND METHODS
Cloning of VASP, Mena, and Profilin Constructs
The cloning of all Mena constructs is described in Loureiro et al. (2002). Enhanced green fluorescent protein (EGFP)-tagged full-length VASP (Carl et al., 1999) was cloned into the pMSCV vector after introducing EcoRI and ClaI restriction sites by polymerase chain reaction (PCR) by using the following primers: forward, CGGAATTCGCCACCATGGTGAGCAAGGGC; and reverse, GCATCGATTCAGGGAGAACCCCGCTTCCTCAG.
Mutagenesis of the VASP phosphorylation sites S157, S239, and T278 was done using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The following primers were used (mutated codons are underlined): pMSCV+EGFP-VASP AST (S157A), forward, GCACATAGAGCGCCGGGTCGCCAATGCAGGAGGC; and reverse, GCCTCCTGCATTGGCGACCCGGCGCTCTATGTGC; pMSCV+EGFP-VASP DST (S157D), forward, CATAGAGCGCCGGGTCGACAATGCAGGAGGCC; and reverse, GGCCTCCTGCATTGTCGACCCGGCGCTCTATG; pMSCV+EGFP-VASP SAT (S239A), forward, CTCAGGAAAGTCGCCAAGCAGGAGGAGGCC; and reverse, GGCCTCCTCCTGCTTGGCGACTTTCCTGAG; pMSCV+EGFP-VASP SDT (S239D), forward, CTCAGGAAAGTCGACAAGCAGGAGGAGGCC; and reverse, GGCCTCCTCCTGCTTGTCGACTTTCCTGAG; pMSCV+EGFP-VASP SSA (T278A), forward, GGAGAAGGAAAGCCGCGCAAGTTGGGGAGAAAAC; and reverse, GTTTTCTCCCCAACTTGCGCGGCTTTCCTTCTCC; and pMSCV+EGFP-VASP SSD (T278D), forward, GGAGAAGGAAAGCCGACCAAGTTGGGGAGAAA-AC; and reverse, GTTTTCTCCCCAACTTGGTCGGCTTTCCTTCTCC.
EGFP-VASP constructs harboring mutations of both S157 and S239 were generated as follows. EGFP-VASP constructs mutated at S157 or S239 were excised from the pMSCV vector, digested with PstI, and recloned into the pMSCV vector to obtain the desired double mutations.
The triple phosphorylation mutants S157A/S239A/T278A and S157D/S239D/T278D were generated from the corresponding double phosphorylation mutants by site-directed mutagenesis of T278.
Generation of VASP deletion mutants was done using the overlap-extension PCR method using the following primers. First PCR reaction (internal primers; parts of the primers that do not hybridize are underlined): VASP ΔGP5 (Δ162–186), forward, CTCCAATGCAGGAGGCGGTTTGCCCCCTTCG; and reverse, CGAAGGG-GGCAAACCGCCTCCTGCATTGGAG; VASP ΔPRR (Δ 118–122, 162–186, 204–209), forward, GTTGGAAGGAGGTGGGGC-ACTTCCCACCTGG; and reverse, CCAGGTGGGAAGTGCCC-CACCTCCTTCCAAC; forward, GGAGCAGGGGGAGGACTC-CCGGCAGCACAG; and reverse, CTGTGCTGCCGGGAGTCCT-CCCCCTGCTCC (to generate this mutant we used VASP ΔGP5 as the template); VASP ΔFAB (Δ259–277), forward, GAGAGTGGTCGAAGCACGCAAGTTGGGGAG; and reverse, CTCCCCAACTTGCGTGCTTCGACCACTCTC; and VASP ΔCoco (Δ352–373), forward, GTGAAACAGGAGCTTCTGAGGAAGCGGGG; and reverse, CCCCGCT-TCCTCAGAAGCTCCTGTTTCAC.
First PCR reaction (external primers; used for all deletion mutants) was as follows: forward, GCTGTACAAGTCCGGCCGGACTCAGATCTC; and reverse, GTGGGGTCTTTCATTCCCCCCTTTTTCTGG.
Second PCR reaction was as follows: forward, GCTCAAGCTTAGCAGCCATGAGCGAGACGG; and reverse, CTAAATAAAA-TCTTTTATTTTATCGATTCAGG.
All VASP deletion mutants were cloned into the pMSCV vector by using HindIII and ClaI. The correct molecular size of all mutants was verified by Western blotting after staining with a monoclonal antibody against green fluorescent protein (GFP).
Cyan fluorescent protein (CFP)-tagged wild-type VASP and VASP-ΔPRR and Profilin II-yellow fluorescent protein (YFP) were generated as follows. Enhanced cyan fluorescent protein (ECFP) was amplified using the following primers: forward, CGGAATTCACCATGGTGAGCAAGGGCGAGG; and reverse, TTCGAAGCTTTGAGCTCGAGATCTGAGTCCG by using pECFP-C1 (CLONTECH, Palo Alto, CA) as the template. The PCR product was then digested with EcoRI and HindIII and cloned into the same restriction sites of pMSCV+EGFP-VASP and pMSCV+EGFP-VASP ΔPRR.
To generate profilin II-YFP, we first amplified profilin II-GFP by using the following primers: forward, ACGCGGCCGCCCTTCCATGGCCGGTTGGCAGAGCTACG; and reverse, CGCAAGCTTTTACTTGTACAGCTCGTCCATGCC and profilin II-EGFP as the template. The PCR product was then digested with NotI and HindIII and cloned into the same restriction sites of pML2X. pML2X+profilin II-EYFP was obtained after amplification of EYFP (forward primer, CCGGGATCCACCGGTCGCCACCATGGTGAGC; and reverse primer, CGCGGAAGCTTTACTTGTACAGCTCGTCCATGC; template pEYFP-N1; CLONTECH), digestion with BamHI and HindIII, and cloning into the same restriction sites of pML2X+profilin II-EGFP.
Bacterial Culture
The wild-type weakly hemolytic L. monocytogenes strain EGD (serotype 1/2) and its isogenic Listeria mutants ActA5 and ActA12 (Domann et al. 1992; Niebuhr et al., 1997; Pistor et al., 2000) were grown in brain heart infusion broth (Difco, Detroit, MI) at 37°C with agitation.
Cell Culture and Infection
MVD7 cells and G7 mouse fibroblasts were grown in DMEM supplemented with 15% fetal calf serum, 2 mM l-glutamine, and 50 U/ml mouse interferon-γ at 32°C in the presence of 5% CO2. All media and supplements were obtained from Invitrogen (Carlsbad, CA). Infection of MVD7 cells with L. monocytogenes was done according to Sechi et al. (1997) by using a final bacterial concentration of 109–1010 colony-forming units/ml and an incubation time for bacterial entry of 90 min at 37°C.
Cell Transfection and Sorting
MVD7 cells were transfected using a retroviral transfection system. Briefly, pMSCV plasmids harboring the EGFP-VASP constructs and the helper plasmid pCL-Eco (Imgenex, San Diego, CA) were introduced into BOSC23 cells by using a calcium phosphate transfection procedure. Two days later, the cell medium containing the retroviral particles released by the BOSC23 cells was collected and used to transfect MVD7 cells. Afterward, MVD7 cells were sorted according to low, medium, and high levels of EGFP expression using a fluorescence-activated cell sorting (FACS) sorter (MoFlo; Cytomation, Ft. Collins, CO). After cell thawing, the correct expression levels of all GFP-tagged constructs were confirmed using an FACS Calibur device (BD Biosciences, San Jose, CA).
Immunofluorescence Microscopy
Four hours after the beginning of the infection, cells were fixed with 4% paraformaldehyde in cytoskeleton buffer (10 mM MES, 150 mM NaCl, 5 mM EGTA, 5 mM glucose, and 5 mM MgCl2, pH 6.1) for 20 min at room temperature and then extracted with 0.1% Triton X-100 in cytoskeleton buffer for 1 min at room temperature. Bacteria were labeled with the polyclonal antibody K52 followed by Alexa 488-conjugated goat anti-rabbit IgG (Dianova, Hamburg, Germany). The actin cytoskeleton was labeled with Alexa 594-conjugated phalloidin (Molecular Probes, Eugene, OR). Coverslips were mounted in Prolong (Molecular Probes).
Fluorescence Video Microscopy
For fluorescence video microscopy, cells were plated onto 40-mm round coverslips. Four hours after beginning the infection, coverslips carrying infected cells were mounted in a Focht Chamber System (FCS2; Bioptechs, Butler, PA). An objective heater (Bioptechs) was used to eliminate the temperature gradient between chamber and objective. The cells were observed by phase contrast or epifluorescence with an Axiovert 135 TV microscope (Carl Zeiss, Thornwood, NY) equipped with a Plan-Apochromat 100×/1.40 numerical aperture oil immersion objective in combination with 1.6× or 2.5× optovar optics. Images were recorded with a cooled, back-illuminated charge-coupled device camera (TE/CCD-1000 TKB; Princeton Instruments, Trenton, NJ) driven by IPLab Spectrum software (Scanalytics, Fairfax, VA). Digital handling of the images was done using IPLab Spectrum and Adobe Photoshop 5.0 (Adobe Systems, Mountain View, CA).
Analysis of Bacterial Speed
All motile bacteria within a single cell were scored according to the following criteria: 1) they did not interact with other motile or stationary bacteria; 2) they never stopped or started to move during the observation period; and 3) they did not move within cellular extensions (pseudopodia). Paths of motile bacteria (observed in at least 20 infected cells; 3 independent experiments) were generated after marking the bacterial poles proximal to the actin tails using the Dynamic Imaging Analysis system (Solltech, Oakdale, IA). To smooth out sudden speed oscillations, the instantaneous speed of the bacteria was calculated according to the central difference method. Analysis of the bacterial speed was done using MiniTab 10.5 (MiniTab, State College, PA) and DeltaGraph 3.5 (Delta Point, SSPS Inc., Chicago, IL). Because the measured values of Listeria speed were not normally distributed as determined using the Anderson-Darling test, we analyzed differences in bacterial speed using the Mann-Whitney nonparametric U test and rejected the null hypothesis (the two groups have the same median value, i.e., they are not different) when p < 0.05.
RESULTS
Motility of Listeria monocytogenes Is Impaired in MVD7 Fibroblasts
An essential prerequisite for studying the function of Ena/VASP protein domains in Listeria motility is the availability of a cell line that does not express any of the known Ena/VASP proteins to avoid any interference with the function of the ectopically expressed Mena and VASP mutants. We have recently isolated one clonal cell line (MVD7 fibroblasts) from mena/vasp-null mouse embryos, which does not express detectable levels of EVL (Bear et al., 2000).
To test whether MVD7 cells are suitable for studying the intracellular motility of Listeria, we analyzed these cells after infection with the wild-type strain EGD of L. monocytogenes. Immunofluorescence microscopy revealed that, in all cells analyzed, Listeria induced the formation of very short actin comet tails (Figure 1A). To verify whether the formation of such short tails was due to the lack of Ena/VASP proteins, we infected a fibroblast cell line (G7 cells; Lommel et al., 2001), which, like MVD7 fibroblasts, was immortalized with a temperature-sensitive version of the simian virus 40 large-T antigen and grown under the same culture conditions (see MATERIALS AND METHODS). In G7 mouse fibroblasts, which express Ena/VASP proteins (our unpublished data), wild-type Listeria were associated with normal actin tails (Figure 1C). The same result was obtained using cell lines such as PtK2 and HeLa (our unpublished data). Overall, these observations suggest that the formation of short tails in MVD7 is due to the deficiency in Ena/VASP proteins.
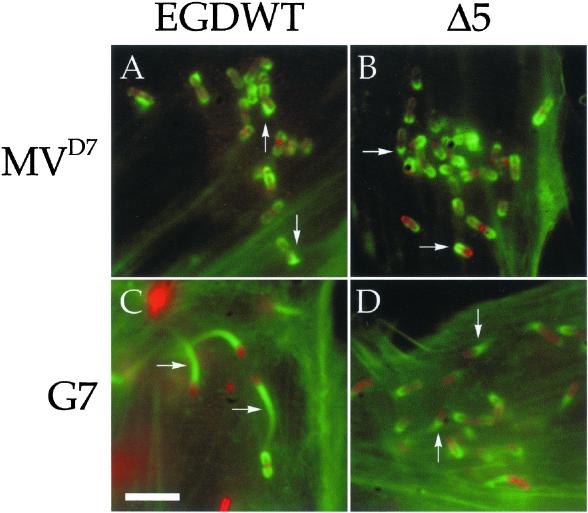
Figure 1.
Motility of L. monocytogenes is impaired in MVD7 cells. MVD7 fibroblasts (A and B) and G7 mouse fibroblasts (C and D) were infected with wild-type Listeria (A and C) or the Listeria mutant Δ5 (B and D), fixed, and stained with antibodies against the bacteria (red) and fluorescent phalloidin (green). In G7 mouse fibroblasts wild-type Listeria induced the formation of long actin tails (arrows in C). In contrast, the actin tails induced by these bacteria in MVD7 cells were much shorter (arrows in A) and closely resembled those induced by the Listeria mutant Δ5 in both cell types (arrows in B and D). Bar, 2 μm.
To corroborate this result, we infected both MVD7 cells and G7 cells with the Listeria mutant Δ5. This mutant expresses on its surface a mutated version of ActA that lacks the central proline-rich region and is, as a consequence, unable to interact with Ena/VASP proteins and therefore leads to the formation of short actin tails (Niebuhr et al., 1997). As expected, Listeria Δ5 was associated with short actin tails in both normal G7 fibroblasts and Ena/VASP-deficient fibroblasts (Figure 1, B and D).
The visual impression that the intracellular motility of Listeria is impaired in MVD7 cells was further confirmed by video microscopy. In particular, the average speed of wild-type Listeria in MVD7 cells was comparable with that of the Listeria mutant Δ5 in the same cell line (Figure 2D) and was 5–10 times lower than their average speed measured in G7 fibroblasts and other cell lines (our unpublished data; Niebuhr et al., 1997). Thus, MVD7 cells represent a suitable system for analyzing the contribution of Ena/VASP protein domains to Listeria motility.
Figure 2.
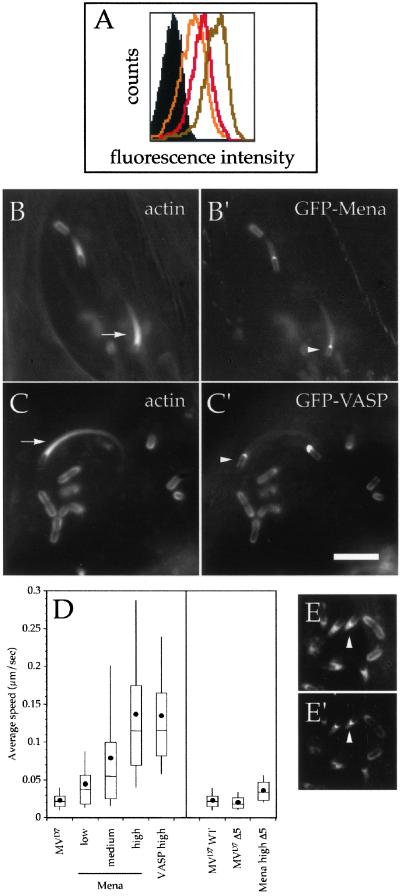
(A) FACS analysis of MVD7 cells expressing low (yellow line), medium (red line), and high (light brown line) levels of GFP-tagged Mena and VASP constructs. The black-filled area indicates the background fluorescence of untransfected MVD7 cells. (B–C′) The expression of wild-type GFP-Mena and GFP-VASP rescues Listeria motility in MVD7 cells in a concentration-dependent manner. MVD7 cells expressing high levels of wild-type GFP-Mena or GFP-VASP were infected with Listeria, fixed, and stained with fluorescent phalloidin. In these cells, Listeria recruited GFP-Mena and GFP-VASP at their surface (arrowheads in B′ and C′) and induced the formation of normal actin tails (arrows in B and C; compare with Figure 1). (D) Box and whiskers plots of bacterial speed. Dot indicates the mean, line in the middle of the box indicates the median, top of the box indicates the 75th quartile, whereas the bottom of the box indicates the 25th quartile, and whiskers indicates the 10th and 90th percentiles, respectively. (E–E′) GFP-tagged wild-type Mena and VASP localize at the actin tails induced by the Listeria mutant Δ5. MVD7 cells expressing high levels of wild-type GFP-Mena were infected with Listeria Δ5, fixed, and stained with fluorescent phalloidin. This Listeria mutant typically induced the formation of short actin tails (arrowhead in E), which were robustly stained with GFP-Mena (arrowhead in E′). Bar (for B–C′ and E–E′), 2 μm.
Efficient Motility of Listeria in MVD7 Cells Is Rescued by Full-Length Ena/VASP Proteins in a Concentration-dependent Manner
Because the impaired movement of Listeria in MVD7 cells seems to be due to the lack of Ena/VASP proteins, we reasoned that normal bacterial motility could be rescued upon expression of Ena/VASP proteins. We therefore infected MVD7 cells with a retrovirus that drives the expression of GFP-tagged full-length Mena or VASP and sorted them by FACS into three populations expressing low, medium, and high levels of the fusion proteins according to the intensity of GFP fluorescence signal (Figure 2A). The relative expression levels of GFP-Mena and GFP-VASP corresponded to 35, 71, and 100% for the low, medium, and high populations, respectively, as calculated after setting the average intensity of the high population to 100%.
These fusion proteins properly localized to subcellular regions in MVD7 cells (our unpublished data; Loureiro et al., 2002) and at the surface of both nonmotile and motile bacteria (Figure 2, B′ and C′). Moreover, in MVD7 fibroblasts expressing high levels of GFP-Mena or GFP-VASP Listeria induced the formation of normal actin tails that were indistinguishable from those induced by these bacteria in G7 cells (Figure 2, B and C; compare with Figure 1C).
We then analyzed the bacterial movement in MVD7 cells expressing low-to-high levels of the GFP fusion proteins by using video microscopy. As shown in Figure 2D, the enhancement of Listeria motility directly correlated with the increase in the cellular levels of the ectopically expressed Ena/VASP proteins (Figure 2D). The expression of high levels of GFP-Mena or GFP-VASP rescued Listeria motility equally well (Figure 2D), suggesting that Mena and VASP are interchangeable in this process. Moreover, the average speed of Listeria in MVD7 fibroblasts expressing high levels of GFP-tagged Mena and VASP was similar to that measured in G7 fibroblasts (our unpublished data).
The rescue of Listeria motility in MVD7 cells clearly depends on the expression of Ena/VASP proteins as indicated by the observation that the speed of the Listeria mutant Δ5 in MVD7 cells expressing high levels of GFP-Mena was comparable with its speed measured in G7 cells (Figure 2D; our unpublished data). However, despite the fact that this Listeria mutant is unable to recruit Ena/VASP proteins at its surface, its speed in MVD7 cells expressing high levels of GFP-Mena was significantly higher than the speed of wild-type Listeria in the parental Ena/VASP-deficient cells (Mann-Whitney U test; p < 0.05; wild type, n = 31; Δ5, n = 28). Because GFP-tagged Ena/VASP proteins localized at the short actin tails induced by the Listeria mutant Δ5 (Figure 2, E and E′), it may be that cytoplasmic Ena/VASP proteins can influence bacterial motility via an EVH1-independent recruitment to actin tails, perhaps mediated by their ability to interact with actin filaments.
Generation of Ena/VASP Mutants and Their Expression in MVD7 Cells
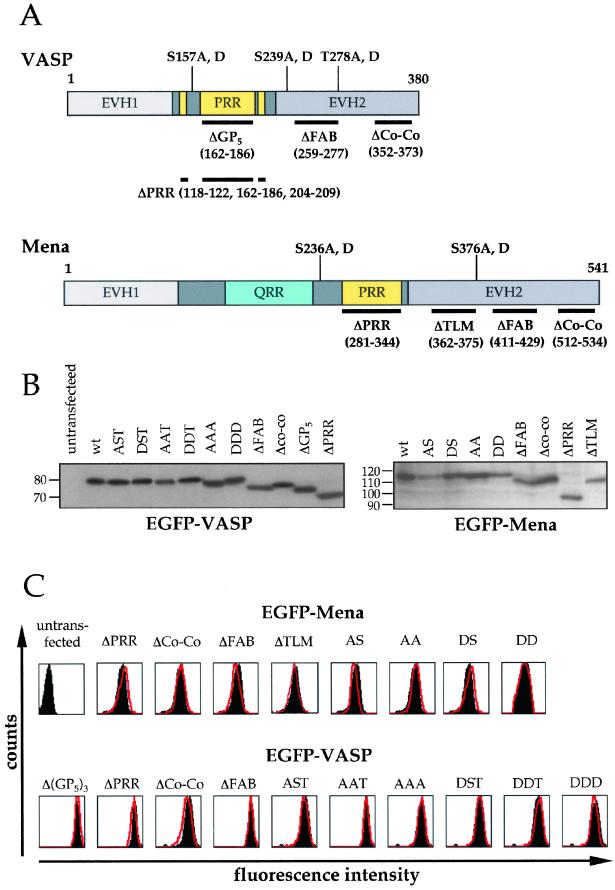
Next, we generated a panel of GFP-tagged fusion proteins in which various domains and phosphorylation sites of Ena/VASP proteins were deleted or mutated, respectively (Figure 3A). The introduction of the GFP moiety at the NH2 terminus in all these fusion proteins did not affect the ability of the EVH1 domain to interact with ActA as indicated by their proper localization at the Listeria surface (Figure 2, B′ and C′; Carl et al., 1999). In addition, Western blot analysis showed that all GFP-tagged constructs migrated at the expected molecular size and that they were not degraded when expressed in MVD7 cells (Figure 3B), suggesting that neither the presence of GFP nor the deletions or point mutations grossly affected the protein stability. We cannot exclude, however, that these deletions or point mutations may affect the overall folding of the proteins.
Figure 3.
(A) Schematic diagram of Mena and VASP constructs. Deletions (thick lines) and phosphorylation sites are shown at the bottom and top of Mena and VASP cartoons, respectively. EVH1, Ena-VASP homology 1; PRR, proline-rich region; QRR, glutamine-rich region; EVH2, Ena-VASP homology 2. (B) Western blot analysis of GFP-tagged Mena and VASP fusion proteins. Cell lysates of MVD7 cells expressing high levels of all Mena, and VASP constructs were resolved by SDS-PAGE, blotted, and probed with an anti-GFP monoclonal antibody. Numbers on the left side of each blot represent molecular weight markers in kilodaltons. (C) Comparison between the expression levels of GFP-tagged wild-type Mena and VASP and their mutated fusion proteins. MVD7 cells expressing high levels of wild-type GFP-Mena and GFP-VASP (black-filled areas) were compared with MVD7 cells expressing high levels of each of the GFP-tagged Mena and VASP constructs (red lines) by FACS. The overlapping between the black-filled areas and the red lines in each FACS scan indicates that all cellular populations are equivalent with respect to the expression levels of all Ena/VASP constructs.
As noted above, normal intracellular Listeria motility can be rescued by expressing high levels of Ena/VASP proteins. We therefore evaluated the influence of these Ena/VASP mutants on Listeria motility in MVD7 cells that expressed high levels of these mutant proteins. To this end, MVD7 fibroblasts transfected with Ena/VASP mutants were sorted by FACS by using MVD7 cells expressing high levels of the corresponding wild-type Ena/VASP protein as reference. As shown in Figure 3C, the overlap between the different pairs of FACS scans clearly indicated that the expression levels of all Ena/VASP mutants were similar to those of the nonmutated counterparts.
Proline-rich Region of Ena/VASP Proteins Is Essential for Efficient Listeria Motility
A combination of biochemical, genetic, and cell biological approaches suggests that the interaction between the proline-rich region of Ena/VASP proteins and profilin serves to recruit polymerization-competent actin monomers to sites of actin assembly (Reinhard et al., 1995; Smith et al., 1996; Lanier et al., 1999; Geese et al., 2000).
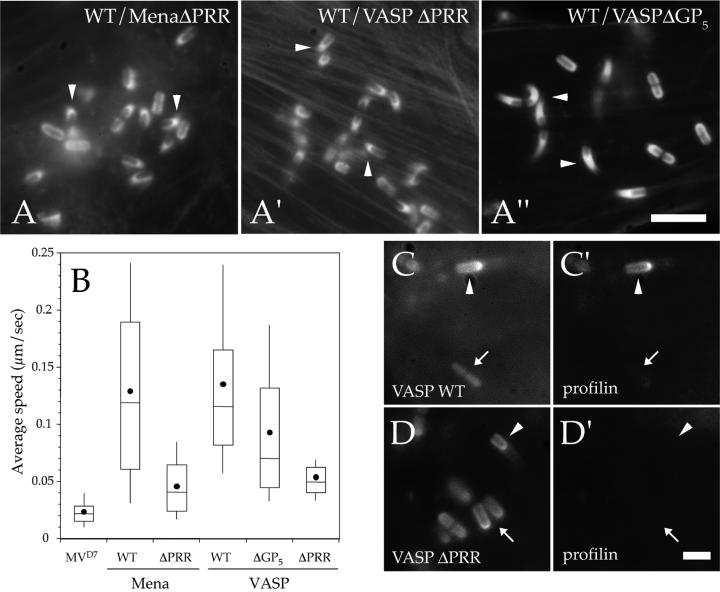
To study the contribution of this region to Listeria motility, we expressed GFP-Mena ΔPRR (for proline-rich region), GFP-VASP ΔGP5, and GFP-VASP ΔPRR in MVD7 cells. The GFP-VASP ΔGP5 construct lacks the central GP5 motifs that has been shown to bind to profilin (Kang et al., 1997), whereas the GFP-VASP ΔPRR construct includes two additional small deletions (also characterized by a GP5 motif) flaking the triple GP5 sequence (Figure 3A). As appraised by fluorescence microscopy, wild-type Listeria were associated with short actin comet tails that were morphologically similar to those induced by these bacteria in the parental MVD7 cells (Figure 4, A–A"). Moreover, video microscopy analysis showed that the speed of Listeria was greatly reduced in cells expressing GFP-Mena ΔPRR and GFP-VASP ΔPRR compared with the full-length counterparts (Figure 4B). The deletion of the proline-rich stretches in the GP5 motif in VASP also resulted in an intermediate but still significant reduction of bacterial motility (Mann-Whitney U test, p < 0.05; GFP-VASP, n = 128; GFP-VASP ΔPRR, n = 111) that was characterized by the formation of actin tails slightly longer than those induced by wild-type Listeria in cells expressing Ena/VASP ΔPRR mutants (Figure 4, A" and B).
Figure 4.
Proline-rich region of Ena/VASP proteins is essential for efficient Listeria motility. (A–A") MVD7 cells expressing high levels of GFP-Mena ΔPRR, GFP-VASP ΔGP5, or GFP-VASP ΔPRR were infected with wild-type Listeria, fixed, and stained with fluorescent phalloidin. In cell lines expressing GFP-Mena ΔPRR and GFP-VASP ΔPRR Listeria induced the formation of very short actin tails (arrowheads in A and A′), which were morphologically similar to those induced by this bacterium in the parental MVD7 cells (compare with Figure 1A), whereas Listeria actin tails were significantly longer in cells expressing GFP-VASP ΔGP5 (arrowheads in A"). Bar, 2 μm. (A) Box and whiskers plots of bacterial speed. (C–D′) Deletion of the proline-rich region of Mena and VASP inhibits the targeting of profilin to the Listeria surface. MVD7 cells were transfected with CFP-tagged wild-type VASP or VASP ΔPRR and profilin II-YFP and then infected with wild-type Listeria. In cells expressing CFP-VASP, profilin colocalized with VASP at the surface of motile bacteria (arrowheads in C and C′) but not stationary ones (arrows in C and C′). In contrast, in cells expressing CFP-VASP ΔPRR, profilin was absent from the surface of both stationary (arrow in D′) and motile Listeria (arrowhead in D′). Bar, 1 μm.
Because the effect of GFP-Mena ΔPRR and GFP-VASP ΔPRR on Listeria motility may be due to their inability to bind profilin, we cotransfected MVD7 cells with CFP-tagged wild-type VASP or VASP ΔPRR and profilin II-YFP, infected them with Listeria and analyzed their localization at the bacterial surface by fluorescence microscopy. As expected, both CFP-tagged VASP constructs properly localized at the Listeria surface (Figure 4, C and D). In agreement with previous findings (Geese et al., 2000), profilin-YFP localized at the bacterial surface of motile, but not stationary Listeria in cells expressing full-length CFP-VASP (Figure 4, C and C′), whereas in cells expressing CFP-VASP ΔPRR, profilin-YFP could be detected neither around motile nor stationary bacteria (Figure 4, D and D′).
Overall, these results clearly indicate that the proline-rich region of Ena/VASP proteins is essential for efficient Listeria motility and that the deletion of the PRR correlated with a lack of profilin recruitment at the bacterial surface.
Deletion of Thymosin β4-like-Motif of Mena Reduces Listeria Motility
The deletion of the proline-rich region in Ena/VASP proteins did not cause a reduction in speed to the values measured in the parental MVD7 cells (Figure 4B), suggesting that Ena/VASP proteins play additional roles in Listeria motility other than recruiting profilin–G-actin complexes at the bacterial surface and that other regions of Ena/VASP proteins contribute to Listeria motility. Therefore, we analyzed the contribution of three regions contained in the EVH2 domain to this process.
The amino-terminal part of the EVH2 domain of VASP and Mena harbors a motif that is similar to the G-actin-binding site KLKR found in thymosin β4 (Gertler et al., 1996; van Troys et al., 1996). Similar sequences are also found in the headpiece of the actin-binding proteins villin and dematin (van Troys et al., 1996). In MVD7 cells expressing GFP-Mena ΔTLM (for thymosinβ4-like-motif) Listeria moved significantly slower (Mann-Whitney U test, p < 0.05; GFP-Mena, n = 93; GFP-Mena ΔTLM, n = 73) than in cells expressing full-length GFP-Mena and induced the formation of short actin comet tails (Figure 5, A and B). Based on these results and on the observation that VASP rescues the ability of an ActA mutant, which lacks the G-actin–binding site, to support both the actin-nucleating activity of the Arp2/3 complex and the accumulation of actin at the Listeria surface (Skoble et al., 2001), we speculated that the TLM motif substitutes for the activity of the G-actin–binding site of ActA in these events.
Figure 5.
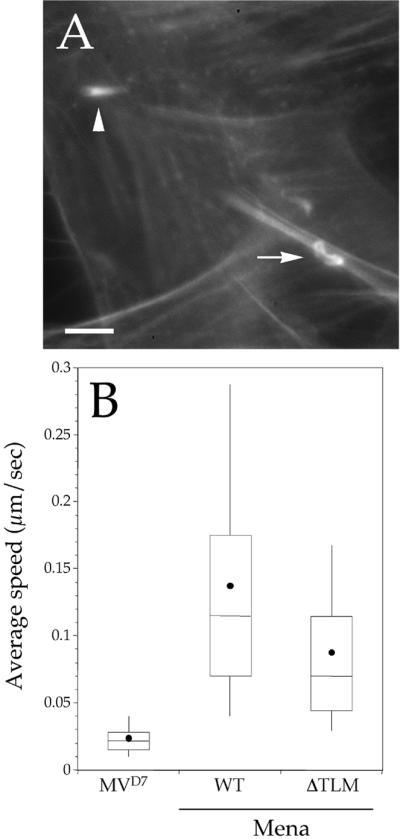
(A) Deletion of the thymosin β4-like motif of Mena causes a significant decrease of Listeria motility. MVD7 cells expressing high levels of GFP-Mena ΔTLM were infected with wild-type Listeria, fixed, and stained with fluorescent phalloidin. In these cells, Listeria induced the formation of short actin tails (arrowheads). Bar, 2 μm. (B) Box and whiskers plots of bacterial speed.
To test this hypothesis, we infected MVD7 cells and MVD7 cells expressing GFP-Mena or GFP-Mena ΔTLM with wild-type bacteria or with a Listeria mutant that expresses a mutated version of ActA that lacks the G-actin–binding site (Δ12; deletion spanning amino acids 68–109; Pistor et al., 2000). In all these cell lines, wild-type bacteria induced a normal accumulation of actin at their surface as judged after labeling with fluorescent phalloidin. In contrast, the ability of the Δ12 mutant to induce a normal actin accumulation at the bacterial surface was impaired only in MVD7 cells and in cells expressing GFP-Mena ΔTLM (our unpublished data). The number of Δ12 bacteria associated with deficient actin accumulation was higher than that of wild-type Listeria in MVD7 cells and in MVD7 cells expressing GFP-Mena ΔTLM (Table 1, compare with MVD7 cells expressing GFP-Mena). Because Ena/VASP proteins may rescue the ability of the Δ12 mutant to induce actin accumulation at its surface also via a PRR-dependent recruitment of G-actin, we infected with this mutant MVD7 cells expressing GFP-Mena or GFP-Mena ΔPRR. As shown in Table 1, the deficient actin accumulation at the surface of the Δ12 mutant was much less pronounced in MVD7 cells expressing GFP-Mena ΔPRR, indicating that the effect observed was mainly due to the deletion of the TLM motif. Thus, these results suggest that the TLM motif is implicated in the regulation of actin filament nucleation at the bacterial surface.
Table 1.
TLM motif contributes to the regulation of actin filament nucleation at the bacterial surface
| Wild type
|
Δ12
|
|
|---|---|---|
| % bacteria with deficient actin accumulation/(total bacteria) | % bacteria with deficient actin accumulation/(total bacteria) | |
| MVD7 | 38 (125) | 96 (91) |
| MVD7+ Mena WT | 9 (47) | 13 (52) |
| MVD7+ Mena ΔTLM | 11 (55) | 81 (48) |
| MVD7+ Mena ΔPRR | 9 (68) | 16 (67) |
Deletion of F-Actin–binding Site of Ena/VASP Proteins Enhances Listeria Motility
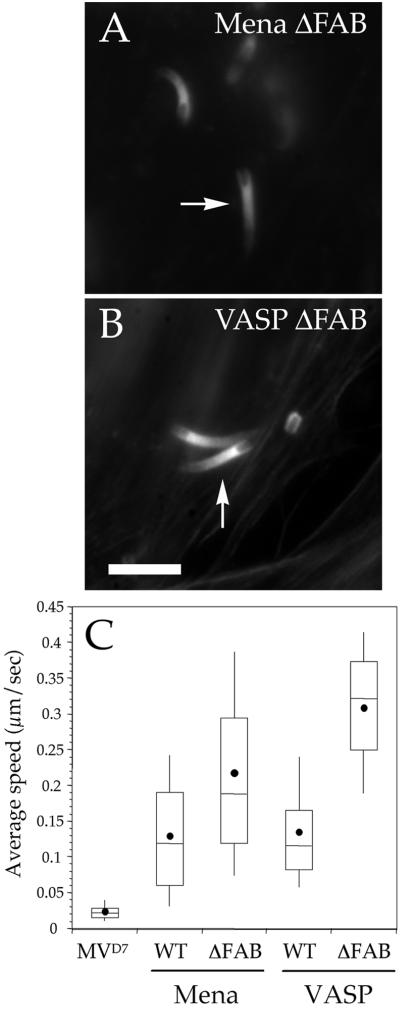
The EVH2 domain of Ena/VASP proteins has also been implicated in the interaction of this protein family with F-actin in vitro (Bachmann et al., 1999; Hüttelmaier et al., 1999; Harbeck et al., 2000). Moreover, Ena/VASP proteins stimulate actin polymerization by shortening the lag phase of actin filament formation, an effect that can be reversed by adding a peptide (corresponding to amino acids 261–283 of EVL) that has been shown to interact with F-actin in sedimentation assays (Laurent et al., 1999; Harbeck et al., 2000; Lambrechts et al., 2000). To test whether the binding of Ena/VASP proteins to F-actin is required for Listeria motility, we expressed GFP-Mena ΔFAB (for F-actin binding) or GFP-VASP ΔFAB in MVD7 cells. These fusion proteins did not cause morphological changes in actin comet tails, as appraised by fluorescence microscopy (Figure 6, A and B). Unexpectedly, the speed of Listeria in MVD7 cells expressing these fusion proteins was significantly higher than that of bacteria in MVD7 cells expressing full-length Ena/VASP proteins (Figure 6C; Mann-Whitney U test, p < 0.05; GFP-Mena, n = 93; GFP-Mena ΔFAB, n = 78; GFP-VASP, n = 128; GFP-VASP ΔFAB, n = 85), indicating that the interaction between Ena/VASP proteins and actin filaments is not required for intracellular Listeria motility.
Figure 6.
(A and B) Deletion of the F-actin–binding site of Mena and VASP increases Listeria motility. MVD7 cells expressing high levels of GFP-Mena ΔFAB (A) or GFP-VASP ΔFAB (B) were infected with wild-type Listeria, fixed, and stained with fluorescent phalloidin. In both cases, Listeria induced the formation of actin tails (arrows in A and B) that were not distinguishable from those induced by the same bacterium in MVD7 cells expressing wild-type Ena/VASP proteins (compare to Figure 2). Bar, 2 μm. (C). Box and whiskers plots of bacterial speed.
A Coiled-Coil Motif in EVH2 Domain of Ena/VASP Proteins Is Not Required for Listeria Motility
Various experimental approaches indicate that the most C-terminal 35 amino acids in the EVH2 domain, which is predicted to form coiled-coil structures, can mediate the formation of Ena/VASP multimers (Haffner et al., 1995; Ahern-Djamali et al., 1998; Bachmann et al., 1999; Carl et al., 1999). In the Listeria context, it has been hypothesized that the formation of Ena/VASP multimers at the bacterial surface increases the recruitment of profilin–G-actin complexes and, as a consequence, Listeria motility (Kang et al., 1997).
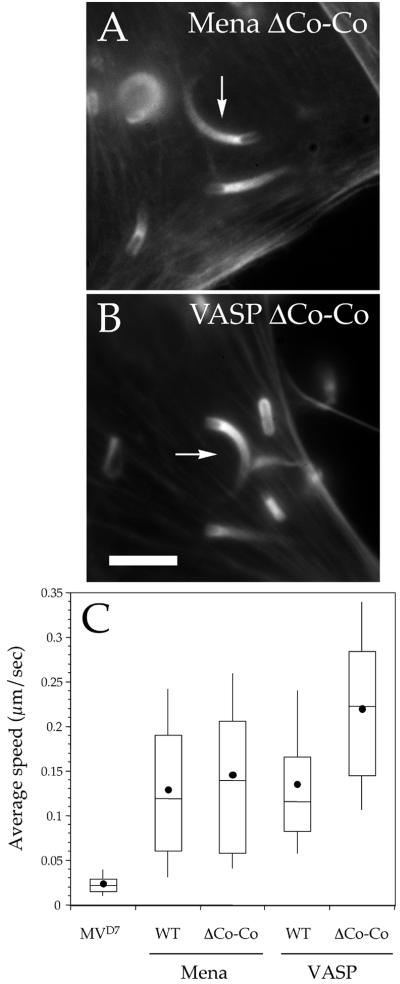
In MVD7 cells expressing GFP-Mena ΔCo-Co (for coiled-coil) and GFP-VASP ΔCo-Co Listeria induced the formation of normal actin comet tails (Figure 7, A and B). The visual impression that Listeria motility is not grossly altered in the presence of Ena/VASP ΔCo-Co proteins was confirmed by video microscopy analysis. The average speed of the bacteria in cells expressing GFP-Mena ΔCo-Co was not different from that measured in cells expressing full-length Mena (Figure 7C; Mann-Whitney U test, p = 0.24; GFP-Mena, n = 93; GFP-Mena ΔCo-Co, n = 90), whereas Listeria moved at a higher average speed in MVD7 cells expressing GFP-VASP ΔCo-Co (Figure 7B; Mann-Whitney U test, p < 0.05; GFP-VASP, n = 128; GFP-VASP ΔCo-Co, n = 85). Thus, the formation of Ena/VASP multimers is not required for Listeria motility.
Figure 7.
(A and B) Potential multimerization of Ena/VASP proteins is not required for Listeria motility. MVD7 cells expressing high levels of GFP-Mena ΔCo-Co (A) or GFP-VASP ΔCo-Co (B) were infected with wild-type Listeria, fixed, and stained with fluorescent phalloidin. In both cell lines, the bacteria induced the formation of actin tails (arrows in A and B) that were not distinguishable from those induced by the same bacterium in MVD7 cells expressing wild-type Ena/VASP proteins (compare to Figure 2). Bar, 2 μm. (B) Box and whiskers plots of bacterial speed.
Phosphorylation of Serine and Threonine Residues of Ena/VASP Proteins Increases Listeria Motility
VASP and Mena harbor three (S157, S239, and T278) and two (S236 and S376) phosphorylation sites, respectively, that can be phosphorylated in a cAMP- and cGMP-dependent manner (Halbrugge et al., 1990). In vitro, the phosphorylation state of Ena/VASP proteins influences their ability to interact with F-actin and some SH3-containing proteins (Laurent et al., 1999; Harbeck et al., 2000; Lambrechts et al., 2000).
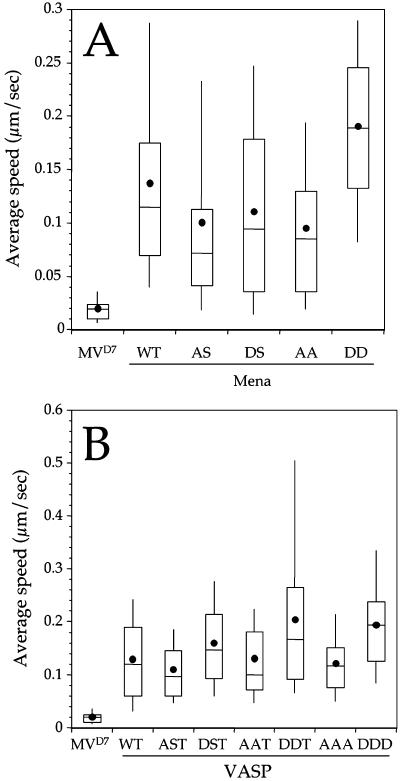
To assess whether the phosphorylation of Ena/VASP proteins plays a role in Listeria motility, we mutated all phosphorylation sites of VASP and Mena to alanine to block phosphorylation or to aspartic acid to mimic the constitutively phosphorylated forms of these proteins, respectively (Figure 3A). None of these phosphorylation mutants grossly affected the formation of actin comet tails as judged by fluorescence microscopy (our unpublished data). Compared with MVD7 cells expressing wild-type GFP-Mena, GFP-Mena AA caused a slight but significant reduction in Listeria motility, whereas the speed of the bacteria in MVD7 cells expressing GFP-Mena DD was significantly increased (Figure 8A; Mann-Whitney U test, p = 0.04 for WT vs. AA, p < 0.05 for WT vs. DD; GFP-Mena, n = 93; GFP-Mena, AA, n = 67; GFP-Mena DD, n = 72). The slight reduction in Listeria speed is mainly due to the first Ser → Ala mutation as indicated by the observation that the speed of Listeria in MVD7 cells expressing GFP-Mena AS is not different from that measured in cells transfected with GFP-Mena AA (Figure 8A; Mann-Whitney U test, p = 0.058 for AS vs. AA; GFP-Mena AS, n = 126; GFP-Mena, AA, n = 67). Moreover, the increase in bacterial speed caused by GFP-Mena DD seems to be mostly dependent on the second Ser → Asp mutation as suggested by observation that there is no significant difference between Listeria motility measured in GFP-Mena WT and GFP-Mena DS (Figure 8A; Mann-Whitney U test, p > 0.05 for WT vs. DS; GFP-Mena, n = 93; GFP-Mena DS, n = 69).
Figure 8.
(A and B) Phosphorylation of the serine and threonine residues of Mena (A) and VASP (B) increases Listeria motility. Box and whiskers plots of Listeria speed showing the influence of Mena and VASP phosphorylation state on bacterial motility.
The effect of similar mutations in VASP was slightly different. In particular, in MVD7 cells expressing GFP-VASP AAA the average bacterial speed was comparable with that measured in cells expressing wild-type GFP-VASP. Conversely, the expression of GFP-VASP DDD caused a significant enhancement of Listeria motility (Figure 8B; Mann-Whitney U test, p = 0.48 for WT vs. AAA, p < 0.05 for WT vs. DDD; GFP-VASP, n = 128; GFP-VASP AAA, n = 88; GFP-VASP DDD, n = 92). Because the first two Ser → Asp mutations mainly cause the enhancement of Listeria motility, these residues seems to be critical for this process (Figure 8B).
DISCUSSION
In this study we have characterized the contributions of four Ena/VASP protein domains to the intracellular motility of L. monocytogenes. In particular, we showed that the interaction of Ena/VASP proteins with F-actin and their potential ability to form multimers are both dispensable for their function in actin-based Listeria movement, whereas the proline-rich region, the putative G-actin binding site and the Ser/Thr phosphorylation of Ena/VASP proteins contribute to efficient Listeria motility.
Ena/VASP proteins were originally thought to regulate actin filament remodeling through their ability to interact with the G-actin–binding protein profilin (Reinhard et al., 1995; Gertler et al., 1996; Lambrechts et al., 2000). This notion is clearly supported by genetic studies, which suggest a physiological role for the interaction between Mena and profilin during the actin-based process of neurulation (Lanier et al., 1999). Moreover, the injection of the proline-rich region of VASP into Listeria- and Shigella-infected cells causes the arrest of bacterial movement (Zeile et al., 1996; Kang et al., 1997), whereas a similar VASP peptide favors the disassociation of profilin–G-actin complexes leading to the enhancement of nucleation and elongation of actin filaments, in vitro (Jonckheere et al., 1999). On the other hand, Ena/VASP proteins enhance Listeria motility in cell-free systems in absence of profilin (Loisel et al., 1999), and Ena/VASP-profilin interaction is not required for the function of this protein family in whole cell motility (Loureiro et al., 2002). Thus, although this study demonstrates that the interaction between Ena/VASP proteins and profilin at the Listeria surface is important for supporting the efficient bacterial motility, it is possible that the binding of Ena/VASP proteins to profilin is not required for, or plays a minor role in, the contribution of these proteins to other actin-based processes. Although none of the known proteins that contain SH3 and WW domains has been involved in Listeria motility, at present we cannot rule out that the reduced bacterial motility we observed in cells expressing Ena/VASP ΔPRR is in part due to the lack of recruitment of these proteins.
Ena/VASP proteins harbor a short sequence that is similar to the G-actin–binding motif of the actin-sequestering molecule thymosin β4 (van Troys et al., 1996). Although a direct binding between G-actin and Ena/VASP proteins has not yet been demonstrated, we show that the deletion of this site in Mena causes a small but still significant reduction in Listeria motility. It has recently been shown that VASP exerts a weak actin nucleating activity in vitro (Harbeck et al., 2000), suggesting that the deletion of the putative G-actin–binding site in Mena could result in a decrease in actin filament formation. This possibility seems unlikely in light of many observations demonstrating that Listeria mutants that are not able to bind to the Arp2/3 complex, but are still fully competent for interacting with Ena/VASP proteins, cannot induce the formation of actin clouds at their surface (Lasa et al., 1995, 1997; Pistor et al., 1995, 2000; Smith et al., 1996; Skoble et al., 2000). Although Ena/VASP proteins are not able to nucleate actin, recent data suggest that they may support the nucleation activity of the Arp2/3 complex. In particular, Skoble et al. (2001) showed that VASP can rescue the ability to activate the Arp2/3 complex of an ActA mutant that lacks the G-actin–binding site, and suggested that the F-actin-binding site of VASP is required for this process. In contrast with their conclusion and based on our data showing that the deletion of the F-actin–binding site of Ena/VASP proteins does not affect Listeria motility and that the deletion of the TLM site clearly impairs actin accumulation at the bacterial surface, we suggest that Ena/VASP proteins may stimulate actin filament nucleation at the bacterial surface by supplying actin monomers to the Arp2/3 complex. Our hypothesis is consistent with the observation that stimulators of the Arp2/3 complex such as WASp/Scar proteins require binding to G-actin to activate this complex, and that Ena/VASP proteins enhance Listeria motility in cell-free systems in the absence of profilin (Loisel et al., 1999; Machesky et al., 1999). It will be important to characterize the function of the TLM motif in detail and test whether it actually binds G-actin.
The EVH2 domain of Ena/VASP proteins has been implicated in the interaction of this protein family with F-actin in vitro (Bachmann et al., 1999; Hüttelmaier et al., 1999). Kuo and McGrath (2000) have recently demonstrated that Listeria are tightly linked to their own actin tails raising the possibility that this tight interaction limits bacterial motility. Accordingly, we show herein that the deletion of the FAB site in Ena/VASP proteins seems to remove this physical constraint and thereby increase bacterial speed. In vitro, VASP seems to protect actin filaments from the actin-severing activity of gelsolin (Bearer et al., 2000). In addition, gelsolin, which localizes at the interfaces between bacteria and actin tails, enhances Listeria motility when overexpressed or injected in fibroblasts (Laine et al., 1998). The conclusion that could be made from these results is that the deletion of the FAB region of Ena/VASP proteins makes actin filaments more susceptible to gelsolin's action, resulting in higher bacterial speed. Laurent et al. (1999) reported that a GST-tagged EVH2 domain inhibits Listeria motility in platelets extracts concluding that the interaction between F-actin and Ena/VASP proteins is essential for this process. We believe, however, that their interpretation was more likely to arise by the interference with both binding activities of thymosin β4-like motif and F-actin-binding site of endogenous VASP. Alternatively, other regions within the EVH2 domain of hitherto unknown function could also be responsible for such effect.
The binding of Ena/VASP proteins to F-actin in vitro seems to be dependent on their phosphorylation state (Laurent et al., 1999; Harbeck et al., 2000; Lambrechts et al., 2000). We have shown that deletion of the F-actin–binding site of Mena and VASP results in an enhancement of Listeria motility. Similarly, the expression of Ena/VASP mutants that mimics the full phosphorylation state of these proteins increases bacterial speed. Thus, it is possible that the phosphorylation of Ena/VASP proteins weakens their binding to F-actin, resulting in faster Listeria motility. This possibility would be in agreement with the findings of Lambrechts et al. (2000) and Harbeck et al., (2000), who showed that fully phosphorylated EVL and VASP bind less efficiently to F-actin. The possibility that Ena/VASP phosphorylation plays a role in actin-based processes is supported by the observation that VASP phosphorylation directly correlates with spreading of neutrophils (Lawrence and Pryzwansky, 2001). Moreover, Mena phosphorylation is required for its function as negative regulator of cell motility in fibroblasts (Loureiro et al., 2002).
A number of studies indicate that the EVH2 domain can mediate the formation of Ena/VASP multimers and that they may be required for the function of this protein family in vivo (Ahern-Djamali et al., 1998; Bachmann et al., 1999; Carl et al., 1999). In particular, Ena/VASP multimers seem to be required for the function of Ena/VASP proteins as suggested by the observation that a truncated form of Ena lacking the EVH2 domain caused lethality of Drosophila embryos (Ahern-Djamali et al., 1998). Moreover, the Mena mutant ΔCo-Co is only partially able to rescue normal motile properties of MVD7 fibroblasts (Loureiro et al., 2002), suggesting that Ena/VASP multimerization could play a role in this process. In the context of Listeria motility, the formation of Ena/VASP multimers at the bacterial surface has been proposed to increase the availability of polymerization-competent actin monomers and, as a consequence, bacterial motility. Therefore, we expected that the inhibition of Ena/VASP multimerization would result in the decrease of Listeria motility due to the limited availability of actin monomers. In contrast, we found that the expression of Ena/VASP ΔCo-Co proteins did not reduce bacterial movement but, in VASP, increased it. Based on our results that deletion of FAB also causes an increase of bacterial motility, it is conceivable that the deletion of the multimerization site in VASP, by causing a reduction in the number of F-actin–binding sites, weakens the interaction of the bacteria with the actin tails and, as a consequence, augments bacterial speed. This hypothesis is consistent with the observation that the deletion of the multimerization motif from the EVH2 domain of VASP decreases its ability to interact with F-actin, in vitro (Bachmann et al., 1999).
CONCLUSION
Our data clearly indicate that the proline-rich core, the putative G-actin–binding site, and the phosphorylation state of Ena/VASP proteins are important for Listeria motility, whereas, in contrast with previous models for Listeria motility, the F-actin–binding and multimerization regions of this protein family are dispensable for this actin-based process. Finally, in light of this study and that of Loureiro et al. (2002), it is clear that Ena/VASP protein domains can contribute to different extents in distinct actin-based processes.
How then, can Ena/VASP proteins influence the dynamics of the actin cytoskeleton? Other than affecting the nucleation/elongation of actin filaments, Ena/VASP proteins may influence the architecture of the actin cytoskeleton. This view is consistent with the observation that VASP seems to influence the branching of actin filaments induced by the Arp2/3 complex (Skoble et al., 2001) and that the expression of Mena in the MVD7 background affects the organization of the actin filaments in lamellipodia (Bear et al., 2002). Based on these and our study, we propose that Ena/VASP proteins act as multifunctional organizers of the actin cytoskeleton that regulate both the nucleation/elongation and the architecture of actin networks.
ACKNOWLEDGMENTS
We thank David A. Monner, Matthias Krause, and Adam Kwiatkowski for critical reading of the manuscript and helpful discussions. We thank Petra Hagendorff and Maria Höxter for excellent technical assistance. J.W. was supported by the DFG grant JO 55/15-3 and by the Fonds der Chemischen Industrie. J.J.L. was supported by the Anna Fuller Molecular Oncology Fund. J.E.B. is supported by a Special Fellow award from the Leukemia and Lymphoma Society (3476-02). F.B.G. was supported by National Institutes of Health grant GM-58801 and by funds from the WM Keck Distinguished Young Scholar Award.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–01–0058. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–01–0058.
REFERENCES
- Ahern-Djamali SM, Comer AR, Bachmann C, Kastenmeier AS, Reddy SK, Beckerle MC, Walter U, Hoffmann FM. Mutations in Drosophila Enabled and rescue by human vasodilator-stimulated phosphoprotein (VASP) indicate important functional roles for Ena/VASP homology domain 1 (EVH1) and EVH2 domains. Mol Biol Cell. 1998;9:2157–2171. doi: 10.1091/mbc.9.8.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann C, Fischer L, Walter U, Reinhard M. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J Biol Chem. 1999;274:23549–23557. doi: 10.1074/jbc.274.33.23549. [DOI] [PubMed] [Google Scholar]
- Bear JE, Loureiro JJ, Libova I, Fassler R, Wehland J, Gertler FB. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 2000;101:717–728. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- Bear JE, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Bearer EL, Prakash JM, Manchester RD, Allen PG. VASP protects actin filaments from gelsolin: an in vitro study with implications for platelet actin reorganizations. Cell Motil Cytoskeleton. 2000;47:351–364. doi: 10.1002/1097-0169(200012)47:4<351::AID-CM8>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle NP, Holt MR, Davies JE, Price CJ, Critchley DR. The focal-adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem J. 1996;318:753–757. doi: 10.1042/bj3180753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carl UD, Pollmann M, Orr E, Gertler FB, Chakraborty T, Wehland J. Aromatic and basic residues within the EVH1 domain of VASP specify its interaction with proline-rich ligands. Curr Biol. 1999;9:715–718. doi: 10.1016/s0960-9822(99)80315-7. [DOI] [PubMed] [Google Scholar]
- Castellano F, Le Clainche C, Patin D, Carlier MF, Chavrier P. A WASp-VASP complex regulates actin polymerization at the plasma membrane. EMBO J. 2001;20:5603–5614. doi: 10.1093/emboj/20.20.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty T, et al. A focal adhesion factor directly linking intracellularly motile Listeria monocytogenes and Listeria ivanovii to the actin-based cytoskeleton of mammalian cells. EMBO J. 1995;14:1314–1321. doi: 10.1002/j.1460-2075.1995.tb07117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppolino MG, Krause M, Hagendorff P, Monner DA, Trimble W, Grinstein S, Wehland J, Sechi AS. Evidence for a molecular complex consisting of Fyb/SLAP, SLP-76, Nck, VASP and WASP that links the actin cytoskeleton to Fcγ receptor signaling during phagocytosis. J Cell Sci. 2001;114:4307–4318. doi: 10.1242/jcs.114.23.4307. [DOI] [PubMed] [Google Scholar]
- Cossart P, Bierne H. The use of host cell machinery in the pathogenesis of Listeria monocytogenes. Curr Opin Immunol. 2001;13:96–103. doi: 10.1016/s0952-7915(00)00188-6. [DOI] [PubMed] [Google Scholar]
- Domann E, Wehland J, Rohde M, Pistor S, Hartl M, Goebel W, Leimeister-Wachter M, Wuenscher M, Chakraborty T. A novel bacterial virulence gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 1992;11:1981–1990. doi: 10.1002/j.1460-2075.1992.tb05252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees B, Friederich E, Fradelizi J, Louvard D, Beckerle MC, Golsteyn RM. Characterization of the interaction between zyxin and members of the Ena/vasodilator-stimulated phosphoprotein family of proteins. J Biol Chem. 2000;275:22503–22511. doi: 10.1074/jbc.M001698200. [DOI] [PubMed] [Google Scholar]
- Ermekova KS, Zambrano N, Linn H, Minopoli G, Gertler F, Russo T, Sudol M. The WW domain of neural protein FE65 interacts with proline-rich motifs in Mena, the mammalian homolog of Drosophila Enabled. J Biol Chem. 1997;272:32869–32877. doi: 10.1074/jbc.272.52.32869. [DOI] [PubMed] [Google Scholar]
- Frischknecht F, Way M. Surfing pathogens and the lessons learned for actin polymerization. Trends Cell Biol. 2001;11:30–38. doi: 10.1016/s0962-8924(00)01871-7. [DOI] [PubMed] [Google Scholar]
- Geese M, Schluter K, Rothkegel M, Jockusch BM, Wehland J, Sechi AS. Accumulation of profilin II at the surface of Listeria is concomitant with the onset of motility and correlates with bacterial speed. J Cell Sci. 2000;113:1415–1426. doi: 10.1242/jcs.113.8.1415. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Doctor JS, Hoffmann FM. Genetic suppression of mutations in the Drosophila abl proto-oncogene homolog. Science. 1990;248:857–860. doi: 10.1126/science.2188361. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- Haffner C, Jarchau T, Reinhard M, Hoppe J, Lohmann SM, Walter U. Molecular cloning, structural analysis and functional expression of the proline-rich focal adhesion and microfilament-associated protein VASP. EMBO J. 1995;14:19–27. doi: 10.1002/j.1460-2075.1995.tb06971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbrugge M, Friedrich C, Eigenthaler M, Schanzenbacher P, Walter U. Stoichiometric and reversible phosphorylation of a 46-kDa protein in human platelets in response to cGMP- and cAMP-elevating vasodilators. J Biol Chem. 1990;265:3088–3093. [PubMed] [Google Scholar]
- Harbeck B, Hüttelmaier S, Schluter K, Jockusch BM, Illenberger S. Phosphorylation of the vasodilator-stimulated phosphoprotein regulates its interaction with actin. J Biol Chem. 2000;275:30817–30825. doi: 10.1074/jbc.M005066200. [DOI] [PubMed] [Google Scholar]
- Hüttelmaier S, Harbeck B, Steffens O, Messerschmidt T, Illenberger S, Jockusch BM. Characterization of the actin binding properties of the vasodilator-stimulated phosphoprotein VASP. FEBS Lett. 1999;451:68–74. doi: 10.1016/s0014-5793(99)00546-3. [DOI] [PubMed] [Google Scholar]
- Jonckheere V, Lambrechts A, Vandekerckhove J, Ampe C. Dimerization of profilin II upon binding the (GP5)3 peptide from VASP overcomes the inhibition of actin nucleation by profilin II and thymosin beta4. FEBS Lett. 1999;447:257–263. doi: 10.1016/s0014-5793(99)00293-8. [DOI] [PubMed] [Google Scholar]
- Kang F, Laine RO, Bubb MR, Southwick FS, Purich DL. Profilin interacts with the Gly-Pro-Pro-Pro-Pro-Pro sequences of vasodilator-stimulated phosphoprotein (VASP): implications for actin-based Listeria motility. Biochemistry. 1997;36:8384–8392. doi: 10.1021/bi970065n. [DOI] [PubMed] [Google Scholar]
- Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- Krause M, Sechi AS, Konradt M, Monner D, Gertler FB, Wehland J. Fyn-binding protein (Fyb)/SLP-76-associated protein (SLAP), Ena/vasodilator-stimulated phosphoprotein (VASP) proteins and the Arp2/3 complex link T cell receptor (TCR) signaling to the actin cytoskeleton. J Cell Biol. 2000;149:181–194. doi: 10.1083/jcb.149.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo SC, McGrath JL. Steps and fluctuations of Listeria monocytogenes during actin-based motility. Nature. 2000;407:1026–1029. doi: 10.1038/35039544. [DOI] [PubMed] [Google Scholar]
- Laine RO, Phaneuf KL, Cunningham CC, Kwiatkowski D, Azuma T, Southwick FS. Gelsolin, a protein that caps the barbed ends and severs actin filaments, enhances the actin-based motility of Listeria monocytogenes in host cells. Infect Immun. 1998;66:3775–3782. doi: 10.1128/iai.66.8.3775-3782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts A, Kwiatkowski AV, Lanier LM, Bear JE, Vandekerckhove J, Ampe C, Gertler FB. cAMP-dependent protein kinase phosphorylation of EVL, a Mena/VASP relative, regulates its interaction with actin and SH3 domains. J Biol Chem. 2000;275:36143–36151. doi: 10.1074/jbc.M006274200. [DOI] [PubMed] [Google Scholar]
- Lanier LM, Gates MA, Witke W, Menzies S, Wehman AM, Macklis JD, Kwiatkowski D, Soriano P, Gertler FB. Mena is required for neurulation and commissure formation. Neuron. 1999;22:313–325. doi: 10.1016/s0896-6273(00)81092-2. [DOI] [PubMed] [Google Scholar]
- Lasa I, David V, Gouin E, Marchand JB, Cossart P. The amino-terminal part of ActA is critical for the actin-based motility of Listeria monocytogenes; the central proline-rich region acts as a stimulator. Mol Microbiol. 1995;18:425–436. doi: 10.1111/j.1365-2958.1995.mmi_18030425.x. [DOI] [PubMed] [Google Scholar]
- Lasa I, Gouin E, Goethals M, Vancompernolle K, David V, Vandekerckhove J, Cossart P. Identification of two regions in the N-terminal domain of ActA involved in the actin comet tail formation by Listeria monocytogenes. EMBO J. 1997;16:1531–1540. doi: 10.1093/emboj/16.7.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent V, Loisel TP, Harbeck B, Wehmann A, Gröbe L, Jockusch BM, Wehland J, Gertler FB, Carlier MF. Role of proteins of the Ena/VASP family in actin based motility of Listeria monocytogenes. J Cell Biol. 1999;144:1245–1258. doi: 10.1083/jcb.144.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence DW, Pryzwansky KB. The vasodilator-stimulated phosphoprotein is regulated by cyclic GMP-dependent protein kinase during neutrophil spreading. J Immunol. 2001;166:5550–5556. doi: 10.4049/jimmunol.166.9.5550. [DOI] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Lommel S, Benesch S, Rottner K, Franz T, Wehland J, Kuhn R. Actin pedestal formation by enteropathogenic Escherichia coli and intracellular motility of Shigella flexneri are abolished in N-WASP-defective cells. EMBO Rep. 2001;2:850–857. doi: 10.1093/embo-reports/kve197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro, J.J., Rubinson, D.A., Bear, J.E., Baltus, G.A., Kwiatkowski, A.V., and Gertler, F.B. (2002). Critical roles of phosphorylation and actin-binding motfits but not the central proline-rich region, for Ena/VASP function during cell migration. Mol. Biol. Cell 13, in press. [DOI] [PMC free article] [PubMed]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci USA. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machner MP, Urbanke C, Barzik M, Otten S, Sechi AS, Wehland J, Heinz DW. ActA from Listeria monocytogenes can interact with up to four Ena/VASP homology 1 domains simultaneously. J Biol Chem. 2001;276:40096–40103. doi: 10.1074/jbc.M104279200. [DOI] [PubMed] [Google Scholar]
- May RC, Hall ME, Higgs HN, Pollard TD, Chakraborty T, Wehland J, Machesky LM, Sechi AS. The Arp2/3 complex is essential for the actin-based motility of Listeria monocytogenes. Curr Biol. 1999;9:759–762. doi: 10.1016/s0960-9822(99)80337-6. [DOI] [PubMed] [Google Scholar]
- Mykkanen OM, Gronholm M, Ronty M, Lalowski M, Salmikangas P, Suila H, Carpen O. Characterization of human palladin, a microfilament-associated protein. Mol Biol Cell. 2001;12:3060–3073. doi: 10.1091/mbc.12.10.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebuhr K, Ebel F, Frank R, Reinhard R, Domann E, Carl UD, Walter U, Gertler FB, Wehland J, Chakraborty T. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J. 1997;16:5433–5444. doi: 10.1093/emboj/16.17.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistor S, Chakraborty T, Walter U, Wehland J. The bacterial actin nucleator protein ActA of Listeria monocytogenes contains multiple binding sites for host microfilament proteins. Curr Biol. 1995;5:517–525. doi: 10.1016/s0960-9822(95)00104-7. [DOI] [PubMed] [Google Scholar]
- Pistor S, Grobe L, Sechi AS, Domann E, Gerstel B, Machesky LM, Chakraborty T, Wehland J. Mutations of arginine residues within the 146-KKRRK-150 motif of the ActA protein of Listeria monocytogenes abolish intracellular motility by interfering with the recruitment of the Arp2/3 complex. J Cell Sci. 2000;113:3277–3287. doi: 10.1242/jcs.113.18.3277. [DOI] [PubMed] [Google Scholar]
- Reinhard M, Halbrugge M, Scheer U, Wiegand C, Jockusch BM, Walter U. The 46/50 kDa phosphoprotein VASP purified from human platelets is a novel protein associated with actin filaments and focal contacts. EMBO J. 1992;11:2063–2070. doi: 10.1002/j.1460-2075.1992.tb05264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K, Behrendt B, Small JV, Wehland J. VASP dynamics during lamellipodia protrusion. Nat Cell Biol. 1999;1:321–322. doi: 10.1038/13040. [DOI] [PubMed] [Google Scholar]
- Sechi AS, Wehland J, Small JV. The isolated comet tail pseudopodium of Listeria monocytogenes: a tail of two actin filament populations, long and axial and short and random. J Cell Biol. 1997;137:155–167. doi: 10.1083/jcb.137.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoble J, Portnoy DA, Welch MD. Three regions within ActA promote Arp2/3 complex-mediated actin nucleation and Listeria monocytogenes motility. J Cell Biol. 2000;150:527–538. doi: 10.1083/jcb.150.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoble J, Auerbuch V, Goley ED, Welch MD, Portnoy DA. Pivotal role of VASP in Arp2/3 complex-mediated actin nucleation, actin branch-formation, and Listeria monocytogenes motility. J Cell Biol. 2001;155:89–100. doi: 10.1083/jcb.200106061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GA, Theriot JA, Portnoy DA. The tandem repeat domain in the Listeria monocytogenes ActA protein controls the rate of actin based motility, the percentage of moving bacteria and the localization of VASP and profilin. J Cell Biol. 1996;135:647–660. doi: 10.1083/jcb.135.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Troys M, Dewitte D, Goethals M, Carlier MF, Vandekerckhove J, Ampe C. The actin binding site of thymosin beta 4 mapped by mutational analysis. EMBO J. 1996;15:201–210. [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- Zalevsky J, Grigorova I, Mullins RD. Activation of the Arp2/3 complex by the Listeria acta protein. Acta binds two actin monomers and three subunits of the Arp2/3 complex. J Biol Chem. 2001;276:3468–3475. doi: 10.1074/jbc.M006407200. [DOI] [PubMed] [Google Scholar]
- Zeile WL, Purich DL, Southwick FS. Recognition of two classes of oligoproline sequences in profilin-mediated acceleration of actin-based Shigella motility. J Cell Biol. 1996;133:49–59. doi: 10.1083/jcb.133.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]