Abstract
We have used double-stranded RNA-mediated interference (RNAi) to study Drosophila cytokinesis. We show that double-stranded RNAs for anillin, acGAP, pavarotti, rho1, pebble, spaghetti squash, syntaxin1A, and twinstar all disrupt cytokinesis in S2 tissue culture cells, causing gene-specific phenotypes. Our phenotypic analyses identify genes required for different aspects of cytokinesis, such as central spindle formation, actin accumulation at the cell equator, contractile ring assembly or disassembly, and membrane behavior. Moreover, the cytological phenotypes elicited by RNAi reveal simultaneous disruption of multiple aspects of cytokinesis. These phenotypes suggest interactions between central spindle microtubules, the actin-based contractile ring, and the plasma membrane, and lead us to propose that the central spindle and the contractile ring are interdependent structures. Finally, our results indicate that RNAi in S2 cells is a highly efficient method to detect cytokinetic genes, and predict that genome-wide studies using this method will permit identification of the majority of genes involved in Drosophila mitotic cytokinesis.
INTRODUCTION
Cytokinesis is the complex process by which the daughter cells separate at the end of cell division. In animal cells cytokinesis involves at least four subprocesses that must be tightly coordinated to ensure the fidelity of chromosome segregation (reviewed by Glotzer, 1997; Straight and Field, 2000). First, interactions between the spindle and the cortex determine the site of cleavage furrow formation. Second, an actomyosin-based contractile ring assembles at this cortical site. Third, the actomyosin ring constricts, leading to furrow ingression. Fourth, during both furrow ingression and the completion of cytokinesis new membrane is added to allow separation of the daughter cells.
The spindle plays a crucial role in both the coordination and execution of these subprocesses. In large cells such as those in early marine invertebrate embryos the positioning of the cleavage furrow seems to be determined by interactions between the cortex and the spindle's astral microtubules (Rappaport, 1985). However, in smaller cells, such as vertebrate and Drosophila cells, the site of cleavage furrow formation is specified by the central spindle (Cao and Wang, 1996; Bonaccorsi et al., 1998; Giansanti et al., 2001), the bundle of antiparallel, interdigitating microtubules that assembles during ana-telophases between the daughter nuclei. Moreover, in all the systems thus far analyzed the integrity of the central spindle is an essential requirement for completion of cytokinesis (reviewed by Gatti et al., 2000).
Many proteins have been identified that are required for central spindle formation and, thus, for the execution of cytokinesis. These molecules include several plus-end directed kinesin-like proteins that accumulate at the central spindle midzone. Examples of these proteins are the orthologs mammalian CHO1/MKLP1, Drosophila Pavarotti (Pav), and Caenorhabditis elegans ZEN-4, as well as Drosophila Klp3A (Nislow et al., 1992; Williams et al., 1995; Adams et al., 1998; Powers et al., 1998; Raich et al., 1998). The activities of these kinesin-like proteins are regulated by kinases of both the Polo and Aurora families. For example, the Drosophila Polo kinase and its human homolog Plk coimmunoprecipitate and phosphorylate Pav and CHO1/MKLP1, respectively (Lee et al., 1995; Adams et al., 1998), whereas the C. elegans Aurora B kinase (AIR-2) interacts with ZEN-4 (Severson et al., 2000). Finally, studies on Drosophila male meiosis have suggested that central spindle formation depends on cooperative interactions between the central spindle microtubules and elements of the actomyosin ring (Giansanti et al., 1998; Gatti et al., 2000).
The actomyosin-based contractile ring assembles at the equatorial cortex during late anaphase and constricts while remaining anchored to the plasma membrane, until cytokinesis is completed. Although many components of the contractile ring machinery have been identified, its precise molecular composition and its regulation during cytokinesis are still poorly understood (reviewed by Goldberg et al., 1998; Robinson and Spudich, 2000). Contractile ring assembly and function are regulated by the Rho GTPase and its upstream activators and downstream effectors (reviewed by Prokopenko et al., 2000). These effectors include several cytokinesis-specific kinases and the formin-homology proteins, such as Drosophila Diaphanous, mouse p140Dia1, and C. elegans CYK-1 (Swan et al., 1998; Wassermann, 1998; Prokopenko et al., 2000). The formins bind and regulate profilin (Wassermann, 1998), a small actin-binding protein that promotes actin polymerization and is required for contractile ring assembly (Giansanti et al., 1998).
The contractile ring interacts with a number of additional proteins that play regulatory and structural roles. One of these proteins is cofilin, a polypeptide with actin filament-severing activity; Drosophila cofilin encoded by the tsr gene is required for contractile ring disassembly at the end of cell division (Gunsalus et al., 1995). Other contractile ring-associated proteins are anillin and the septins. Anillin contains an actin-binding domain and a pleckstrin homology (PH) domain and may mediate the anchoring of the contractile ring to the plasma membrane (Field and Alberts, 1995; Giansanti et al., 1999; Oegema et al., 2000). Septins are a group of conserved proteins that interact with components of the exocist complex and may be involved in membrane–contractile ring interactions (reviewed by Field and Kellogg, 1999; Straight and Field, 2000).
The accomplishment of cytokinesis requires deposition of new membrane at the ingressing furrow (reviewed by Straight and Field, 2000). This new membrane is thought to arise from Golgi-derived vesicles that are targeted to the furrow through a microtubule-dependent transport. Once at the furrow, these vesicles fuse with the invaginating plasma membrane, thus creating new membrane surface for cleavage (Straight and Field, 2000; Skop et al. 2001). Membrane-vesicle fusion may be mediated by syntaxin, a member of the t-soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) family of proteins. t-SNAREs are associated with the target membrane and interact with v-SNAREs that reside on vesicles, mediating the process of membrane fusion (Chen and Scheller, 2001). Studies in C. elegans and Arabidopsis thaliana have shown that mutations in syntaxin-encoding genes abrogate cytokinesis, strongly supporting a role of this protein in membrane addition during cytokinesis (Lauber et al., 1997; Jantsch-Plunger and Glotzer, 1999).
Although molecular genetic analyses in model systems have led to the discovery of many gene products involved in cytokinesis, it is clear that the inventory of cytokinetic proteins is still largely incomplete. Herein, we have used double-stranded RNA-mediated interference (RNAi) to ablate genes required for cytokinesis in Drosophila S2 tissue culture cells. Our phenotypic analyses of RNAi cells define the functions of several cytokinetic proteins and provide new insight into the interplay among microtubules, microfilaments, and membranes during the assembly and functioning of the cytokinetic machinery.
MATERIALS AND METHODS
Cell Cultures and RNAi Treatments
S2 cells were cultured at 25°C in Shields and Sang M3 medium (Sigma-Aldrich, St. Louis, MO) supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich). RNAi treatments were carried out according to Clemens et al. (2000). Cells were suspended in serum-free Shields and Sang medium at a concentration of 1 × 106 cells/ml, and plated, 1 ml/well, in a six-well culture dish (Nalge Nunc, Naperville, IL). To perform RNAi, each culture was inoculated with 15 μg of double-stranded (ds)RNA. After 1-h incubation at 25°C, 2 ml of medium supplemented with 15% fetal bovine serum was added to each culture. Control cultures were prepared in the same way but without addition of dsRNA. Both RNA-treated and control cells were grown for 72 h at 25°C and then processed for either fluorescence-activated cell sorting (FACS), biochemical, or cytological analysis.
dsRNA Production
Individual gene sequences were amplified by polymerase chain reaction (PCR) from a pool of cDNAs obtained from five different libraries: four embryonic libraries from 0–4-, 4–8-, 8–12-, and 12–24-h embryos and one imaginal disc library, all kindly provided by N. Brown (Brown and Kafatos, 1988). The primers used in the PCR reactions were 35 nucleotides long and contained a 5′ T7 RNA polymerase-binding site (5′-TAATACGACTCACTATAGGGAGG-3′) flanked by a gene-specific sequence. The GenBank accession number (an), the sense and antisense gene-specific sequences, and the position (pos.) of their 5′ nucleotide were as follows: acGAP, an AJ251502, sense AACCACACCTTC pos. 1110, antisense TGCATATAGCGA pos. 1961; ani, an X89858, sense GCTCGAGAAGGC pos. 249, antisense AGCTTCATCCGC pos. 1270; chic, an M84529, sense TAAAGCAACAGC pos. 139, antisense TTCGCTCTTATC pos. 704; fwd, an AE003467, sense TCGGTAGTCGCG pos. 289291, antisense ATCCTCCGGGTC pos. 288198; klp3A, an AF132186, sense AGCTGGAAATGC pos. 2642, antisense TCTGGGGCTCGT pos. 3595; pav, an AF005853, sense TCAAAATCCGCG pos. 1151, antisense CACTCCACATCG pos. 2081; pnut, an U08103, sense CGCCTCCAACGG pos. 337, antisense TCCTGAAGGTGC pos. 1266; pbl, an AF136492, sense AGGCCTGAAGG pos. 2127, antisense CAGGTGTTAGAG pos. 2834; rho1, an AF177871, sense CGCCATAAGAAT pos. 75, antisense TTGTTCAGCTCG pos. 821; sqh, an M67494, sense CATTCGGCAGCT pos. 250, antisense CAGCTGGCTAGT pos. 1890; syx1A, an L37732, sense TGGCCGTCAATG pos. 358, antisense CAGATCAGTATC pos. 1114; and tsr, an U24490, sense TTGTTCGTGAAA pos. 13, antisense ATACGTGTTTCC pos. 629.
The PCR products were purified by using the Microcon kit (Millipore, Bedford, MA) and used as templates to produce dsRNA with the Megascript transcription kit (Ambion, Austin, TX). The RNA products were treated with DNase I (Ambion) to digest template DNA, extracted with phenol/chloroform, ethanol-precipitated, and resuspended in water. To ensure that most of the RNA products were in a double-stranded form, the RNA solutions were heated at 65°C for 30 min and then slowly cooled to room temperature. The quality and concentration of dsRNAs were checked by 1% agarose gel electrophoresis. dsRNAs were stored at −20°C before use.
Immunoblotting
Cells were harvested by centrifugation at 800 × g for 5 min. Pellets were lysed in 50 μl of Laemmli buffer. Lysate (20 μl) was electrophoresed on a 10% SDS-polyacrylamide gel and transferred to an Immobilion membrane (Millipore) by using a semidry transfer apparatus (Bio-Rad, Hercules, CA). The membrane was blocked for 1 h in a 5% dry milk in TBS-T (20 mM Tris-HCl, 150 mM NaCl, and 0.1% Tween 20, pH 7.4) and then incubated overnight with any of the following primary antibodies diluted in TBS-T: anti-anillin raised in rabbit against amino acids 401–828 (1:1000; Field and Alberts, 1995); anti-Chic monoclonal from cell line 6F (1:10; Verheyen and Cooley, 1994); anti-Klp3A rabbit antibody (1:1000; Williams et al., 1995); anti-Pav rabbit antibody (1:1000; Adams et al., 1998); and anti-Pnut monoclonal (1:40; Neufeld and Rubin, 1994). To check for loading each membrane was also incubated with the C1A9 monoclonal antibody to heterocromatin protein 1 (HP1; 1:500; James et al., 1989). Membranes were washed in TBS-T, incubated for 1 h with either anti-mouse or both anti-mouse and anti-rabbit horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences, Piscataway, NJ), and then washed again in TBS-T. Signals were detected using the ECL kit (Amersham Biosciences) following the manufacturer's protocol.
FACS Analysis
To perform FACS analysis 1 ml of 72-h cultures was centrifuged at 800 × g for 5 min. The pelleted cells were washed in 10 ml of phosphate-buffered saline (PBS) and resuspended in 500 μl of PBT (PBS with 0.1% Triton-X) containing 25 μg/ml propidium iodide. FACS was performed on an FACStar Plus machine (BD Biosciences, San Jose, CA).
Cytological Procedures
Cells from 3-ml cultures were harvested by centrifugation at 800 × g for 5 min and washed in 10 ml of PBS. The pelleted cells were resuspended in 3 ml of 3.7% formaldehyde in PBS and fixed for 5 min. Cells were then spin down by centrifugation, resuspended in 500 μl of PBS, and cytocentrifuged using a cytocentrifuge (Shandon Scientific, Cheshire, England) at 900 rpm for 4 min. The slides were immersed in liquid nitrogen for at least 5 min, transferred to PBT for 15 min, and then to PBT containing 3% bovine serum albumin for 20 min. These preparations were stained for tubulin and either anillin, Pav, or actin. For tubulin plus anillin or tubulin plus Pav staining, slides were incubated overnight with both an anti-tubulin mAb (1:50; Amersham Biosciences) and either an anti-anillin (1:100) or an anti-Pav (1:100) antibody (see above), all diluted in PBS. These primary antibodies were detected by incubation for 1 h with both fluorescein isothiocyanate-conjugated anti-mouse IgG (Jackson Laboratories, Bar Harbor, ME) and Cy3-conjugated anti-rabbit IgG (Jackson Laboratories). For tubulin plus F actin staining slides were first immunostained for tubulin and then with rhodamine-phalloidin as described previously (Giansanti et al., 1999). All slides were mounted in Vectashield with 4,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA) to stain DNA and reduce fluorescence fading.
Immunostained preparations were examined with an Axioplan fluorescence microscope (Carl Zeiss, Oberkochen, Germany) equipped with a cooled charge-coupled device (Photometrics, Tucson, AZ) as described previously (Giansanti et al., 1999). Gray scale digital images were collected using the IPLab Spectrum software, converted to Photoshop 3.0 (Adobe Systems, Mountain View, CA) and merged in pseudocolors.
RESULTS
Disruption of Cytokinesis by RNAi
We treated Drosophila S2 tissue culture cells with dsRNA for 12 genes implicated in cytokinesis. Nine of these genes (chickadee [chic], four wheel drive [fwd], klp3A, pebble [pbl], rho1, pavarotti [pav], peanut [pnut], spaghetti squash [sqh], and twinstar [tsr]) are identified by Drosophila mutations that have been shown to disrupt cytokinesis in either somatic cells, or male meiotic cells, or both (Table 1). For the other three genes (anillin [ani], acGAP, and syntaxin1A [syx1A]) an involvement in Drosophila cytokinesis has never been demonstrated by mutational/phenotypical analysis.
Table 1.
Genes required for cytokinesis in D. melanogaster
| Gene name | Protein encoded | Protein localization | Required for cytokinesis in
|
Phenotype of mutant | References | ||
|---|---|---|---|---|---|---|---|
| Embryonic cellsa | Larval brain cells | Male meiosis | |||||
| chickadee (chic) | Profilin | Cortex and c.f. | nd | nob | yes | Absence of both c.s. and c.r. | Cooley et al., 1992; Giansanti et al., 1998 |
| four wheel drive (fwd) | Phospholipid kinase | nd | no | no | yes | Abnormal c.r. morphology | Brill et al., 2000 |
| klp3A | Kinesin-like | c.s. midzone | nd | no | yes | Absence of both c.s. and c.r. | Giansanti et al., 1998; Williams et al., 1995 |
| pebble (pbl) | Rho GEF | c.f. | yes | nd | nd | Absence of both c.s. and c.r. | Prokopenko et al., 1999 |
| rho1 | GTPase | nd | yes | nd | nd | ndc | Prokopenko et al., 1999 |
| pavarotti (pav) | Kinesin-like | c.s. midzone | yes | nd | nd | Absence of both c.s. and c.r. | Adams et al., 1998 |
| peanut (pnut) | Septin | c.f. | nd | yes | nod | nde | Neufeld and Rubin, 1994 |
| spaghetti squash (sqh) | Regulatory light chain of myosin II | c.f.f | nd | yes | nd | nd | Karess et al., 1991 |
| twinstar (tsr) | Cofilin | nd | nd | yes | yes | Failure of c.r. disassembly | Gunsalus et al., 1995 |
nd, not determined; c.f., cleavage furrow; c.s., central spindle; c.r., contractile ring.
Embryonic cells of cycles 14–16.
chic lethal mutants are severely defective in meiotic cytokinesis of Drosophila males but not in cytokinesis of larval brain cells, suggesting that chic is not essential for cytokinesis of somatic cells (Giansanti, Bonaccorsi, Gatti, unpublished data).
rho1 has been shown to be involved in cytokinesis of embryonic cells but the primary defect leading to the failure of cytokinesis has not been determined (Prokopenko et al., 1999).
In testes of larvae homozygous for pnut null mutations, meiotic cytokinesis is normal (Bonaccorsi, Giansanti, Gatti, unpublished data).
The primary defect that causes cytokinesis failures in larval brain cells of pnut mutants has not been defined (Neufeld and Rubin, 1994).
It has recently been shown that Sqh-GFP accumulates in the cleavage furrow (Karess, personal communication).
We prepared dsRNA for each of the 12 genes described above. These RNAs, which ranged in size from 610 to 1020 base pairs, were added to the medium of fresh S2 cultures at a final concentration of ∼5 μg/ml. After 72 h of dsRNA treatment cells were harvested by centrifugation and subjected to two different analyses. The cells of one of the treated cultures and those of a parallel control were stained with propidium iodide and analyzed with a FACS. The cells of another dsRNA-treated culture and its control were stained for DNA, tubulin, and F-actin and examined under a fluorescence microscope. For some of the dsRNAs tested (chic, klp3A, pav, pnut, and ani) we used additional treated and control cultures for Western blotting analysis.
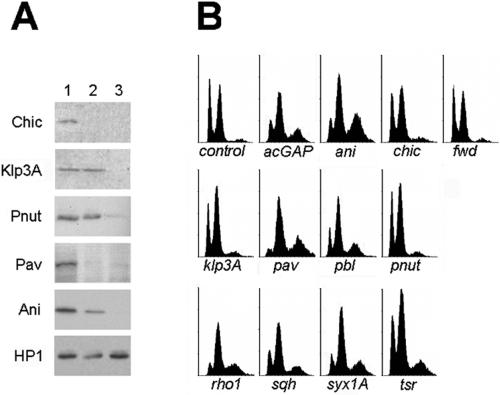
The overall results of our experiments are reported in Figures 1 and 2. Western blotting analysis of cells treated with either ani, chic, klp3A, pav, or pnut dsRNA shows that each of these dsRNAs caused a dramatic depletion of the corresponding gene product (Figure 1A). We could not obtain comparable results for the other six genes studied due to the unavailability of specific antibodies. However, the strong phenotypical effects observed after treatments with acGAP, pbl, rho1, sqh, syx1A, and tsr dsRNAs (see below) strongly suggest that these genes were also silenced by RNAi.
Figure 1.
Effects of RNAi with Drosophila genes involved in cytokinesis. (A) Western blots from cells grown for 72 h in the presence of mock (plasmid) RNA (lane 1) or incubated for either 24 h (lane 2) or 72 h (lane 3) with the dsRNA for the protein indicated. The HP1 protein was used as a loading control in all cases. Note that 72-h treatments cause a dramatic depletion of all gene products. (B) FACS profiles of cells treated for 72 h with dsRNA; abscissa, DNA content; ordinate, cell number. Note that the control profile exhibits two main peaks of 2C and 4C cells and a very small peak of 8C cells. FACS profiles of chic (profilin), fwd (phospholipid kinase), klp3A (kinesin-like), and pnut (septin) (RNAi) cells do not differ from control, whereas the profiles of acGAP (rhoGAP), ani (anillin), pav (kinesin-like), pbl (rhoGEF), rho1, sqh (myosin II regulatory light chain), syx1A (t-SNARE), and tsr (cofilin) (RNAi) cells display a decrease of the 2C peak (G1 diploid cells) and a relative increase of both the 4C (G2-M diploid cells and G1 tetraploid cells) and the 8C peak (G2-M tetraploid cells and G1 octoploid cells).
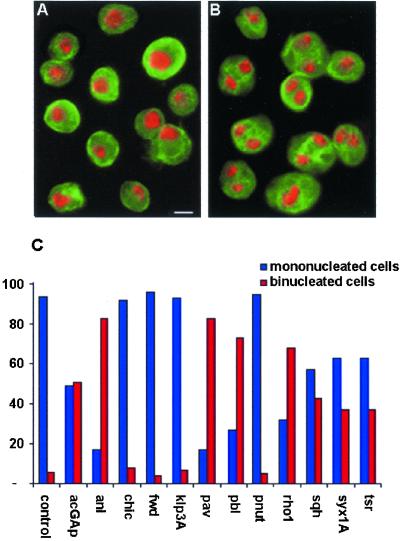
Figure 2.
Binucleated cells in cultures treated with dsRNA of genes involved in cytokinesis. (A and B) Control cells (A) and cells treated for 72 h with pav dsRNA (B) stained for tubulin (green) and DNA (with 4,6-diamidino-2-phenylindole, red). (C) Frequencies of binucleated cells after 72-h treatments with dsRNAs. Bar, 10 μm.
An examination of the FACS profiles shown in Figure 1B reveals that control cells are distributed into two main peaks corresponding to cells containing 2C and 4C DNA, and in a very small peak of 8C cells. FACS profiles of cells treated with either chic, fwd, klp3A, or pnut dsRNA did not exhibit appreciable differences from controls. However, in samples treated with either acGAP, ani, pav, pbl, rho1, sqh, syx1A, or tsr dsRNA there is a clear increase of the 8C peak at the expense of the 2C and 4C peaks, suggesting that a substantial fraction of the cells have become polyploid. To estimate the levels of cell death caused by the dsRNA treatments, we also determined the frequencies of hypodiploid cells showing a side scatter higher than G1 cells; cells displaying these features are considered to be apoptopic (Darzynkiewicz et al., 1997). In most treated samples these frequencies were comparable with those of the corresponding control. We observed slight increases of apoptotic cells only in cultures treated with acGAP, pav, or tsr dsRNA (our unpublished data).
The observed increases in 8C cells (Figure 1) could result from either metaphase arrest or a failure in cytokinesis. RNAi treatments causing metaphase arrest, followed by reversion to interphase and DNA duplication, should give rise to 8C mononucleated tetraploid cells. In contrast, treatments that suppress cytokinesis should produce G1 binucleated 4C cells that, upon completing DNA synthesis, will become 8C binucleated tetraploid. Thus, an increase of binucleated cells in RNAi-treated cultures is diagnostic of errors in cytokinesis. Our examination of fixed cells stained for DNA, tubulin, and actin revealed that binucleated cells are indeed very frequent in all RNAi-treated cells that display major 8C peaks (Figure 2). Thus, our combined FACS and cytological analyses indicate that RNAi for either acGAP, ani, pav, pbl, rho1, sqh, syx1A, or tsr causes frequent failures of cytokinesis in S2 cells, whereas treatment with dsRNA of either chic, fwd, klp3A, or pnut does not impair the cytokinetic process.
To determine the fate of the 8C cells produced by failures in cytokinesis, we examined metaphase spreads from cultures treated for 110 h with pav dsRNA. In these (RNAi) cultures the frequencies of octoploid (∼40 chromosomes) and highly polyploid (>60 chromosomes) cells were 40 and 50% (n = 186), respectively. These results indicate that S2 cells do not possess a stringent checkpoint that prevents progression through the cell cycle of cells that have failed to undergo cytokinesis.
Cytological Phenotypes of RNAi-induced Cytokinesis Mutants
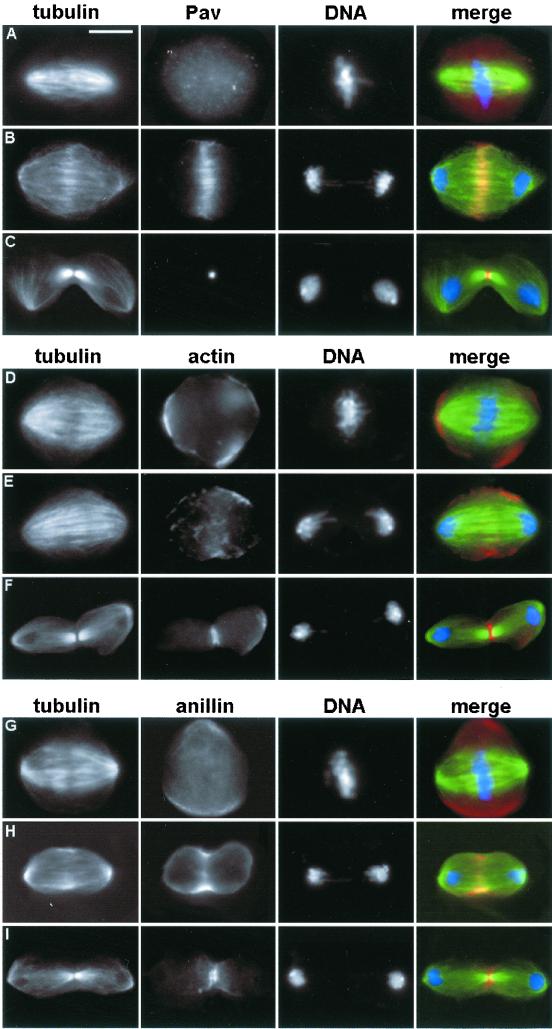
S2 cells are a highly suitable material for the cytological analysis of cytokinesis. These cells can be successfully stained for several proteins involved in cytokinesis such as actin, anillin, and Pav that can be used to track critical cytokinetic structures (Figure 3). At metaphase the Pav kinesin is diffused throughout the cytoplasm but during anaphase and telophase it concentrates at the plus ends of central spindle microtubules (Figure 3, A–C). Anillin and actin are not associated with central spindle microtubules and instead exhibit a similar localization that marks the equatorial region of the cortex (Figure 3, D–I). At metaphase both proteins display a rather diffuse cortical localization. During anaphase both anillin and actin accumulate at the cell equator, forming wide cortical bands. As cells proceed through telophase the anillin and actin bands narrow down, and the two proteins fully colocalize in the contractile ring.
Figure 3.
Localization of Pav (A–C), actin (D–F), and anillin (G–I) during mitotic division of S2 cells. (A, D, and G) Metaphases. (B, E, and H) Anaphases. (C, F, and I) Telophases. In the merged figures Pav, actin, and anillin are colored in red, tubulin in green, and DNA in blue. Bar, 10 μm.
It is worth noting that the behavior of anillin and actin in S2 cells is rather different from that described previously in meiotic cells of Drosophila males. In metaphase and anaphase A, anillin and actin do not exhibit any detectable cortical accumulations in spermatocytes. During anaphase B, anillin abruptly concentrates in a narrow equatorial band before the assembly of the actin-based contractile ring. In late anaphase, spermatocytes assemble a narrow actomyosin ring that precisely colocalizes with the anillin band throughout meiotic division (Giansanti et al., 1999). In contrast, the two proteins arrive at the equatorial cortex simultaneously in S2 cells, forming wide bands. The reasons why spermatocytes and S2 cells concentrate anillin and actin in the cleavage furrow with different dynamics are not understood. However, the pattern of anillin and actin accumulation seen in S2 cells seems to be typical of mitotic cells, because it has been observed also in dividing larval neuroblasts (Giansanti et al., 2001).
With these cytological techniques in hand, we examined cell division in cultures treated for 72 h with dsRNA. Cells collected from these cultures were fixed and stained for both DNA and tubulin, and for either Pav, F-actin, or anillin. The analysis of these preparations revealed that (RNAi) cells display gene-specific cytological phenotypes and allowed definition of the primary defects that cause cytokinesis failures.
Phenotypes of acGAP, pav, pbl, rho1, and sqh Mutants
In cells treated with dsRNA for either acGAP (rhoGAP), pav (kinesin-like), pbl (rhoGEF), rho1, or sqh (myosin II regulatory light chain) the metaphase and anaphase figures are morphologically normal and the frequencies of anaphases relative to metaphases are comparable with those of untreated controls. However, in all these RNAi-induced mutants, telophases are severely affected (Table 2 and Figure 4). In addition to a very few, morphologically normal telophases with a fully developed central spindle (henceforth defined as “long” telophases; Figure 3, C, F, and I), these mutants display many characteristic telophases shorter in length than normal counterparts (henceforth defined as “short” telophases; Figure 4). These peculiar mitotic figures can be easily distinguished from anaphases because they exhibit typical telophase nuclei with fully decondensed chromosomes. Yet, they are very different from regular telophases, because they lack the central spindle and are substantially shorter than normal telophases. In control S2 cells undergoing anaphase A, the pole-to-pole distance is 17.6 μm (n = 42). This distance increases during anaphase B, so that telophase figures are 23.7 μm (n = 110) long. The short telophases observed in acGAP, pav, pbl, rho1, and sqh RNAi-induced mutants have pole-to-pole lengths ranging from 18.9 to 19.9 μm (Table 2), and are thus only slightly longer than control anaphase A figures.
Table 2.
Relative frequencies of mitotic figures in RNAi-induced cytokinesis mutants
| Gene/mutant | No. of prometaphases plus metaphases | Frequencies relative to metaphases (×100)
|
Average length of short telophases | ||
|---|---|---|---|---|---|
| Anaphases | Long telophases | Short telophasesa | |||
| control | 648 | 25 | 65 | 0 | |
| acGAP | 381 | 22 | 12 | 20 | 19, 6 |
| ani | 201 | 21 | 72b | 0 | |
| pbl | 164 | 22 | 4 | 20 | 19, 9 |
| pav | 297 | 26 | 1 | 30 | 19, 9 |
| rho1 | 229 | 24 | 2 | 18 | 19, 7 |
| sqh | 266 | 26 | 14 | 25 | 19, 0 |
| syx1A | 319 | 22 | 47 | 40 | 21, 2 |
| tsr | 256 | 29 | 52 | 0 | |
Only diploid cells have been scored. To calculate the relative frequencies of anaphases, long telophases and short telophases, the observed number of each class of mitotic figures has been divided by the total number of prometaphases plus metaphases.
In counting the short telophases in acGAP, pav, pbl, rho1, and sqh (RNAi) cells we only recorded those cells where the remnants of the spindle and/or the actin staining clearly identified them as aberrant telophase figures. We did not score cells that had the same shape and nuclear morphology of the short telophases but cannot be unambiguously distinguished from binucleated interphase cells. Most likely this conservative criterion led to an underestimation of the actual number of short telophases.
Seventy-one percent of the 145 telophases scored showed large membrane bulges at the cell equator.
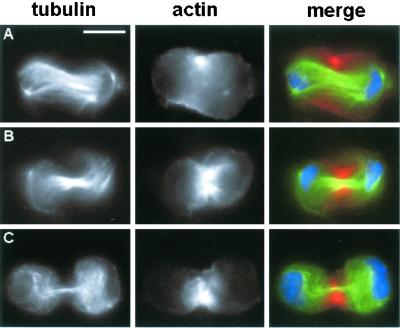
Figure 4.
Examples of aberrant (short) telophases observed in pav (kinesin-like), rho1, and sqh (myosinII regulatory light chain) (RNAi) cells. Cells were stained for tubulin (green), DNA (blue), and either Pav, actin, or anillin (red). (A–C) rho1 (RNAi) telophases. (D–F) sqh (RNAi) telophases. (G and H) pav (RNAi) telophases. (A and D) Pav immunostaining. (B, E, and G) Actin immunostaining. (C, F, and H) Anillin staining. Note that all these aberrant telophases exhibit severe defects in the central spindle, which is either absent or very poorly organized. In addition, in all telophases actin does not form a contractile ring but displays characteristic cortical localizations (see text). Pav localization is also severely disrupted; Pav staining never traverses the cells as occurs in controls but it is either absent or associated with the small and irregular microtubules bundles that occasionally form between the two daughter nuclei. Bar, 10 μm.
Staining for either anillin, actin, or Pav revealed that the few normal, long telophases observed in acGAP, pbl, rho1, and sqh (RNAi) cells exhibit normal accumulations of these proteins in the cleavage furrow. However, in the short telophases of all these (RNAi) cultures the localization of actin, anillin, and Pav is disrupted. Immunostaining with anti-Pav antibody did not detect any signal in the short telophases of both acGAP and pav RNAi-induced mutants. In the short telophases of all the other mutants, Pav staining is either absent or associated with small irregular bundles of microtubules laying between the two daughter nuclei (Figure 4). Staining with rhodamine-phalloidin showed that the short telophases of all mutants lack a regular actin-enriched contractile ring. In rho1 short telophases actin is enriched at the polar cortex but is excluded from the equatorial region (Figure 4B). In pbl aberrant telophases F actin exhibits a uniform cortical localization. A uniform F actin distribution is also observed in 20–30% of acGAP, pav, and sqh short telophases, whereas in the remaining aberrant telophases seen in these (RNAi) cells F actin concentrates in a wide equatorial cortical band (Figure 4, E and G). In the aberrant telophases of all these RNAi cells the pattern of anillin localization resembles that of F actin (Figure 4).
We would like to point out that in acGAP, pav, pbl, rho1, and sqh (RNAi) cells we never observed telophase figures with a normal central spindle and a poorly organized contractile ring, or with a normal actin ring and a defective central spindle. Moreover, in these (RNAi) cultures morphologically normal telophases either occur at very low frequencies (acGAP and sqh) or are virtually absent (pav, pbl, and rho1) (Table 2). Taken together, these observations strongly suggest that the short telophases do not arise from regular telophases that have failed to maintain their cytokinetic structures. Rather, it is likely that these aberrant telophases result from anaphases that have failed to form both the central spindle and the contractile ring and to undergo normal spindle elongation.
To summarize, our results show that in acGAP, pav, pbl, rho1, and sqh (RNAi) cultures most telophases are poorly elongated and lack both the central spindle and the actin-based contractile ring. In addition, anillin fails to concentrate properly in the contractile ring of all these aberrant short telophases. Due to the absence of a contractile apparatus these mutants telophases fail to undergo cytokinesis, giving rise to binucleated cells.
Phenotypes of syx1A, tsr, and ani Mutants
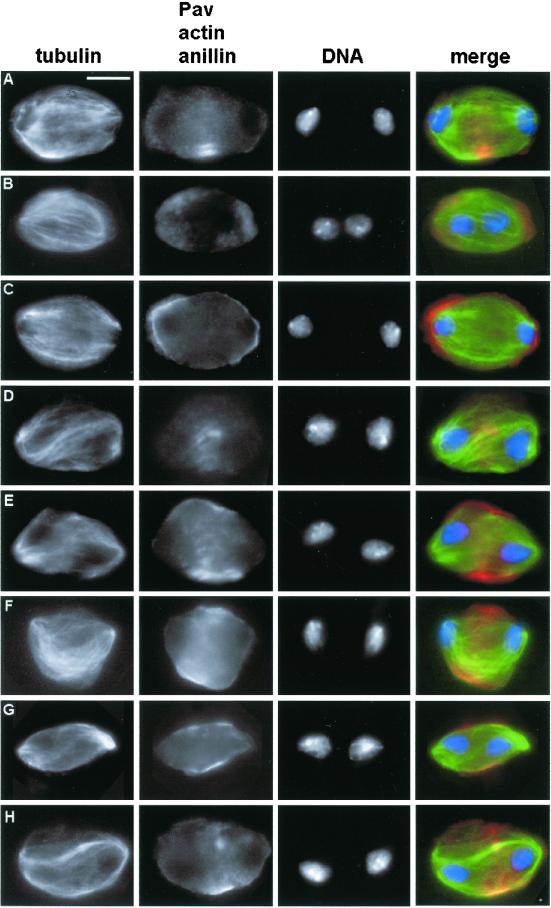
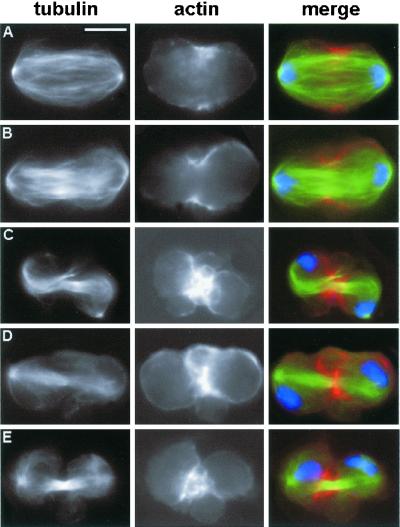
In cells treated with dsRNA for the syx1A gene (encoding a t-SNARE) the anaphase/metaphase ratio is comparable with that of controls (Table 2). However, the telophase/metaphase ratio in these cultures is higher than in untreated cells (Table 2), suggesting that depletion of Syx1A increases the duration of telophase. About half of the telophases observed in syx1A mutants are morphologically normal and exhibit normal accumulations of actin, anillin, and Pav. The other half of syx1A mutant telophases are shorter than their normal counterparts (21.2 μm [n = 95] vs. 23.7 μm [n = 110]) and exhibit poorly organized central spindles that contain fewer microtubules than those of normal telophases (Figure 5, A–C). The abnormality of these central spindles is underlined by the irregular distribution of the Pav protein, which, instead of accumulating in the midzone, concentrates in a few patches associated with regions of higher microtubule density (Figure 5A). The short syx1A telophases always display actin and anillin in wide equatorial bands similar to those present in late anaphases of control cells (Figure 5, B and C). Thus, in syx1A mutants a substantial fraction of telophases fail to assemble a normal central spindle and to undergo full elongation. Although these cells accumulate both actin and anillin in the equatorial region, they fail to form a morphologically normal contractile apparatus and cannot undergo the cytokinetic process.
Figure 5.
Abnormal telophases observed in syx1A (RNAi) cells. Cells were stained for tubulin (green), DNA (blue), and either Pav (A, red), actin (B, red), or anillin (C, red). Note the defective central spindle and contractile ring, and the abnormal distribution of Pav. Bar, 10 μm.
Cells treated with dsRNA for the tsr gene have regular frequencies of anaphases and telophases (Table 2). tsr mutant ana-telophases display regular central spindles and normal accumulations of Pav and anillin, but they exhibit an excess of F-actin with respect to normal cells (Figure 6). The actin excess is particularly evident in late telophases, which often contain very prominent and misshaped actin rings (Figure 6B). These abnormal accumulations of F actin persist even in the terminal stages of cell division when the two daughter cells are connected only by a very thin intercellular bridge (Figure 6C). A very similar phenotype has been observed in late meiotic telophases of tsr mutant males, where the contractile ring overgrows and fails to disassemble (Gunsalus et al., 1995; Giansanti et al., 1999). tsr encodes a polypeptide homologous to cofilins (Gunsalus et al., 1995), a family of low molecular mass actin-binding proteins that can severe and depolymerize actin filaments in vitro (Moon and Drubin, 1995). This suggest that in Tsr-depleted cells the actin filaments of the contractile ring are not properly degraded, leading to the formation of large and persistent F-actin aggregates that are likely to create a physical obstruction to the completion of cytokinesis.
Figure 6.
Excessive actin accumulation in the contractile apparatus of tsr (RNAi) cells. Cells were stained for tubulin (green), DNA (blue), and actin (red). (A) Late anaphase/early telophase. (B and C) Late telophases. Bar, 10 μm.
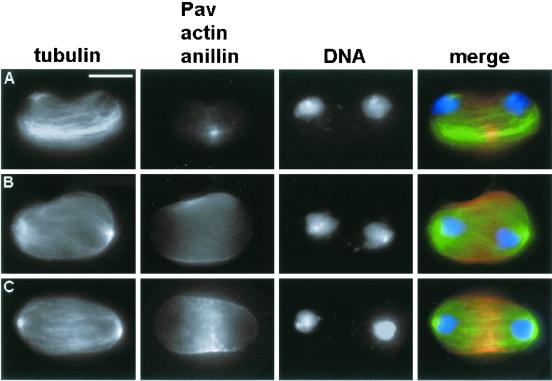
Cells treated with ani dsRNA also exhibit normal frequencies of anaphases and telophases (Table 2). Both types of mitotic figures have completely normal spindles and normally concentrate the Pav protein in the central spindle midzone (Figure 7). Anillin-depleted late anaphases and early telophases also assemble a morphologically normal contractile ring (Figure 7). However, mutant late telophases display severe disruptions of both the contractile ring and membrane organization in the cleavage area. Approximately 70% of these cells contain prominent, aberrant membrane bulges at the cell equator (Figure 7). In telophases with membrane protrusions the actin ring displays an aberrant morphology and F-actin diffuses along the irregular membrane bulges (Figure 7). The absence of anillin thus seems to disrupt the membrane–contractile ring interactions that mediate completion of cytokinesis.
Figure 7.
Abnormal membrane behavior during late telophase of ani (RNAi) cells. Cells were stained for tubulin (green), DNA (blue), and actin (red). (A and B) Anaphase (A) and early telophase (B) figures with normal actin accumulations. (C–E) Late telophases showing large membrane bulges in the cleavage area. The actin-associated fluorescence of these cells has been artificially enhanced to visualize the membrane bulges; these aberrant telophases do not seem to contain more actin than their control counterparts. Bar, 10 μm.
DISCUSSION
Efficiency of RNAi in Detecting Genes Involved in Cytokinesis
Our data show that treatment with dsRNA for either acGAP, ani, pav, pbl, rho1, sqh, syx1A, or tsr disrupts cytokinesis in S2 tissue culture cells. Mutations in pav, pbl, rho1, sqh, and tsr have been reported to disrupt Drosophila mitotic cytokinesis (Table 1). Herein, we show for the first time that the acGAP, ani, and syx1A genes are also required for cytokinesis in flies. Our experiments indicate that the penetrance of the RNAi effect is very high; in cultures exposed for 72 h to either ani, pav, pbl, rho1, sqh, or tsr dsRNA >70% of the telophases are affected, whereas RNAi of acGAP and syx1A produces 46 and 63% abnormal telophases, respectively.
The finding that chic, fwd, and klp3A are not required for cytokinesis in S2 cells is not surprising, because previous studies pointed toward a specific involvement of these genes in meiotic cytokinesis of males. Null mutations in klp3A, a gene encoding a kinesin-like protein expressed both in testes and somatic tissues, disrupt meiotic cytokinesis but have no effect on larval neuroblast division (Williams et al., 1995). Similarly, flies homozygous for null mutations in fwd, which encodes a phosphatidyl-inositol kinase, are viable but male sterile, and are specifically defective in male meiotic cytokinesis (Brill et al., 2000). In contrast with fwd and klp3A that are not required for viability, chic is an essential gene that specifies a Drosophila homolog of profilin (Cooley et al., 1992). However, both male sterile chic mutants and heteroallelic chic combinations resulting in lethality, display severe disruptions in meiotic cytokinesis but have no defects in neuroblast cytokinesis (Giansanti et al., 1998; Giansanti, Bonaccorsi, and Gatti, unpublished data).
We were initially surprised to find that RNAi depletion of the Pnut protein, which shares homology with the yeast septins, did not markedly affect cytokinesis in S2 cells. This protein concentrates in the cleavage furrow of several Drosophila cell types; null pnut mutants die at the larval/pupal boundary and exhibit polyploid cells in their brains, consistent with a defect in cytokinesis (Neufeld and Rubin, 1994). It is possible that the lack of an effect in pnut (RNAi) cells reflects a small amount of residual Pnut protein in these cells, as seen in Figure 1A. However, we instead believe that Pnut's role in cytokinesis is not fundamental to the process. We have reexamined the larval brains of null pnut mutants and have confirmed the presence of polyploid cells. However, we found that polyploid cells represent only 10.5% of the mitotic figures (n = 1558), indicating that most neuroblasts can undergo cytokinesis even in the absence of Pnut (Bonaccorsi, Giansanti, and Gatti, unpublished data). In addition, Pnut is not required for cytokinesis during either male meiosis (Bonaccorsi, Giansanti, and Gatti, unpublished data) or the cystoblast divisions in the female germline (Adam et al., 2000). Taken together, these findings indicate that the Pnut function is either partially or totally dispensable for cytokinesis in Drosophila.
In summary, our data indicate that S2 cells are a highly suitable model system for molecular dissection of cytokinesis by RNAi. We have shown that treatments of S2 cells with dsRNA for eight different genes implicated in cytokinesis result in severe disruptions of the process. Moreover, it has been recently reported that RNAi experiments with either aurora B or INCENP dsRNA cause frequent failures of S2 cell cytokinesis (Adams et al., 2001; Giet and Glover, 2001). In contrast, S2 cells do not respond to the ablation of genes that are either specifically involved in meiotic cytokinesis of males (chic, fwd, and klp3A) or that play only a peripheral role during Drosophila cytokinesis (pnut). Taken together, these results predict that genome-wide studies using RNAi in S2 cells will permit identification of the majority of genes that govern cytokinesis in Drosophila mitotic cells.
Phenotypes of acGAP, pav, pbl, rho1, and sqh RNAi Cells Suggest Microtubule–Contractile Ring Interactions
Our phenotypical analyses of RNAi-induced mutants in the acGAP, rho1, and sqh genes provide the first description of the cytological defects that lead to cytokinesis failures when the function of these genes is ablated. Previous studies have shown that mutations in rho1 and sqh disrupt mitotic cytokinesis but have not defined the cytological phenotypes elicited by these mutations (Karess et al., 1991; Prokopenko et al., 1999). In addition, we have characterized pav and pbl (RNAi) cells; the phenotypes of these (RNAi) cells are consistent with those previously observed in animals homozygous for mutations in these genes (Lehner, 1992; Adams et al., 1998; Prokopenko et al., 1999).
Cells in which the acGAP, pav, pbl, rho1, and sqh genes are ablated by RNAi normally undergo anaphase A, but they then fail to elongate and to undergo anaphase B. After anaphase A, mutant cells proceed toward telophase and decondense their chromosomes, forming typical telophase nuclei. However, these cells fail to develop a central spindle, to assemble an actomyosin contractile ring and to concentrate anillin in the cleavage furrow. This results in the formation of short, aberrant telophases that are unable to undergo cytokinesis and will thus give rise to binucleated cells.
We emphasize that the functional ablation of genes influencing either the actin or the microtubule cytoskeleton have similar effects on cytokinesis. The genes pbl, rho1, and sqh likely play primary roles in controlling the actin cytoskeleton. The sqh gene encodes a regulatory light chain of myosin II (Karess et al., 1991). Rho1 is a member of the Rho family GTPases that cycle from an inactive GDP-bound state to an active GTP-bound state under the regulation of guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) (reviewed by Prokopenko et al., 2000). GEFs enhance the exchange of bound GDP for GTP, whereas GAPs increase the GTPase activity of Rho (Prokopenko et al., 2000). Rho proteins (Mabuchi et al., 1993; Drechsel et al., 1997, Nishimura et al., 1998) and Rho GEFs, such as Drosophila Pbl (Prokopenko et al., 1999) and human ECT2 (Tatsumoto et al., 1999), localize to the cleavage furrow and are required for contractile ring assembly. In contrast, the activities of acGAP and pav are likely to primarily influence the function of the central spindle. The Pav kinesin-like protein, a homolog of the C. elegans ZEN-4, is localized in the central spindle, and is thought to mediate microtubule cross-linking at the central spindle midzone (Adams et al., 1998). The acGAP gene encodes a Rho GAP, and it is orthologous to the cyk-4 gene of C. elegans. CYK-4 interacts with ZEN-4, and the two proteins are mutually dependent for their localization to the central spindle (Jantsch-Plunger et al., 2000). The complete absence of Pav immunostaining in acGAP (RNAi) telophases suggests a similar interaction between AcGAP and Pav, pointing to a role of AcGAP in central spindle assembly. In summary, the cytological phenotypes of pbl, rho1, and sqh (RNAi) cells indicate that a primary defect in acto-myosin ring formation results in a secondary defect in central spindle assembly. The phenotypes of AcGAP- and Pav-depleted cells suggest the converse: that a primary defect in the central spindle can secondarily disrupt contractile ring formation. Thus, taken together, these data indicate that the central spindle and the actomyosin ring are interrelated structures. Although we do not currently understand the molecular mechanisms underlying the cross talk between these structures, we can envisage two possibilities. The formation and maintenance of both the central spindle and the actomyosin ring could be mediated by physical interactions between interzonal microtubules and components of the contractile ring. Alternatively, the central spindle and the contractile ring could be coupled by a checkpoint-like regulatory mechanism, which would inhibit the formation of either of these structures when the other is not properly assembled.
Although acGAP, pav, pbl, rho1, and sqh (RNAi) cells display similar terminal phenotypes, the aberrant telophases observed in these cultures differ in both actin and anillin distribution. In rho1 telophases these proteins are excluded from the cell equator, in pbl they are uniformly distributed, and in acGAP, pav, and sqh they concentrate in a wide equatorial band. This suggests that rho1 and pbl are required for actin and anillin accumulation in the equatorial region of the dividing cell. In contrast acGAP, pav, and sqh seem to be required for the assembly of the contractile machinery from proteins already concentrated at the cell equator. In sqh (RNAi) cells the failure to assemble an actomyosin ring is likely to be a direct consequence of the depletion of an essential component of the ring. In acGAP and pav cells this failure is instead likely to be a secondary effect of problems in central spindle assembly.
An interplay between the central spindle and the contractile ring was previously suggested by studies on Drosophila male meiosis. Mutant spermatocytes in the chic, and dia loci, which encode products thought to be involved in contractile ring formation, and mutants in the kinesin-encoding gene klp3A, all display severe defects in both structures (Giansanti et al., 1998; Gatti et al. 2000). Although all the extant results on Drosophila cells strongly suggest an interdependence of the central spindle and the contractile ring, it is currently unclear whether this is true in all animal cells. Studies on mammalian cells have shown that central spindle plays an essential role during cytokinesis (Wheatley and Wang, 1996; Eckley et al., 1997). However, these experiments have provided limited information on whether perturbations in the actomyosin ring assembly disrupt the central spindle (reviewed by Gatti et al., 2000). The best evidence of an interplay between the central spindle and the contractile ring has been provided by Cao and Wang (1996) in rat kidney cells. By puncturing these cells with a blunt needle they created a physical barrier between the central spindle and the equatorial cortex. This barrier not only abrogated actomyosin ring assembly on the side of perforation facing the cortex, but also disrupted the organization of central spindle microtubules on the opposite side.
In contrast, studies on C. elegans embryos indicate that, at least in the early stages of cytokinesis, the actomyosin ring and the central spindle can assemble independently (Powers et al., 1998; Raich et al., 1998; Jantsch-Plunger et al., 2000). Why do Drosophila, and possibly mammalian cells, differ from C. elegans in the interactions between the central spindle and the contractile ring? We believe that the answer to this question reflects differences in the distance between the central spindle and the equatorial cortex. In Drosophila and mammalian cells during central spindle assembly the equatorial cortex is very close to the interzonal microtubules. In contrast, in C. elegans embryos the central spindle assembles in the center of the cell when the cleavage furrow has just began to ingress, so that during their assembly the actomyosin ring and the central spindle lie a considerable distance apart. Only later in cell division, after substantial furrow ingression, can the actomyosin ring and the central spindle come into contact. We thus hypothesize that in embryonic cells of C. elegans the cytokinetic process consists of two steps: an early step, where the central spindle and the contractile ring assemble independently in distant cellular regions, and a late step that begins when the central spindle and the contractile ring have come into contact. The early stage might be mediated by interactions between astral (rather than central spindle) microtubules and the contractile ring. The late step of C. elegans cytokinesis may then require that the contractile ring and the central spindle interact cooperatively to complete cytokinesis successfully. This two-step hypothesis also applies to other large cells, such as echinoderm eggs, where the central spindle and the cortex are separated by large masses of cytoplasmic material and seem to assemble independently (Rappaport, 1985)
Phenotypes of syx1A and ani (RNAi) Cells Reveal Membrane–Contractile Ring Interactions
The syx1A gene, which encodes a t-SNARE, plays an essential role in embryonic cellularization (Burgess et al., 1997), but its direct role in cytokinesis has not been demonstrated. In syx1A (RNAi) cells approximately half of the telophases are shorter that those of control cells and display severe defects in both the central spindle and the contractile ring. These findings are rather surprising, because there is abundant evidence that syntaxins are specifically involved in membrane fusion processes (reviewed by Chen and Scheller, 2001). Thus, our observations on syx1A (RNAi) cells raise the question of how a defect in membrane formation can affect both the central spindle and contractile ring assembly. Studies of C. elegans embryos depleted of the cytokinesis-specific Syntaxin-4 protein by RNAi have shown that in some of these embryos there is a complete failure of cleavage furrow ingression, suggesting an underlying defect in the contractile ring machinery. It has been thus proposed that formation of new membrane may positively regulate contractile ring assembly (Jantsch-Plunger and Glotzer, 1999). In agreement with this hypothesis, we suggest that RNAi-induced Syx1A depletion in S2 cells disrupts membrane formation at the site of cleavage furrow, causing a secondary defect in contractile ring formation and thus also in central spindle assembly.
Although anillin localizes in the cleavage furrow of a variety of Drosophila cell types its involvement in Drosophila cytokinesis has never been demonstrated (Field and Alberts, 1995; Giansanti et al., 1999). The human homolog of anillin is required for cytokinesis but its role in the process is unclear (Oegema et al., 2000). In ani (RNAi) late anaphases and early telophases the central spindle and the contractile ring are both morphologically normal. However, in many late telophases the cleavage area displays large membrane protrusions and an aberrant morphology of the actin-based ring. Anillin is thus not required for the initial formation and contraction of the actomyosin ring. Rather, it seems that this protein plays an essential role in regulating membrane behavior during the late steps of cytokinesis. Anillin contains an actin-binding domain and a PH domain (Field and Alberts, 1995; Oegema et al., 2000); PH domains are found in many membrane-associated proteins and have been implicated in protein–protein and protein–phospholipid interactions (reviewed by Rebecchi and Scarlata, 1998). Based on these biochemical properties, we suggest that anillin interacts with both the plasma membrane and the actin-based contractile apparatus, regulating the membrane-contractile ring interactions that mediate completion of cytokinesis.
ACKNOWLEDGMENTS
We thank L. Cooley for anti-Chic antibody, S. Elgin for anti-HP1 antibody, C. Field for anti-anillin antibody, D. Glover for anti-Pav antibody, M. Goldberg for anti-KLP3A antibody, and T. Neufeld for anti-Pnut antibody. We also thank M. Goldberg, S. Bonaccorsi and M. G. Giansanti for critical readings of the manuscript. This article is dedicated to the memory of Franco Tatò.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–12–0589. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–12–0589.
REFERENCES
- Adam JC, Pringle JR, Peifer M. Evidence for functional differentiation among Drosophila septins in cytokinesis and cellularization. Mol Biol Cell. 2000;11:3123–3135. doi: 10.1091/mbc.11.9.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams RR, Maiato H, Earnshaw WC, Carmena M. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J Cell Biol. 2001;153:865–880. doi: 10.1083/jcb.153.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams RR, Tavares AA, Salzberg A, Bellen HJ, Glover DM. pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 1998;12:1483–1494. doi: 10.1101/gad.12.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi S, Giansanti MG, Gatti M. Spindle self-organization and cytokinesis during male meiosis in asterless mutants of Drosophila melanogaster. J Cell Biol. 1998;142:751–761. doi: 10.1083/jcb.142.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill JA, Hime GR, Scharer-Schuksz M, Fuller MT. A phospholipid kinase regulates actin organization and intercellular bridge formation during germline cytokinesis. Development. 2000;127:3855–3864. doi: 10.1242/dev.127.17.3855. [DOI] [PubMed] [Google Scholar]
- Brown NH, Kafatos FC. Functional cDNA libraries from Drosophila embryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- Burgess RW, Deitcher DL, Schwarz TL. The synaptic protein syntaxin1 is required for cellularization of Drosophila embryos. J Cell Biol. 1997;138:861–875. doi: 10.1083/jcb.138.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L-G, Wang Y-L. Signals from the spindle midzone are required for the stimulation of cytokinesis in cultured epithelial cells. Mol Biol Cell. 1996;7:225–232. doi: 10.1091/mbc.7.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Scheller RH. SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci USA. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L, Verheyen E, Ayers K. chickadee encodes a profilin required for intercellular cytoplasm transport during Drosophila oogenesis. Cell. 1992;69:173–184. doi: 10.1016/0092-8674(92)90128-y. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Juan G, Li X, Gorczyca W, Murakami T, Traganos F. Cytometry in cell necrobiology: analysis of apoptosis and accidental cell death (necrosis) Cytometry. 1997;27:1–20. [PubMed] [Google Scholar]
- Drechsel DN, Hyman AA, Hall A, Glotzer M. A requirement for Rho and Cdc42 during cytokinesis in Xenopus embryos. Curr Biol. 1997;7:12–23. doi: 10.1016/s0960-9822(06)00023-6. [DOI] [PubMed] [Google Scholar]
- Eckley DM, Ainsztein AM, Mackay AM, Goldberg IG, Earnshaw WC. Chromosomal proteins and cytokinesis: patterns of cleavage furrow formation and inner centromere protein positioning in mitotic heterokaryons and mid-anaphase cells. J Cell Biol. 1997;136:1169–1183. doi: 10.1083/jcb.136.6.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field C, Alberts BM. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J Cell Biol. 1995;131:165–178. doi: 10.1083/jcb.131.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CM, Kellogg D. Septins: cytoskeletal polymers or signaling GTPases? Trends Cell Biol. 1999;9:387–394. doi: 10.1016/s0962-8924(99)01632-3. [DOI] [PubMed] [Google Scholar]
- Gatti M, Giansanti MG, Bonaccorsi S. Relationships between the central spindle, and the contractile ring during cytokinesis in animal cells. Microsc Res Tech. 2000;49:202–208. doi: 10.1002/(SICI)1097-0029(20000415)49:2<202::AID-JEMT13>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Giansanti MG, Bonaccorsi S, Gatti M. The role of anillin in meiotic cytokinesis of Drosophila males. J Cell Sci. 1999;112:2323–2334. doi: 10.1242/jcs.112.14.2323. [DOI] [PubMed] [Google Scholar]
- Giansanti MG, Bonaccorsi S, Williams B, Williams EV, Santolamazza C, Goldberg ML, Gatti M. Mutations in the profilin-encoding gene chickadee reveal interactions between the central spindle and the contractile ring during meiotic cytokinesis in Drosophila melanogaster males. Genes Dev. 1998;12:396–410. doi: 10.1101/gad.12.3.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti MG, Gatti M, Bonaccorsi S. The role of centrosomes and astral microtubules during asymmetric division of Drosophila neuroblasts. Development. 2001;128:1137–1145. doi: 10.1242/dev.128.7.1137. [DOI] [PubMed] [Google Scholar]
- Giet R, Glover DM. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. The mechanism and control of cytokinesis. Curr Opin Cell Biol. 1997;9:815–823. doi: 10.1016/s0955-0674(97)80082-8. [DOI] [PubMed] [Google Scholar]
- Goldberg ML, Gunsalus K, Karess RE, Chang F. Cytokinesis, or breaking up is hard to do. In: Endow S, Glover D, editors. Mechanics of Cell Division. Oxford, England: Oxford University Press; 1998. pp. 270–316. [Google Scholar]
- Gunsalus K, Bonaccorsi S, Williams E, Vernì F, Gatti M, Goldberg ML. Mutations in twinstar, a Drosophila gene encoding a Cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J Cell Biol. 1995;131:1243–1259. doi: 10.1083/jcb.131.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TC, Eissenberg JC, Craig C, Dietrich V, Hobson A, Elgin SC. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur J Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- Jantsch-Plunger V, Glotzer M. Depletion of syntaxins in the early Caenorhabditis elegans embryo reveals a role for membrane fusion events in cytokinesis. Curr Biol. 1999;9:738–745. doi: 10.1016/s0960-9822(99)80333-9. [DOI] [PubMed] [Google Scholar]
- Jantsch-Plunger V, Gonczy P, Romano A, Schnabel H, Hamill D, Schnabel R, Hyman AA, Glotzer M. CYK-4: A Rho family gtpase activating protein (GAP) required for central spindle formation and cytokinesis. J Cell Biol. 2000;149:1391–1404. doi: 10.1083/jcb.149.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess RE, Edwards KA, Kulkarni S, Aguilera I, Kiehart DP. The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell. 1991;65:1177–1189. doi: 10.1016/0092-8674(91)90013-o. [DOI] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jurgens G. The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol. 1997;139:1485–1493. doi: 10.1083/jcb.139.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Yuan YL, Kuriyama R, Erikson RL. Plk is an M-phase-specific protein kinase and interacts with a kinesin-like protein, CHO1/MKLP-1. Mol Cell Biol. 1995;15:7143–7151. doi: 10.1128/mcb.15.12.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner CF. The pebble gene is required for cytokinesis in Drosophila. J Cell Sci. 1992;103:1021–1030. doi: 10.1242/jcs.103.4.1021. [DOI] [PubMed] [Google Scholar]
- Mabuchi I, Hamaguchi Y, Fujimoto H, Morii N, Mishima M, Narumiya S. A rho-like protein is involved in the organization of the contractile ring in dividing sand dollar eggs. Zygote. 1993;1:325–331. doi: 10.1017/s0967199400001659. [DOI] [PubMed] [Google Scholar]
- Moon A, Drubin DO. The ADF/cofilin proteins: stimulus responsive modulators of actin dynamics. Mol Biol Cell. 1995;6:1423–1431. doi: 10.1091/mbc.6.11.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP, Rubin GM. The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell. 1994;77:371–379. doi: 10.1016/0092-8674(94)90152-x. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Nakano K, Mabuchi I. Localization of Rho GTPase in sea urchin eggs. FEBS Lett. 1998;441:121–126. doi: 10.1016/s0014-5793(98)01531-2. [DOI] [PubMed] [Google Scholar]
- Nislow C, Lombillo VA, Kuriyama R, McIntosh JR. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 1992;359:543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- Oegema K, Savoian MS, Mitchison TJ, Field C. Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J Cell Biol. 2000;150:539–551. doi: 10.1083/jcb.150.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J, Bossinger O, Rose D, Strome S, Saxton W. A nematode kinesin required for cleavage furrow advancement. Curr Biol. 1998;8:1133–1136. doi: 10.1016/s0960-9822(98)70470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopenko SN, Brumby A, O'Keefe L, Prior L, He Y, Saint R, Bellen HJ. A putative exchange factor for Rho1 GTPase is required for initiation of cytokinesis in Drosophila. Genes Dev. 1999;13:2301–2314. doi: 10.1101/gad.13.17.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopenko SN, Saint R, Bellen HJ. Untying the Gordian knot of cytokinesis. Role of small G proteins and their regulators. J Cell Biol. 2000;148:843–848. doi: 10.1083/jcb.148.5.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raich WB, Moran AN, Rothman JH, Hardin J. Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol Biol Cell. 1998;9:2037–2049. doi: 10.1091/mbc.9.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport R. Repeated furrow formation from a single mitotic apparatus in cylindrical sand dollar eggs. J Exp Zool. 1985;234:167–171. doi: 10.1002/jez.1402340120. [DOI] [PubMed] [Google Scholar]
- Rebecchi MJ, Scarlata S. Pleckstrin homology domains: a common fold with diverse functions. Annu Rev Biophys Biomol Struct. 1998;27:503–528. doi: 10.1146/annurev.biophys.27.1.503. [DOI] [PubMed] [Google Scholar]
- Robinson DN, Spudich JA. Towards a molecular understanding of cytokinesis. Trends Cell Biol. 2000;10:228–237. doi: 10.1016/s0962-8924(00)01747-5. [DOI] [PubMed] [Google Scholar]
- Severson AF, Hamill DR, Carter JC, Schumacher J, Bowerman B. The aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr Biol. 2000;10:1162–1171. doi: 10.1016/s0960-9822(00)00715-6. [DOI] [PubMed] [Google Scholar]
- Skop AR, Bergmann D, Mohler WA, White JG. Completion of cytokinesis in C. elegans requires a brefeldin A-sensitive membrane accumulation at the cleavage furrow apex. Curr Biol. 2001;11:735–746. doi: 10.1016/s0960-9822(01)00231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Field CM. Microtubules, membranes and cytokinesis. Curr Biol. 2000;10:760–770. doi: 10.1016/s0960-9822(00)00746-6. [DOI] [PubMed] [Google Scholar]
- Swan KA, Severson AF, Carter JC, Martin PR, Schnabel H, Schnabel R, Bowerman B. cyk-1: a C. elegans FH gene required for a late step in embryonic cytokinesis. J Cell Sci. 1998;111:2017–2027. doi: 10.1242/jcs.111.14.2017. [DOI] [PubMed] [Google Scholar]
- Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J Cell Biol. 1999;147:921–928. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen EM, Cooley L. Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development. 1994;120:717–728. doi: 10.1242/dev.120.4.717. [DOI] [PubMed] [Google Scholar]
- Wassermann S. FH proteins as cytoskeletal organizers. Trends Cell Biol. 1998;8:111–115. doi: 10.1016/s0962-8924(97)01217-8. [DOI] [PubMed] [Google Scholar]
- Wheatley SP, Wang Y. Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J Cell Biol. 1996;135:981–989. doi: 10.1083/jcb.135.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BW, Riedy MF, Williams EV, Gatti M, Goldberg ML. The Drosophila Kinesin-like protein KLP3A is a midbody component required for central spindle assembly and initiation of cytokinesis. J Cell Biol. 1995;129:709–723. doi: 10.1083/jcb.129.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]