Abstract
Digestive and psychiatric disorders tend to co-occur, yet mechanisms remain unclear. Leveraging genetic and transcriptomic data integration, we conduct multi-trait analysis of GWAS (MTAG) and weighted gene co-expression network analysis (WGCNA) to explore shared mechanism between psychiatric and gastrointestinal disorders. Significant genetic correlations were found between these disorders, especially in irritable bowel syndrome (IBS), gastroesophageal reflux disease (GERD), depression (DEP), and neuroticism (NE). MTAG identify 60 novel pleiotropic loci for IBS and 14 for GERD, predominantly located near genes associated with neurological pathways. Further WGCNA identifies multiple co-expression modules enriched with genes involved in neurological pathways in digestive tissues, with some modules strongly preserved across brain and digestive tissues. Moreover, our network analysis suggests BSN, CELF4, and NRXN1 as central players in the regulation of the gut-brain axis (GBA). This study enhances our understanding of the GBA and underscores BSN, CELF4, and NRXN1 as crucial targets for future research.
Subject terms: Computational biology and bioinformatics, Genetics
Genetics and transcriptomics reveal shared mechanisms of psychiatric and gastrointestinal disorders, highlighting neurological pathways and pleiotropic loci. Genes BSN, CELF4, NRXN1 regulate gut-brain axis, revealing potential therapeutic targets.
Introduction
The intricate interplay of genetics, lifestyle factors, and environmental influences contributes to the prevalence and diversity of gastrointestinal tract diseases, making them a notable concern in global public health1–4. Patients with psychiatric disorders, like depression (DEP) and anxiety (ANX), often manifest a higher prevalence of digestive symptoms5,6. Similarly, individuals with digestive diseases are more prone to experiencing psychiatric comorbidities7,8. In line with these observations, a Mendelian randomization study has provided compelling evidence for the genetic predisposition of DEP to an elevated risk of a spectrum of digestive tract diseases9, suggesting the presence of shared genetic risk factors underlying these two disease categories.
With increasing sample sizes across phenotypes, genome-wide association studies (GWAS) have become an invaluable tool for uncovering the genetic basis of digestive and psychiatric disorders10–13. However, conventional GWAS approaches only concentrate on individual traits in isolation. In recent years, there has been a growing focus on investigating the genetic overlap between psychiatric disorders and digestive diseases using genome-wide cross-trait analyses14–16. The study by Markos Tesfaye et al. used bivariate MiXeR analysis to find extensive polygenic overlap between irritable bowel syndrome (IBS) and psychiatric disorders, and will apply conditional FDR (condFDR) to identify 70 unique loci shared between IBS and psychiatric disorders. Genome-wide pleiotropy analyses of four gastrointestinal disorders (IBS etc.) and six psychiatric disorders (major depressive disorder etc.) identified a total of 2910 unique loci under the composite null hypothesis (PLACO), with 83 pleiotropic loci and 24 co-localized loci. Although the above studies were only at the genomic level, these advanced statistical methods enhance the ability of GWAS to identify pleiotropic genetic variants that commonly influence both digestive and psychiatric disorders. Furthermore, despite consistent findings that the gut–brain axis (GBA) plays a crucial role in the etiology of digestive disorders, its underlying mechanisms remain elusive. Further investigation is needed to address critical questions such as: (1) Is the presence of GBA-associated genes a primary and determining factor influencing the development of digestive tract diseases? (2) Does the impact of GBA genes on these disorders remain consistent across various digestive tract diseases? (3) In addition to GBA genes, what other genetic factors play an important role in digestive tract diseases? Identifying these factors is crucial for constructing a comprehensive picture of diseases development. (4) Is there a mutual connection between GBA genes at the transcriptional level in both brain and intestinal tissue? Investigating this aspect can shed light on potential gene expression interactions that bridge the gap between the gut and the brain.
To uncover shared genetic underpinnings among digestive tract diseases and psychiatric disorders, we collected GWAS summary statistics for 14 digestive conditions and ten psychiatric disorders. Through multi-trait GWAS analyses, we uncovered numerous pleiotropic genetic loci associated with digestive and psychiatric conditions. In addition, we employed Tissue co-regulation score regression (TCSC) to identify tissue-specific contributions to digestive diseases. Furthermore, a comprehensive gene co-expression network analysis was conducted to pinpoint key genes potentially linked to the mechanisms of GBA underlying digestive and psychiatric diseases.
Results
The overview for our study is provided in Supplementary Fig. 1.
Global genetic correlations among psychiatric disorders and digestive conditions
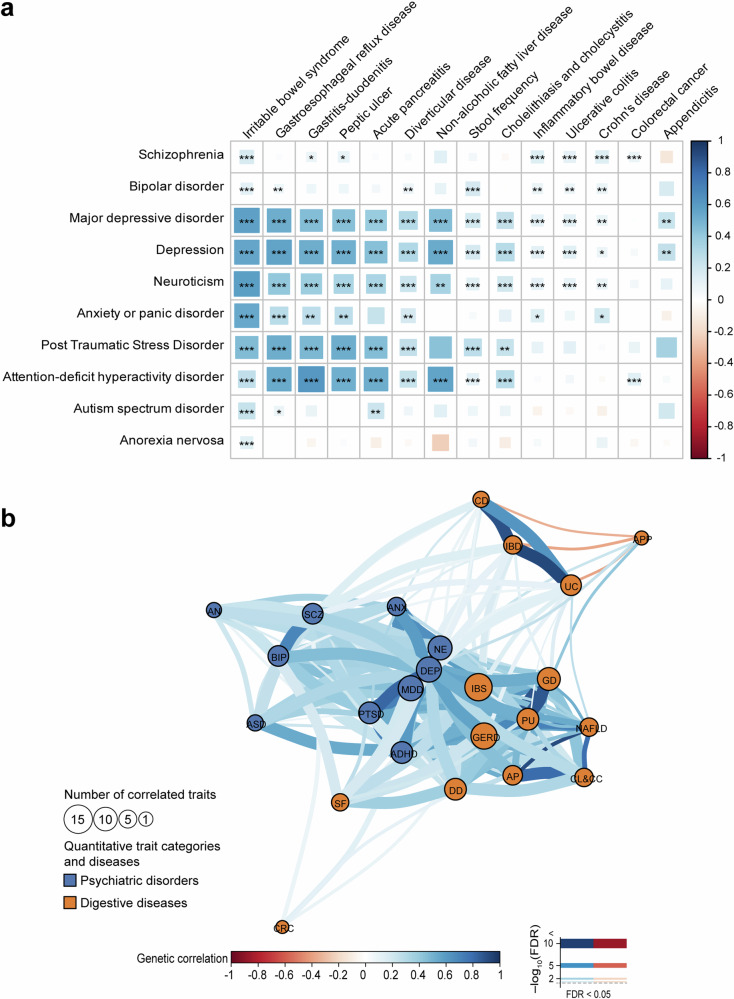
We observed substantial variability in SNP-based heritability for digestive tract diseases, spanning from less than 1–44.42%. Psychiatric disorders also demonstrate a similar pattern, ranging from 63.76 to 3.77% (Supplementary Table 1). We next studied the genetic correlation between psychiatric disorders and digestive diseases. As previously reported9, DEP and major depressive disorder (MDD) exhibit robust correlations with most digestive diseases. The most significant correlation (Rg = 0.57, P = 2.07 × 10−206) was observed between DEP and gastroesophageal reflux disease (GERD) (Supplementary Table 2 and Fig. 1a). In addition, neuroticism (NE), attention-deficit hyperactivity disorder (ADHD), post-traumatic stress disorder (PTSD) and ANX are also associated with multiple digestive diseases (Supplementary Table 2 and Fig. 1a). Among digestive disorders, irritable bowel syndrome (IBS) displayed a notably high degree of correlation with all psychiatric disorders, with the most substantial correlation (Rg = 0.56, P = 3.79 × 10−131) observed with DEP. Additionally, GERD, gastritis–duodenitis (GD), peptic ulcer (PU), acute pancreatitis (AP), diverticular disease (DD) and non-alcoholic fatty liver disease (NAFLD) also demonstrated strong associations with multiple psychiatric disorders (Supplementary Table 2 and Fig. 1a).
Fig. 1. Global genetic correlations among digestive diseases and psychiatric disorders.
a Heatmap of genetic correlation estimates between digestive diseases and psychiatric disorders. Positive genetic correlations are depicted in blue, while negative correlations are represented in red. Larger squares denote more significant FDR values, and asterisks indicate correlations that are statistically significant (FDR < 0.05). ∗FDR < 0.05, ∗∗FDR < 0.01, ∗∗∗FDR < 0.001. b Genetic correlation networks across digestive diseases and psychiatric disorders. Each circle within the network represents a disease or trait, and edges depict significant genetic correlations (FDR < 0.05), with positive correlations in blue and negative correlations in red. Thicker edges on the network correspond to a lower false discovery rate (FDR). IBS irritable bowel syndrome, GERD gastroesophageal reflux disease, PU peptic ulcer, AP acute pancreatitis, NAFLD non-alcoholic fatty liver disease, APP appendicitis, CRC colorectal cancer, IBD inflammatory bowel disease,CD Crohn’s disease, UC ulcerative colitis, SF stool frequency, DD diverticular disease, GD gastritis–duodenitis, ANX anxiety, NE neuroticism, MDD major depressive disorder, DEP depression, SCZ schizophrenia, ADHD attention-deficit hyperactivity disorder, BIP bipolar disorder, PTSD post-traumatic stress disorder.
To further explore the complex relationships among all the examined diseases, we constructed a correlation network based on the pairwise genetic correlations between them (Supplementary Tables 2–4 and Fig. 1b). In the network, the proximity between related phenotypes is determined by the weight of their correlation17. Our observations revealed that psychiatric disorders, including MDD, DEP, and NE were tightly interconnected. These psychiatric disorders further extended their links to other psychiatric conditions including ANX, PTSD, and ADHD as well as digestive diseases like IBS and GERD. For digestive diseases, IBS, GERD, GD, DD, PU, AP, NAFLD and “cholelithiasis and cholecystitis” (CL&CC) were clustered together, while inflammatory bowel disease (IBD), crohn’s disease (CD), ulcerative colitis (UC), appendicitis (APP) and colorectal cancer (CRC) appeared to be separated from the primary cluster.
Local genetic correlations among psychiatric disorders and digestive conditions
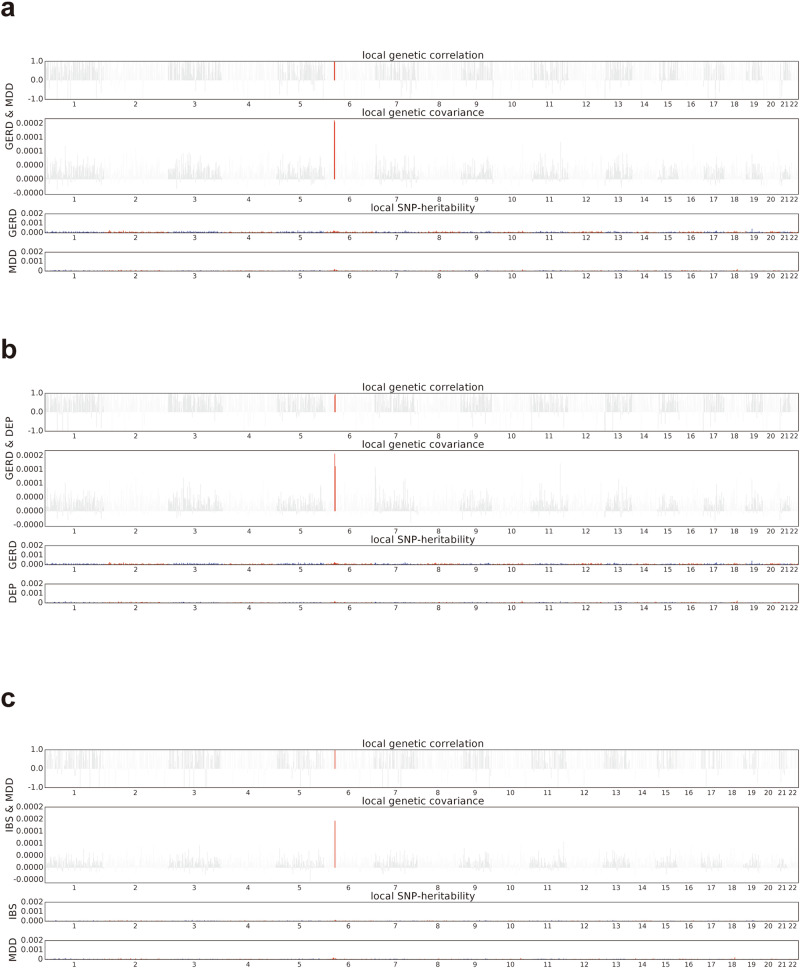
We next conducted a comprehensive genome-wide scan to explore whether specific genomic regions play a role in the shared heritability of genetically related traits. Following correction for multiple testing, we identified a significant correlation between GERD and MDD within the major histocompatibility complex (MHC) region (Fig. 2a). This discovery mirrors the local genetic correlation observed between GERD and DEP (Fig. 2b). Similarly, the local genetic correlation region between IBS and MDD was also identified at MHC region (Fig. 2c). It’s worth noting that in pairs of digestive diseases such as GERD and IBS, which exhibited significant global genetic correlations, no specific genomic region was detected to contribute to heritability (Supplementary Fig. 2a–c).
Fig. 2. Local genetic correlations among digestive diseases and psychiatric disorders.
a Manhattan plot showing the estimates of local genetic correlation, genetic covariance, and SNP heritability between GERD and MDD in Europeans. b Manhattan plot showing the estimates of local genetic correlation, genetic covariance, and SNP heritability between GERD and DEP in Europeans. c Manhattan plot showing the estimates of local genetic correlation, genetic covariance, and SNP heritability between IBS and MDD in Europeans. Red bars represent loci showing significant local genetic correlation after multiple testing adjustment. GERD gastroesophageal reflux disease, IBS irritable bowel syndrome, MDD major depressive disorder, DEP depression.
Cell-type-specific enrichment of SNP heritability
We further investigated the correlated psychiatric disorders and digestive diseases by dividing the SNP heritability based on six chromatin marks and nine cell types (Supplementary Figs. 3–8). GERD, IBS, stool frequency (SF), and psychiatric disorders exhibited similar patterns on the six chromatin marks and are all enriched in the central nervous system (CNS) related tissues or cell types (Supplementary Figs. 3–8). In contrast, IBD, UC, and CD displayed significant enrichment in blood/immune-related tissues or cell types across the six chromatin marks (Supplementary Figs. 3–8). Almost all chromatin marks showed significant enrichment of CRC, CL&CC, DD, and PU in digestive-related tissues or cell types, particularly in tissues from the stomach, small intestine, and large intestine (Supplementary Figs. 3–8). In addition, DD and SF exhibited genetic enrichment in musculoskeletal/connective tissues. CL&CC and AP exhibited different levels of enrichment in liver (H3K9ac, H3K27ac, H3K4me3, Supplementary Figs. 5–7) and pancreas (H3K4me1, H3K4me3, H3K9ac, H3K27ac, Supplementary Figs. 4–7). Furthermore, NAFLD was characterized by an abundance of chromatin marks in adipose tissues, including H3K4me1, H3K9ac, H3K27ac, and H3K36me3 (Supplementary Figs. 4 and 6–8).
Tissue-specific contributions to 14 digestive diseases/traits
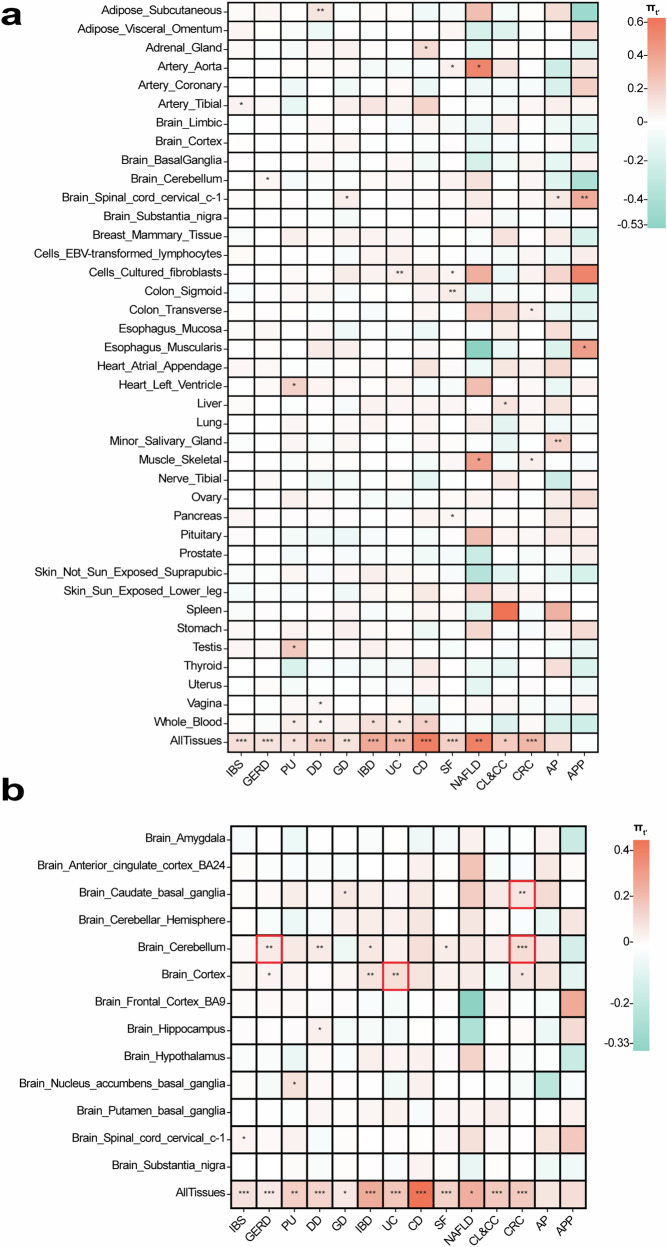
We next employed TCSC to examine the role of specific tissues in the heritability of 14 digestive diseases/traits at the transcriptome level18. This method primarily focuses on uncovering the proportion of disease heritability explained by the cis-genetic component of tissue-specific gene expression. TCSC analysis identified 27 significant causal tissue-trait pairs that made positive contributions to the heritability of diseases/traits (Supplementary Data 1 and Fig. 3a). For instance, whole blood exhibited associations with several digestive diseases, including DD, PU, UC, CD, and IBD, underlining its role in these conditions (Supplementary Data 1 and Fig. 3a). Sigmoid colon, a clinical indicator in various digestive diseases, was associated with SF, while transverse colon and skeletal muscle showed associations with CRC (Supplementary Data 1 and Fig. 3a). In addition, subcutaneous adipose tissue emerged as a causal tissue for DD, in line with clinical observations linking adipose tissue ratios to diverticulitis severity and the impact of obesity on diverticular disease risk (Supplementary Data 1 and Fig. 3a)19,20.
Fig. 3. Estimates of tissue-specific contributions to 14 digestive diseases/traits by TCSC.
TCSC estimates πt′, which denotes the proportion of disease heritability explained by the cis-genetic component of gene expression in tissue t′. a Analyses of all GTEx tissues. b Brain-specific analyses restricting to GTEx brain tissues. Asterisks denotes tissue-trait pairs with an P < 0.05, while red boxes denote tissue-trait pairs with a significant adjusted P (FDR <5%). Tissues are ordered alphabetically. IBS irritable bowel syndrome, GERD gastroesophageal reflux disease, PU peptic ulcer, AP acute pancreatitis, NAFLD non-alcoholic fatty liver disease, APP appendicitis, CRC colorectal cancer, IBD inflammatory bowel disease, CD Crohn’s disease, UC ulcerative colitis, SF stool frequency, DD diverticular disease, GD gastritis–duodenitis.
To enhance TCSC’s power for exploring brain-related digestive diseases/traits, we conducted a brain-specific analysis, focusing on 13 brain tissues from the Genotype-Tissue Expression (GTEx) dataset. This analysis revealed a total of 14 causal tissue-trait pairs, with four pairs showing significant associations (FDR ≤ 0.05). Notably, the cerebellum played a central role in multiple digestive diseases, including GERD, CRC, DD, IBD, and SF (Supplementary Table 5 and Fig. 3b). The cerebral cortex was also identified as the causal tissue for UC, IBD, GERD, and CRC (Supplementary Table 5 and Fig. 3b).
Multi-trait meta-analysis of psychiatric disorders and digestive conditions
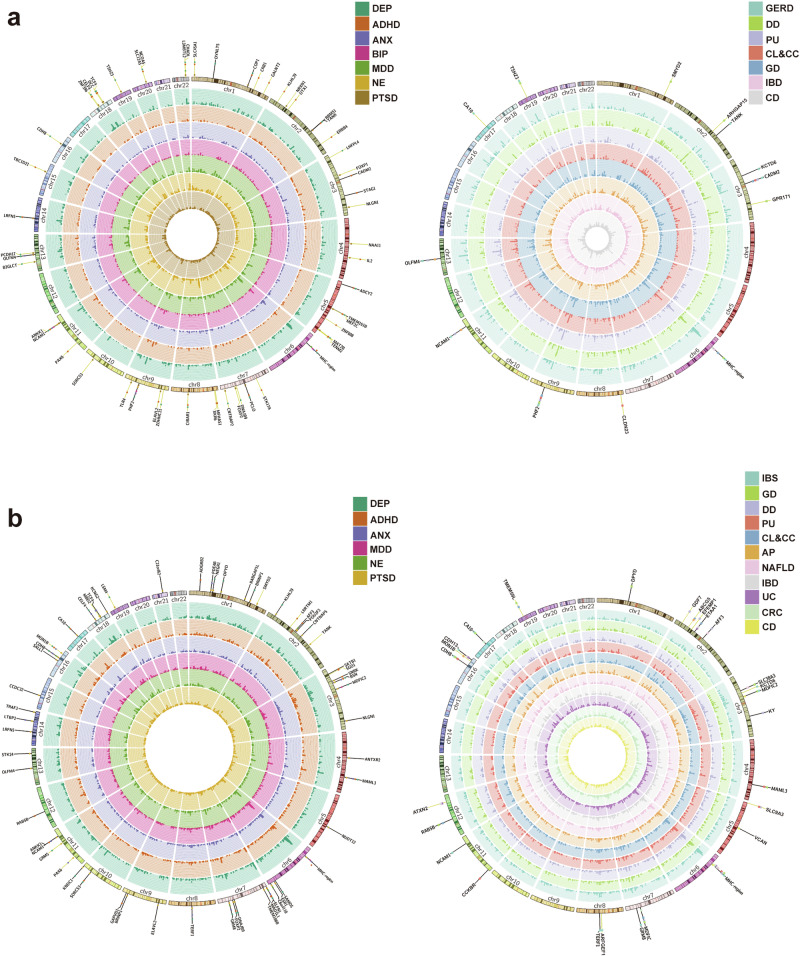
The evidence obtained from genetic correlation analyses between digestive diseases and psychiatric disorders prompted us to further identify the shared pleiotropic loci among them. Given the well-established strong association between IBS and central nervous system (Fig. 1), we initially conducted multi-trait analysis using multi-trait analysis of GWAS (MTAG) to pair IBS with psychiatric disorders that showed significant correlations with IBS, as determined by linkage disequilibrium score regression (LDSC) (Fig. 4a). In total, we identified 94 independent loci, of which 60 were previously unreported in IBS (Supplementary Table 6). As anticipated, most novel loci were discovered in the analysis of IBS and psychiatric conditions showing strong association with IBS, including DEP, MDD, and NE (Supplementary Table 6 and Fig. 4a). Notably, many of the identified loci are situated near genes involved in brain development and synaptic function, such as CAMD2 (3p12.1) and NACM1 (11q23.2), which were also reported in the latest GWAS on IBS21. However, the MTAG analysis revealed more loci potentially linked to neuron development that were previously unknown, such as NRXN1 (2p16.3), NLGN1 (3q26.31), FOXP1 (3p13), and FOXP2 (7q31.1) (Supplementary Table 7 and Fig. 4a). In addition, certain novel pleiotropic loci consistently emerged from the MTAG analysis of IBS and various psychiatric disorders, including CELF4, CIBAR1, DCC, LRFN5, MAT2B, SLC45A1, and TCF4 (Supplementary Table 7). It also worth mentioning that two independent loci (rs2111530 and rs8106322) close to the TSHZ3 were detected from the MTAG analysis of IBS and depression (MDD and DEP) (Supplementary Figs. 9 and 10). In the analysis comparing IBS to other digestive diseases, we found a total of 24 independent signals, with six of them being new discoveries (Supplementary Table 7 and Fig. 4a). In addition to IBS, GERD exhibited strong correlations with multiple psychiatric disorders, as illustrated in Fig. 1. Our MTAG analysis of the association between GERD and psychiatric disorders revealed a total of 93 independent signals, with 14 signals being novel discoveries (Supplementary Table 8 and Fig. 4b). Similar to IBS, certain novel signals were identified in proximity to genes involved in neuron development, such as PAX6 and GRM5 (Supplementary Table 8 and Fig. 4b). Through the analysis of GERD and other digestive diseases, we identified 56 independent signals, 12 of which are novel (Supplementary Table 9 and Fig. 4b). In contrast, MTAG analyses of other digestive disorders with similar genetic correlation patterns yielded far fewer significant loci, particularly those linked to psychiatric traits. These results are included in Supplementary Tables 10–31.
Fig. 4. Overview of the identified pleiotropic loci from multi-trait analyses in IBS and GERD.
a The left circle with dark hues presents the results of MTAG analyses between IBS and psychiatric disorders, while the right circle with light hues displays the results of MTAG analyses between IBS and digestive diseases. b The left circle with dark hues shows the results of MTAG analyses between GERD and psychiatric disorders, and the right circle with light hues illustrates the results for GERD with digestive diseases. Yellow lines indicate novel pleiotropic loci. The color of each dot on the lines corresponds to different diseases analyzed in the meta-analysis. IBS irritable bowel syndrome, GERD gastroesophageal reflux disease, PU peptic ulcer, AP acute pancreatitis, NAFLD non-alcoholic fatty liver disease, CRC colorectal cancer, IBD inflammatory bowel disease, CD Crohn’s disease, UC ulcerative colitis, DD diverticular disease, GD gastritis–duodenitis, CL&CC cholelithiasis and cholecystitis, ANX anxiety, NE neuroticism, MDD major depressive disorder, DEP depression, ADHD attention-deficit hyperactivity disorder, BIP bipolar disorder, PTSD post-traumatic stress disorder.
Weighted gene co-expression network analysis across tissues involved in the gut–brain axis
To explore the potential risk genes underlying the detected genetic variants, we performed gene-based analyses using multi-marker analysis of genomic annotation (MAGMA) on the studied psychiatric and digestive system disorders (Supplementary Table 32). We next conducted gene enrichment analyses using Fisher’s Exact test to assess the significance of gene overlap between psychiatric diseases and digestive diseases. Consistent with results of the genetic correlation analysis, we also observed a significant mutual enrichment of risk genes between psychiatric disorders and digestive diseases (Supplementary Fig. 11).
To prioritize risk genes that may play a crucial role in GBA, we next constructed gene co-expression networks using weighted gene co-expression network analysis (WGCNA) with RNA-Seq data obtained from seven brain tissues and seven digestive tract tissues from the GTEx (v8) database. Based on existing literature, the seven brain tissues were selected due to their potential involvement in psychiatric disorders analyzed in this study, such as DEP, MDD, and NE22. Similarly, the seven digestive tract tissues were chosen for their relevance to the development and progression of GERD and IBS23,24. A total of 4196 tissue samples, including 1426 brain samples, were included in the analysis (Supplementary Table 33). It is important to note that the number of samples (ranging from 152 to 555) and genes (ranging from 16,429 to 18,789) varied across different tissues (Supplementary Table 33). Within each gene network, we observed varying numbers of gene co-expression modules. For instance, the sigmoid colon had 17 modules, while the stomach only had four modules (Supplementary Table 33). The number of genes within each module also displayed considerable variation, ranging from 101 genes (0.06% of total genes) in the amygdala to 11,209 genes (68% of total genes) in the stomach. Gene functional enrichment analysis further revealed that these co-expression networks were significantly enriched in specific biological processes, such as oxidative phosphorylation and immune system process (Supplementary Data 2). These results suggest that the gene co-expression modules within the networks can be viewed as biologically coherent and functionally related units. In addition, we observed an enrichment of risk genes for specific diseases within gene co-expression network modules, as illustrated in the Supplementary Fig. (Supplementary Fig. 12).
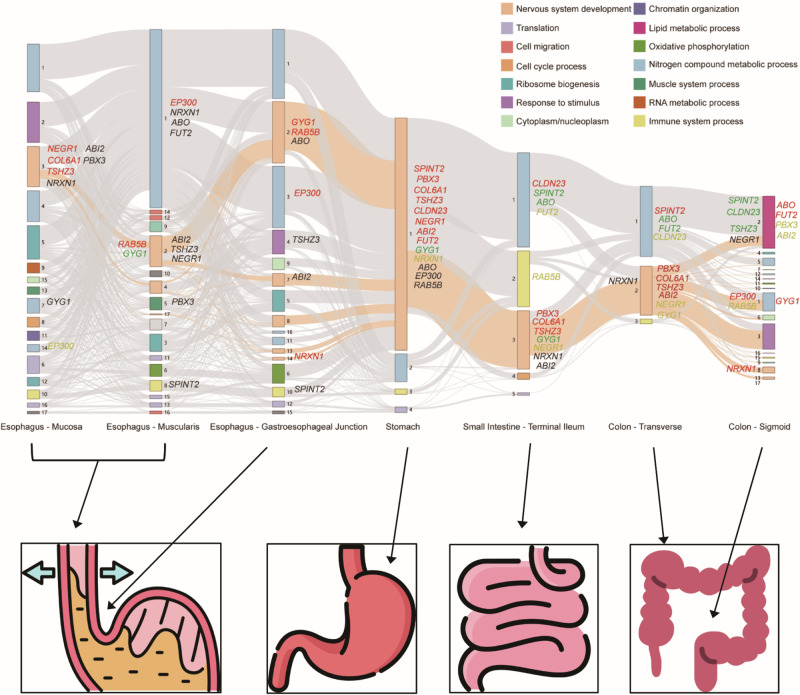
To investigate the relationships between modules across different tissues, we built a Sankey diagram based on the sequential arrangement of digestive tissues in the gastrointestinal tract. Notably, we noted the presence of a sizable module enriched with genes related to nervous system development in both the small intestine (M3 [Small_Intestine]) and transverse colon (M2 [Colon Transverse]), which are contiguous in the digestive tract. Moreover, modules associated with nervous system development exhibited remarkable similarity in their gene compositions between the small intestine and transverse colon (Fig. 5). In stomach, another organ adjacent to the small intestine, we detected a larger module (M1 [Stomach]), which is also linked to nervous system development (Fig. 5). Notably, several potential risk genes identified through MTAG analysis consistently served as hub genes within these modules, such as PBX3, COL6A1, and TSHZ3 (Fig. 5). In contrast to the stomach, small intestine, and transverse colon, there is a significant rise in the number of identified modules in the esophageal and sigmoid colon tissues, indicating more complicated patterns of gene co-expression within these tissues (Fig. 5).
Fig. 5. Co-expression transitions of neurological pathways across digestive tissues.
The Sankey diagram displays the co-expression transitions of genes (or co-expression modules) associated with neurological pathways across digestive tissues. Each rectangle represents a gene co-expression module, with the size corresponding to the number of genes, and different colors indicating distinct functional enrichments. The connection degree of each module is visualized based on the number of shared genes between two modules. Neurological modules and their connections are colored in coral. Hub genes identified by MTAG are highlighted in each module, with red denoting genes ranking within the top 5% of the module, yellow indicating a rank between 5 and 10%, green for a rank between 10 and 20%, and black representing a rank greater than 20%.
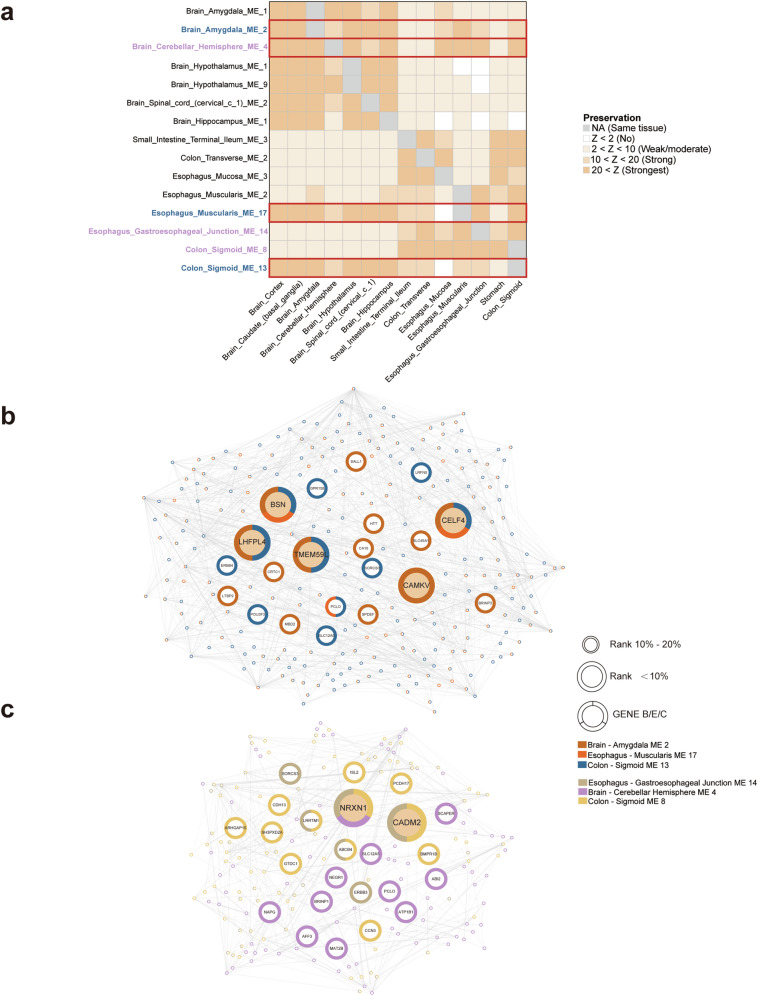
We next selected highly correlated psychiatric disorders (DEP, MDD, and NE) and gastrointestinal diseases (IBS, GERD) according to LDSC, which are potentially influenced by genes involved in GBA. We first conducted a preservation analysis on co-expression modules identified in various brain and digestive tissues, which are enriched with genes related to nervous system development and the aforementioned five disorders (Supplementary Table 34). Our goal was to systematically assess the consistency and reproducibility of these modules across different tissue types. For modules identified in brain tissues, a strong preservation was almost observed (Z > 10) across all brain regions, while a weak to moderate preservation (2 < Z score < 10) was detected in most digestive tissues. It is worth mentioning that modules M2 [Amygdala] and M4 [Cerebellum Hemisphere] not only demonstrated strong preservation within brain tissues but also exhibited comparable conservation in esophageal and sigmoid colon tissues (Fig. 6a). In contrast, while most modules identified in digestive tissues exhibited robust preservation within the digestive system, a predominantly weak to moderate preservation was observed in most brain tissues (Fig. 6a). Notably, modules M17 [Esophagus Muscularis] and M13 [Colon Sigmoid] displayed remarkably high preservation strength within brain tissues (Z > 20), surpassing even their levels observed in digestive tissues (Fig. 6a). These findings provide compelling evidence suggesting substantial regulatory influence from the brain on these gastrointestinal regions and imply the active involvement of these modules in GBA. Furthermore, we observed a significant overlap in the genes within modules (M17 [Esophagus Muscularis] and M13 [Colon Sigmoid]), and both modules exhibited a notable convergence with M2 [Amygdala]. Interestingly, two genes, BSN and CELF4, identified through our MTAG analysis consistently served as intramodular hubs in these three modules (Fig. 6b). In addition, we identified a substantial overlap between M4 [Cerebellum Hemisphere] and two overlapping intestinal modules (ME 14 [Esophagus Gastroesophageal Junction] and M8 [Colon Sigmoid] (Fig. 6c). Remarkably, these modules are primarily interconnected through the NRXN1 gene (ranking second in intramodular connectivity in both modules) (Fig. 6c). Moreover, NRXN1 was found to play a crucial hub role in modules across various brain tissues, as illustrated in Supplementary Table 35.
Fig. 6. Preservation of co-expression modules across brain and digestive tissues.
a Co-expression modules enriched in nervous system development and five closely correlated disorders (DEP, MDD, NE, IBS and GERD) are analyzed. Colors of the heatmap indicate module preservation, with Z values > 20 representing the strongest evidence, >10 denoting strong evidence, 2–10 suggesting weak to moderate preservation, and <2 indicating no preservation. Modules with labels colored in bule are further visulized in (b), while modules with labels colored in purple are visulized in (c). b The PPI network shows the shared genes from modules M2 [Amygdala], M17 [Esophagus Muscularis] and M13 [Colon Sigmoid] and c displays the shared genes from modules M4 [Cerebellum Hemisphere], M14 [Esophagus Gastroesophageal Junction], and M8 [Colon Sigmoid], with the size of the circles reflecting the gene’s rank in at least one of the three modules. The surrounding color shows if the gene is presented in each module.
Discussion
To uncover the shared genetic mechanism contributing to psychiatric and gastrointestinal disorders, recent studies have detected multiple pleiotropic loci at the genomics level, suggesting an important role of GBA in these conditions15,16,25. Despite these advancements, the specific genes participating in GBA and their regulatory mechanisms remain elusive. Here, we broaden our investigations by integrating multi-omics data from various sources, enabling the identification of pivotal genes actively participating in GBA and revealing their potential regulatory mechanisms at both genomic and transcriptomic levels.
By leveraging LDSC, we explored a wide range of psychiatric disorders and digestive conditions. Our investigation unveiled widespread genetic correlations across the majority of these two categories of disorders, suggesting the potentially pervasive influence of GBA in both psychiatric and digestive conditions. In consistent with previous findings25, IBS was most profoundly affected by psychiatric conditions, particularly in its correlation with depression and neuroticism. In addition, we observed a strong association between GERD and IBS, along with their correlation with depression and neuroticism, indicating the crucial role of GBA in these disorders and its significant impact on their shared mechanisms. These findings were further validated through our cell-specific analysis using S-LDSC. We observed similar patterns of SNP heritability across the six chromatin marks for IBS, GERD and psychiatric disorders with all showing enrichment in CNS-related tissues or cell types. In contrast, other digestive diseases were influenced by tissues/cells involved in different functional pathways other than CNS. For instance, IBD was markedly affected by cells participating immune responses, while DD exhibited significant impacts from musculoskeletal/connective tissues. Moreover, we uncovered additional evidence at the transcriptomic level using the TCSC method, which differentiates causal tissues from tagging tissues and divides disease heritability into tissue-specific components.
To identify candidate genes implicated in GBA underlying digestive and psychiatric disorders, we conducted MTAG analyses for genetically correlated psychiatric and digestive diseases. This approach led to the identification of numerous shared genetic loci between digestive and psychiatric disorders. Specifically, our analyses revealed more shared loci between psychiatric disorders and IBS or GERD than other digestive disorders. In addition, many genes situated near these loci are recognized for their associations with mental disorders, potentially participating in shared biological pathways, such as NRXN1, NLGN1, FOXP1, FOXP2, GRM5, and GRM8. In addition, several pleiotropic loci consistently emerged in MTAG analyses conducted across various digestive diseases and psychiatric disorders. This not only supports our earlier findings but also enhances the body of evidence pointing to shared genetic mechanisms underlying the interconnectedness of these diseases.
To further pinpoint key genes at the transcriptome level, we constructed gene co-expression networks using WGCNA with RNA-Seq data from digestive tract tissues and brain tissues. Tissues with more complex biological roles or those involved in multiple physiological processes may exhibit a greater number of co-expression modules. It suggests that each tissue’s gene co-expression network is tailored to its specific biological functions and regulatory needs. In addition, due to the increased statistical power, tissues with larger sample sizes may exhibit more modules. Notably, we identified a primary co-expression module in the stomach containing over 9000 genes, predominantly enriched with genes associated with nervous system development, immune system processes, and nitrogen compound metabolic processes. Genes of this module involved in nervous system development also form a co-expression module in the small intestine and transverse colon tissues near the stomach, indicating their coordinated activity in these tissues. Moreover, specific genes like PBX3, COL6A1, and TSHZ3 consistently acted as hub genes within modules related to nervous system development in the stomach, small intestine, and transverse colon. TSHZ3 also emerged as a hub gene in a module linked to nervous system development in the mucosa, and two independent loci (rs2111530 and rs8106322) in close proximity to TSHZ3 were detected through MTAG analyses. Indeed, multiple lines of evidence suggest that TSHZ3 encodes a transcription factor regulating neuronal development, and disruptions in TSHZ3 may lead to neurodevelopmental disorders26–28. These findings provide support for the convergence of genetic and transcriptomic aspects, emphasizing the critical role of TSHZ3 in GBA and its involvement in psychiatric and digestive disorders. In contrast, the esophageal and sigmoid colon tissues exhibit a notable increase in the number of identified modules, suggesting more intricate patterns of gene co-expression within these tissues. Interestingly, we also found genes identified by MTAG analyses, such as NRXN1 and EP300, function as hub genes across the esophageal and sigmoid colon tissues. NRXN1 encodes a protein from the neurexin family, crucial for the proper formation and maintenance of synapses. It has been widely implicated in diverse psychiatric disorders, including autism spectrum disorder (ASD), schizophrenia (SCZ), and intellectual disabilities29–31. EP300 encodes a histone acetyltransferase, playing a vital role in chromatin modification and gene expression regulation. EP300 has also been associated with various neurological and psychiatric disorders32,33. In addition to the aforementioned genes potentially involved in GBA regulation, our WGCNA also supports the crucial roles of certain genes independent of GBA within the gastrointestinal tract, such as FUT2 and ABO. FUT2 encodes fucosyltransferase 2, a protein controlling the secretion of ABO blood group antigens in the gastrointestinal mucosa and secretory glands34. Recent reports suggest that ABO blood type and FUT2 secretor status may indirectly influence gastrointestinal health and disease states by affecting the expression of gastrointestinal proteins35. Moreover, both ABO and FUT2 have been previously associated with the risk of PU and gastric cancer36,37. In our analysis, FUT2 and ABO were identified as hub genes in the module associated with lipid metabolic processes in the sigmoid colon. However, only FUT2 was recognized as a hub gene in the stomach, indicating variations in the regulatory roles of FUT2 and ABO across different gastrointestinal tissues.
To gain deeper insights into the mechanisms of GBA in digestive and psychiatric disorders, we performed a preservation analysis of co-expression modules enriched in nervous system development and five closely correlated disorders (DEP, MDD, NE, IBS and GERD) across diverse brain and digestive tissues. Strong conservation was observed across all brain regions for modules identified in brain tissues, while weak to moderate preservation was still evident in digestive tissues. Notably, modules M2 [Amygdala] and M4 [Cerebellum Hemisphere] displayed robust preservation in both brain and digestive tissues. Similarly, most modules identified in digestive tissues exhibited strong preservation within the digestive system, while weak to moderate preservation in brain tissues, including the pivotal modules associated with nervous system process M3 [Small_Intestine] and M2 [Colon Transverse]. Nonetheless, modules M17 [Esophagus Muscularis] and M13 [Colon Sigmoid] exhibited exceptional preservation strength in brain tissues. Interestingly, genes within these two modules and M2 [Amygdala] overlapped significantly, and genes BSN and CELF4, detected through our MTAG analysis, consistently served as intramodular hubs in these three modules. The BSN gene encodes the Bassoon protein, a crucial component of the presynaptic cytomatrix at the active zone that facilitates synaptic vesicle trafficking. It is prominently expressed in the cerebral cortex and hippocampus regions of the mammalian brain and linked to multiple psychiatric disorders38. CELF4 encodes an mRNA-binding protein that regulates excitatory neurotransmission39. It is primarily expressed in glutamatergic neurons and associated with psychiatric disorders such as neuroticism and depression21,39. In addition, a substantial overlap between M4 [Cerebellum Hemisphere] and two overlapping intestinal modules (M14 [Esophagus Gastroesophageal Junction] and M8 [Colon Sigmoid]) was detected with NRXN1 acting as a pivotal hub. These results indicated the intricate mechanisms underlying GBA and suggest BSN, CELF4 and NRXN1 as a central player in the regulatory networks connecting the brain and gastrointestinal system.
This study has several limitations. First, while we identified novel pleiotropic loci and key genes such as BSN, CELF4, and NRXN1, which are implicated in the gut–brain axis, their regulatory mechanisms still require further validation through functional experiments. Second, the use of publicly available GWAS data introduces the possibility of sample overlap between cohorts. To mitigate this, we employed the MTAG framework, which accounts for and reduces potential biases from overlapping samples. Finally, our analysis is predominantly based on individuals of European ancestry, potentially limiting its generalizability to other populations or the exploration of population-specific environmental interactions.
In conclusion, our integrative approach provides a comprehensive understanding of the shared genetic mechanisms linking digestive conditions and psychiatric disorders. Through this integration, our analyses revealed novel genetic associations, and offered additional support from gene co-expression network perspectives. Future functional studies are warranted to validate the key gene targets identified in this study, which will facilitate a deeper understanding of the complex interplay between gut and brain, and potentially improve health outcomes for individuals with digestive disorders influenced by GBA.
Methods
GWAS data
The detailed information of the GWAS datasets analyzed in this study is presented in Supplementary Table 36.
Digestive diseases
GWAS summary statistics were collected from publicly available data sources. Data for IBS were obtained from a large meta-analysis with 486,601 individuals (53,400 cases and 433,201 controls)12. For GERD, data were collected from the UK Biobank (UKBB) study, which included 71,522 cases and 261,079 controls. This dataset was defined based on ICD10 codes, self-reported GERD, and the use of GERD medication. In addition, data from the QSkin study, involving heartburn and GERD medication use as recorded in the Pharmaceutical Benefits Scheme (PBS) medical records, were included40. An additional dataset for PU, involving 16,666 cases, were sourced from the same gastrointestinal tract GWAS conducted on a cohort of 456,327 individuals from the UKBB36. GWAS summary statistics for IBD (25,042 cases, 34,915 controls), UC (12,366 cases, 33,609 controls), and CD (12,194 cases, 28,072 controls) were derived from the same IBD GWAS dataset. GWAS summary statistics for GD and CL&CC were derived from the full European data subset from the Lee Lab (https://www.leelabsg.org/resources). In addition, GWAS summary statistics for AP (10,630 cases, 844,679 controls), APP (4089 cases, 480,509 controls), CRC (78,473 cases, 107,143 controls), DD (31,964 cases, 419,135 controls), SF (167,875 participants) and NAFLD (12,194 cases, 28,072 controls) were sourced from the GWAS catalog.
Psychiatric disorders
GWAS summary statistics for NE (523,783 participants) and ANX (6514 cases, 478,084 controls) were obtained from the GWAS Catalog. GWAS data for eight additional psychiatric disorders were obtained from the Psychiatric Genomics Consortium (PGC), including DEP (294,322 cases, 741,438 controls)13, MDD (170,756 cases, 500,199 controls)41, SCZ (76,755 cases, 243,649 controls)42, bipolar disorder (BIP, 41,917 cases, 371,549 controls)43, ADHD (38,691 cases, 186,843 controls)44, ASD (18,381 cases, 27,969 controls)45, PTSD (32,428 cases, 174,227 controls)46 and anorexia nervosa (AN, 16,992 cases, 55,525 controls)47.
Global genetic correlation analysis
Genetic correlation (rg) between each pair of diseases or traits was assessed using LDSC with GWAS summary statistics48. The equation used in LDSC is as follows: , where βj and γj represent the effect sizes of SNP j on the two traits being tested, N1 and N2 are the sample sizes for the two traits, Ns is the number of overlapping samples between the two traits, r is the phenotypic correlation in the overlapping samples, rg is a calculated parameter for the genomic annotation value of the associated risk allele, and lj is the LD score.
In this analysis, pre-calculated linkage disequilibrium scores for HapMap3 SNPs were utilized. These scores were derived from individuals of European ancestry from the 1000 Genomes Project. SNP markers with an imputation INFO score below 0.9 were excluded from the analysis. LDSC was also employed to estimate the SNP-based heritability of the examined traits48.
Cell-type-specific enrichment of SNP heritability
The Stratified LDSC (S-LDSC) method was employed in this study to identify specific functional categories or cell types that have a significant impact on the heritability of the traits under investigation49,50. The annotation data used in this analysis was obtained from the Roadmap project and included information on six chromatin marks (DHS, H3K27ac, H3K36me3, H3K4me1, H3K4me3, and H3K9ac) across a diverse set of 88 cell types or tissues. These annotations were utilized to partition the SNP heritability of each trait. Additionally, the cell-type annotations were grouped into nine categories, namely adipose, central nervous system (CNS), digestive system, cardiovascular, musculoskeletal and connective tissue, immune and blood, liver, pancreas, and others. The enrichment values specific to each annotation are converted into a color scale and visualized using hierarchical clustering techniques in the ComplexHeatmap package in R 4.2.3. Specifically, hierarchical clustering is performed using Euclidean distance as the distance metric and complete linkage as the clustering method, which are the default settings in the ComplexHeatmap package (internally relying on the hclust function from base R).
Local genetic correlation analysis
Given that genetic correlation estimated by LDSC aggregates information across all variants in the genome, we performed an additional analysis using ρ-HESS (heritability estimation from summary statistics) to evaluate pairwise local genetic correlations51. ρ-HESS is specifically tailored to evaluate the local genetic correlation between pairs of traits within each of the 1703 predefined LD-independent segments, which have an average length of 1.6 Mb. To ascertain statistical significance, we applied a Bonferroni correction, considering a p-value lower than 0.05/1703 as indicative of statistical significance.
Multi-trait analysis of GWAS (MTAG)
MTAG was employed to explore genetic associations between psychiatric disorders and digestive diseases by leveraging combined GWAS summary statistics14. This approach enhances the ability to detect loci from related traits by jointly analyzing GWAS summary statistics. Compared to traditional inverse-variance weighted meta-analysis, it also accounts for sample overlap and incomplete genetic correlation. In the initial step of MTAG, variants were filtered by excluding non-common SNPs, duplicated SNPs, or those with strand ambiguity. Subsequently, the pairwise genetic correlation between traits were estimated using LDSC, and these estimates were utilized to calibrate the variance-covariance matrix of the random-effect component. Following calibration, a random-effect meta-analysis was performed by MTAG to generate SNP-level summary statistics. Pleiotropic SNPs achieving genome-wide significance (P < 5 × 10–8) in multi-trait analysis and suggestive significance (P < 0.01) in single-trait GWAS were prioritized. The MHC region was treated as one locus due to its complex LD structure. The results were visualized using Circos52.
Tissue co-regulation score regression (TCSC)
The TCSC method was used to differentiate between causal tissues and annotated tissues, as well as to dissect the heritability of diseases into tissue-specific components18. GWAS summary statistics and gene expression prediction models for each tissue were used as input data. Initially, disease-specific Transcriptome-Wide Association Study (TWAS) summary statistics were computed for gene expression prediction models of 48 GTEx tissues. Subsequently, the tissue co-regulation score was calculated for each gene-tissue pair, which reflects the correlation between gene expression prediction and tissue. The TWAS χ2 statistics (or the product of z-scores for two diseases/traits) were then regressed on the tissue co-regulation scores in order to estimate the tissue-specific contributions to the disease. Finally, a joint modeling approach was employed to estimate the contributions of each tissue and identify the causal tissues.
Multi-marker analysis of genomic annotation (MAGMA)
To assess the genetic overlap between psychiatric and digestive disorders, we used MAGMA v1.0653 to conduct gene-level analyses on GWAS data for10 psychiatric disorders and 14 digestive system disorders, respectively. In addition, in the subsequent preservation analysis, we conducted gene-level analyses on the MTAG results between IBS, GERD, and psychiatric disorders (DEP, MDD, NE) separately. This method assigned SNPs to their nearest gene within a predefined genomic window, specifically 35 kb upstream or 10 kb downstream of a gene body. The gene-based statistic was calculated by summing the log-transformed P values of the assigned SNPs, taking into account the correlation (linkage disequilibrium) between adjacent SNPs. Multiple testing correction was implemented using Bonferroni correction within the MAGMA framework.
Weighted gene co-expression network analysis (WGCNA)
RNA sequencing data (read count) for seven brain tissues and seven digestive tract tissues were obtained from the Genotype-Tissue Expression project portal (version 8). Initially, unexpressed genes (with a read count value of 0) were removed from the analysis. For normalization, the DESeq2 package was utilized to create a DESeq dataset for subsequent differential expression analysis. Variance stability transformation (VST) was applied to achieve approximate homoscedasticity by fitting the dispersion-mean relationship of the dataset, and read counts were normalized by dividing by the size factor.
Gene co-expression modules for the seven brain tissues and seven digestive tract tissues were independently constructed using the WGCNA package in R. A signed pairwise correlation matrix was calculated using Pearson’s product-moment correlation coefficient, emphasizing strong gene–gene correlations with a selected “soft-thresholding” value. This correlation matrix was transformed into an adjacency matrix, normalized using a topological overlap function. Hierarchical clustering and module segmentation were executed using average linkage and the dynamic tree cut algorithm, respectively. Genes with the highest intramodular connectivity (top 10% highest intramodular connectivity) were considered as hub genes. Gene ontology analysis for each identified co-expressed module was conducted using g:Profiler, with significance for enrichment set at false discovery rate (FDR) < 0.0554.
Module preservation
Preservation of network modules across GTEx tissues was analyzed using the “modulePreservation” R function in WGCNA55. This method, involving “reference” and “test” network modules, calculates statistics for three preservation classes: (i) density-based, assessing gene–gene connectivity pattern similarity; (ii) separability-based, examining if test network modules remain distinct in reference network modules; and (iii) connectivity-based, relying on connectivity pattern similarity between genes in reference and test networks. Four complementary statistics, including median rank, Zdensity, Zconnectivity, and Zsummary, were used to module preservation determination. Zdensity and Zconnectivity are standardized statistics for density and connectivity, respectively, while Zsummary is their average. Preservation was established through Zsummary measure. The use of the Zsummary metric allows us to quantify the extent of module preservation and infer the potential biological significance of the gene co-expression networks across different tissues. Understanding these preservation patterns can provide insights into the robustness and variability of biological processes in different tissue contexts. Zsummary > 20: This threshold indicates the strongest evidence of module preservation. Modules with Zsummary values above 20 are considered highly preserved, suggesting that the gene–gene connectivity patterns within these modules are consistently maintained across different tissues. This strong preservation implies that the biological processes represented by these modules are robust and potentially critical across multiple tissue types. Zsummary > 10: A Zsummary value greater than 10 denotes strong evidence of module preservation. Modules in this range are still well-preserved, indicating that the underlying biological processes are likely important and conserved across the tissues being compared. Zsummary between 2 and 10: This range suggests weak to moderate preservation. Modules with Zsummary values in this range may have some degree of conservation, but the gene–gene connectivity patterns are not as consistently maintained as those with higher Zsummary values. This could indicate that the biological processes are somewhat variable or context-dependent across different tissues. Zsummary < 2: A Zsummary value below 2 indicates no preservation. Modules with such low values are not preserved across the tissues, suggesting that the gene–gene connectivity patterns are highly variable or specific to certain conditions. This lack of preservation may reflect tissue-specific biological processes or regulatory mechanisms.
Protein–protein interaction (PPI) network analysis
PPI analysis was conducted on genes (MAGMA for MTAG results between IBS, GERD, and psychiatric disorders) that exhibited overlap across co-expression modules related to nervous system development from distinct tissues. The shared genes from modules M17 [Esophagus Muscularis], M13 [Colon Sigmoid], and M2 [Amygdala], along with shard genes from modules M4 [Cerebellum Hemisphere], M14 [Esophagus Gastroesophageal Junction], and M8 [Colon Sigmoid], were subjected to analysis. The PPI network was constructed by retrieving protein interaction data from the STRING database. The Cytoscape software (version 3.9.1) was utilized to generate a comprehensive visual representation of the PPI network. The size of the circles reflects the ranking of the genes in at least one of the three modules. The surrounding color indicates whether the gene appears in each module.
Statistical analysis and reproducibility
The study outlined the use of several publicly accessible software tools to conduct statistical analyses across different programming environments, ensuring the reproducibility and robustness of the research. In the Python 2.7.18 environment, LDSC version 1.0.1 was used to compute genetic correlations and facilitate cell-type-specific investigations. For Multi-Trait Analysis of GWAS, MTAG version 1.0.8 was employed, while HESS version 0.5.4-beta was used for Heritability Estimation from Summary Statistics. Per l5.16.3 was utilized to generate Manhattan plots using the CIRCOS tool version 0.69-9. In R 4.2.3, the TCSC package version 1.0.0 was applied for identifying causal tissues in disease and complex trait research, and the WGCNA package version 1.72-5 was used for gene co-expression network analysis. Java 17.0.5 facilitated PPI network analysis through Cytoscape version 3.10.1, alongside the string App plugin version 2.0.1. Additionally, MAGMA software version 1.10 was used for gene analysis and generalized gene-set analysis of GWAS data, capable of processing both raw genotype information and summary SNP P values from previous GWAS or meta-analyses. These tools collectively enhance the reproducibility of the research and highlight the significance of accessible and well-documented software in scientific studies. To address the issue of multiple testing, we applied the Bonferroni method in LDSC, TCSC, and WGCNA.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The authors are grateful to the contributors and curators of public databases, particularly GWAS Catalog, Psychiatric Genomics Consortium (PGC) and GTEx for their invaluable role in making extensive GWAS data publicly accessible. This work is supported by Major Basic Research Project of Natural Science Foundation of Shandong Province (ZR2020ZD15), the Special Funds of Taishan Scholar Project, China (tsqn202211224), the National Natural Science Foundation of China (32270661, 82472366), the China Postdoctoral Science Foundation (2024M761870), and Shandong Postdoctoral Science Foundation (SDCX-ZG-202400042). The relevant elements in Fig. 5 and Supplementary Fig. 1 are designed by Freepik, Justicon, Backwoods, Vitaly Gorbachev, DinosoftLabs, and UIUX Mall from Flaticon.
Author contributions
H.X.D., Y.J., and Y.C.S. contributed to data extraction, data analyses, and manuscript drafting S.H.D., Q.X., L.Z.H.L., C.X.L., B.J.L., and H.X.J. contributed to data interpretation and manuscript drafting. B.C.P., S.P., C.M.Z., J.K.Z., and M.W.Z. contributed to manuscript drafting. G.Y.Z. and X.C. contributed to study design, data interpretation, and final approval of the manuscript.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Qiao Fan and Tobias Goris. A peer review file is available.
Data availability
The GWAS summary statistics for ten digestive diseases/traits analyses used in this study are deposited in the GWAS Catalog (https://www.ebi.ac.uk/gwas/) and the accession codes are as follows: irritable bowel syndrome (GCST90016564), acute pancreatitis (GCST90255375), non-alcoholic fatty liver disease (GCST90091033), appendicitis (GCST90038695), colorectal cancer (GCST90255675), inflammatory bowel disease (GCST004131), crohn’s disease (GCST004132), ulcerative colitis (GCST004133), stool frequency (GCST90002250) and diverticular disease (GCST008105). The GWAS summary statistics for the gastritis–duodenitis and cholelithiasis-cholecystitis are publicly available at https://www.leelabsg.org/resources. The GWAS summary statistics for peptic ulcer used in this study are available at https://cnsgenomics.com/content/data, and the GWAS summary statistics for Gastroesophageal reflux disease are available at 10.6084/m9.figshare.8986589. The anxiety and neuroticism GWAS summary statistics used in this study are publicly available in the GWAS Catalog under accession code GCST90038651 and GCST007339. The summary statistics for depression are deposited in: https://ipsych.dk/en/research/downloads/. The GWAS summary statistics for seven other psychiatric disorders, including major depressive disorder, schizophrenia, attention-deficit hyperactivity disorder, autism spectrum disorder, bipolar disorder, post-traumatic stress disorder, and anorexia nervosa, are available for download at https://pgc.unc.edu/for-researchers/download-results/. GTEx project v.8 data were publicly available at https://gtexportal.org/home/.
Code availability
Our manuscript details the use of various publicly accessible software tools for conducting our analyses, as described in the Methods section. We performed statistical analyses across multiple programming environments, including Python 2.7.18, Perl 5.16.3, R 4.2.3, and Java 17.0.5. In this study, we utilized publicly available software for our analyses. Below, we provide a list of URLs where some online resources can be found, offering detailed information about the software tools, including computer code where applicable: LDSC version 1.0.1 (available at https://github.com/bulik/ldsc), MTAG version 1.0.8 (accessible at https://github.com/JonJala/mtag), HESS version 0.5.4-beta (found at https://github.com/huwenboshi/hess), CIRCOS tool version 0.69-9 (https://circos.ca/software/installation/), TCSC package version 1.0.0 (https://github.com/TiffanyAmariuta/TCSC/), WGCNA package version 1.72-5 (available at https://cran.r-project.org/web/packages/WGCNA/index.html), Cytoscape version 3.10.1 (downloadable from https://cytoscape.org/download.html), string App plugin version 2.0.1 (https://apps.cytoscape.org/apps/stringapp), MAGMA software version 1.10 (https://ctg.cncr.nl/software/magma), ComplexHeatmap package version 2.13.1 (https://github.com/jokergoo/ComplexHeatmap).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Huanxin Ding, Yue Jiang, Qing Sun.
Contributor Information
Guangyong Zhang, Email: guangyongzhang@hotmail.com.
Xiao Chang, Email: changxiao@sdfmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-025-07481-6.
References
- 1.Wang, Y. et al. Global burden of digestive diseases: a systematic analysis of the global burden of diseases study, 1990 to 2019. Gastroenterology165, 773–783.e15 (2023). [DOI] [PubMed] [Google Scholar]
- 2.Ng, S. C. et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet390, 2769–2778 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Lovell, R. M. & Ford, A. C. Global prevalence of and risk factors for irritable bowel syndrome: a meta-analysis. Clin. Gastroenterol. Hepatol.10, 712–721.e4 (2012). [DOI] [PubMed] [Google Scholar]
- 4.El-Serag, H. B., Sweet, S., Winchester, C. C. & Dent, J. Update on the epidemiology of gastro-oesophageal reflux disease: a systematic review. Gut63, 871–880 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Y. H. & Lin, H. C. Patterns of psychiatric and physical comorbidities associated with panic disorder in a nationwide population-based study in Taiwan. Acta Psychiatr. Scandinavica.123, 55–61 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Kugathasan, P. et al. Increased mortality from somatic multimorbidity in patients with schizophrenia: a Danish nationwide cohort study. Acta Psychiatr. Scandinavica.140, 340–348 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Staudacher, H. M., Mikocka-Walus, A. & Ford, A. C. Common mental disorders in irritable bowel syndrome: pathophysiology, management, and considerations for future randomised controlled trials. lancet Gastroenterol. Hepatol.6, 401–410 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Zhang, A. Z. et al. Prevalence of depression and anxiety in patients with chronic digestive system diseases: a multicenter epidemiological study. World J. Gastroenterol.22, 9437–9444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruan, X. et al. Depression and 24 gastrointestinal diseases: a Mendelian randomization study. Transl. Psychiatry13, 146 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang, M. H., Cordell, H. J. & Van Steen, K. Statistical methods for genome-wide association studies. Semin. Cancer Biol.55, 53–60 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Tam, V. et al. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet.20, 467–484 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Eijsbouts, C. et al. Genome-wide analysis of 53,400 people with irritable bowel syndrome highlights shared genetic pathways with mood and anxiety disorders. Nat. Genet.53, 1543–1552 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Als, T. D. et al. Depression pathophysiology, risk prediction of recurrence and comorbid psychiatric disorders using genome-wide analyses. Nat. Med.29, 1832–1844 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turley, P. et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet.50, 229–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tesfaye, M. et al. Shared genetic architecture between irritable bowel syndrome and psychiatric disorders reveals molecular pathways of the gut-brain axis. Genome Med.15, 60 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong, W. et al. Role of the gut-brain axis in the shared genetic etiology between gastrointestinal tract diseases and psychiatric disorders: a genome-wide pleiotropic analysis. JAMA Psychiatry80, 360–370 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanai, M. et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet.50, 390–400 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Amariuta, T., Siewert-Rocks, K. & Price, A. L. Modeling tissue co-regulation estimates tissue-specific contributions to disease. Nat. Genet.55, 1503–1511 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Docimo, S. Jr., Lee, Y., Chatani, P., Rogers, A. M. & Lacqua, F. Visceral to subcutaneous fat ratio predicts acuity of diverticulitis. Surg. Endosc.31, 2808–2812 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Yuan, S. & Larsson, S. C. Genetically predicted adiposity, diabetes, and lifestyle factors in relation to diverticular disease. Clin. Gastroenterol. Hepatol.20, 1077–1084 (2022). [DOI] [PubMed] [Google Scholar]
- 21.Dmitrzak-Weglarz, M. et al. Expression biomarkers of pharmacological treatment outcomes in women with unipolar and bipolar depression. Pharmacopsychiatry54, 261–268 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Chan, K. L., Poller, W. C., Swirski, F. K. & Russo, S. J. Central regulation of stress-evoked peripheral immune responses. Nat. Rev. Neurosci.24, 591–604 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tack, J. & Pandolfino, J. E. Pathophysiology of gastroesophageal reflux disease. Gastroenterology154, 277–288 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Ohman, L. & Simrén, M. Pathogenesis of IBS: role of inflammation, immunity and neuroimmune interactions. Nat. Rev. Gastroenterol. Hepatol.7, 163–173 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Alemany, S. et al. Genome-wide multi-trait analysis of irritable bowel syndrome and related mental conditions identifies 38 new independent variants. J. Transl. Med.21, 272 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feichtinger R. G. et al. A TSHZ3 frame-shift variant causes neurodevelopmental and renal disorder consistent with previously described proximal chromosome 19q13.11 deletion syndrome. Genes13, 2191 (2022). [DOI] [PMC free article] [PubMed]
- 27.Caubit, X. et al. Targeted Tshz3 deletion in corticostriatal circuit components segregates core autistic behaviors. Transl. Psychiatry12, 106 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caubit, X. et al. TSHZ3 deletion causes an autism syndrome and defects in cortical projection neurons. Nat. Genet.48, 1359–1369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sebastian, R. et al. Schizophrenia-associated NRXN1 deletions induce developmental-timing- and cell-type-specific vulnerabilities in human brain organoids. Nat. Commun.14, 3770 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varghese, M. et al. Autism spectrum disorder: neuropathology and animal models. Acta Neuropathologica.134, 537–566 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taşkıran, E. Z. et al. Diagnostic yield of whole-exome sequencing in non-syndromic intellectual disability. J. Intellect. Disabil. Res.65, 577–588 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Lo, M. T. et al. Modeling prior information of common genetic variants improves gene discovery for neuroticism. Hum. Mol. Genet.26, 4530–4539 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng, W. et al. Electroconvulsive therapy reduces protein expression level of EP300 and improves psychiatric symptoms and disturbance of thought in patients with schizophrenia. Neuropsychiatr. Dis. Treat.19, 1763–1770 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ravn, V. & Dabelsteen, E. Tissue distribution of histo-blood group antigens. APMIS: Acta Pathologica Microbiologica et. Immunologica Scandinavica.108, 1–28 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Sun, B. B. et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature622, 329–338 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, Y. et al. GWAS of peptic ulcer disease implicates Helicobacter pylori infection, other gastrointestinal disorders and depression. Nat. Commun.12, 1146 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duell, E. J. et al. Variation at ABO histo-blood group and FUT loci and diffuse and intestinal gastric cancer risk in a European population. Int. J. cancer136, 880–893 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Chen, C. H. et al. Identification of rare mutations of two presynaptic cytomatrix genes BSN and PCLO in schizophrenia and bipolar disorder. J. Pers. Med.11, 1057(2021). [DOI] [PMC free article] [PubMed]
- 39.Smith, D. J. et al. Genome-wide analysis of over 106 000 individuals identifies 9 neuroticism-associated loci. Mol. Psychiatry21, 749–757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.An, J. et al. Gastroesophageal reflux GWAS identifies risk loci that also associate with subsequent severe esophageal diseases. Nat. Commun.10, 4219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howard, D. M. et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci.22, 343–352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature604, 502–508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mullins, N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet.53, 817–829 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Demontis, D. et al. Genome-wide analyses of ADHD identify 27 risk loci, refine the genetic architecture and implicate several cognitive domains. Nat. Genet.55, 198–208 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet.51, 431–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nievergelt, C. M. et al. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat. Commun.10, 4558 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watson, H. J. et al. Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat. Genet.51, 1207–1214 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet.47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet.47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finucane, H. K. et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet.50, 621–629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi, H., Mancuso, N., Spendlove, S. & Pasaniuc, B. Local genetic correlation gives insights into the shared genetic architecture of complex traits. Am. J. Hum. Genet.101, 737–751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res.19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol.11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kolberg, L. et al. g:Profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res.51, W207–w12 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langfelder, P., Luo, R., Oldham, M. C. & Horvath, S. Is my network module preserved and reproducible? PLoS Comput. Biol.7, e1001057 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The GWAS summary statistics for ten digestive diseases/traits analyses used in this study are deposited in the GWAS Catalog (https://www.ebi.ac.uk/gwas/) and the accession codes are as follows: irritable bowel syndrome (GCST90016564), acute pancreatitis (GCST90255375), non-alcoholic fatty liver disease (GCST90091033), appendicitis (GCST90038695), colorectal cancer (GCST90255675), inflammatory bowel disease (GCST004131), crohn’s disease (GCST004132), ulcerative colitis (GCST004133), stool frequency (GCST90002250) and diverticular disease (GCST008105). The GWAS summary statistics for the gastritis–duodenitis and cholelithiasis-cholecystitis are publicly available at https://www.leelabsg.org/resources. The GWAS summary statistics for peptic ulcer used in this study are available at https://cnsgenomics.com/content/data, and the GWAS summary statistics for Gastroesophageal reflux disease are available at 10.6084/m9.figshare.8986589. The anxiety and neuroticism GWAS summary statistics used in this study are publicly available in the GWAS Catalog under accession code GCST90038651 and GCST007339. The summary statistics for depression are deposited in: https://ipsych.dk/en/research/downloads/. The GWAS summary statistics for seven other psychiatric disorders, including major depressive disorder, schizophrenia, attention-deficit hyperactivity disorder, autism spectrum disorder, bipolar disorder, post-traumatic stress disorder, and anorexia nervosa, are available for download at https://pgc.unc.edu/for-researchers/download-results/. GTEx project v.8 data were publicly available at https://gtexportal.org/home/.
Our manuscript details the use of various publicly accessible software tools for conducting our analyses, as described in the Methods section. We performed statistical analyses across multiple programming environments, including Python 2.7.18, Perl 5.16.3, R 4.2.3, and Java 17.0.5. In this study, we utilized publicly available software for our analyses. Below, we provide a list of URLs where some online resources can be found, offering detailed information about the software tools, including computer code where applicable: LDSC version 1.0.1 (available at https://github.com/bulik/ldsc), MTAG version 1.0.8 (accessible at https://github.com/JonJala/mtag), HESS version 0.5.4-beta (found at https://github.com/huwenboshi/hess), CIRCOS tool version 0.69-9 (https://circos.ca/software/installation/), TCSC package version 1.0.0 (https://github.com/TiffanyAmariuta/TCSC/), WGCNA package version 1.72-5 (available at https://cran.r-project.org/web/packages/WGCNA/index.html), Cytoscape version 3.10.1 (downloadable from https://cytoscape.org/download.html), string App plugin version 2.0.1 (https://apps.cytoscape.org/apps/stringapp), MAGMA software version 1.10 (https://ctg.cncr.nl/software/magma), ComplexHeatmap package version 2.13.1 (https://github.com/jokergoo/ComplexHeatmap).