Abstract
The Ena/vasodilator-stimulated phosphoprotein (VASP) protein family is implicated in the regulation of a number of actin-based cellular processes, including lamellipodial protrusion necessary for whole cell translocation. A growing body of evidence derived largely from in vitro biochemical experiments using purified proteins, cell-free extracts, and pathogen motility has begun to suggest various mechanistic roles for Ena/VASP proteins in the control of actin dynamics. Using complementation of phenotypes in Ena/VASP-deficient cells and overexpression in normal fibroblasts, we have assayed the function of a panel of mutants in one member of this family, Mena, by mutating highly conserved sequence elements found in this protein family. Surprisingly, deletion of sites required for binding of the actin monomer-binding protein profilin, a known ligand of Ena/VASP proteins, has no effect on the ability of Mena to regulate random cell motility. Our analysis revealed two features essential for Ena/VASP function in cell movement, cyclic nucleotide-dependent kinase phosphorylation sites and an F-actin binding motif. Interestingly, expression of the C-terminal EVH2 domain alone is sufficient to complement loss of Ena/VASP function in random cell motility.
INTRODUCTION
Cell motility is a complex and highly regulated process. Many aspects of organismal development and physiology require that cells control their movement in response to diverse arrays of environmental signals. To move, cells must maintain polarity while coordinating membrane extension, changes in adhesiveness, and contractile mechanisms. These processes all depend upon dynamic remodeling of the actin cytoskeleton. Although the basic biochemistry of actin polymerization has been extensively studied, the mechanisms by which cells orchestrate assembly, organization, and disassembly of actin networks during cell movement remain poorly understood.
The Ena/vasodilator-stimulated phosphoprotein (VASP) proteins are a conserved family of molecules known to regulate cell movement and shape change (Gertler et al., 1996; reviewed in Bear et al., 2001). Drosophila Ena regulates axonal growth cone migration in response to several types of signaling pathways (Bashaw et al., 2000; Lanier and Gertler, 2000). The three vertebrate Ena/VASP proteins, VASP, Mammalian Enabled (Mena), and Ena-VASP-like (EVL), negatively regulate fibroblast motility by modulating lamellipodial behavior (Bear et al., 2000; Bear et al., 2002). Other data implicate Ena/VASP proteins in actin-dependent processes, including Jurkat T-cell polarization, inhibition of platelet aggregation, and the motility of the intracellular pathogen Listeria monocytogenes (Smith et al., 1996; Niebuhr et al., 1997; Aszodi et al., 1999; Krause et al., 2000; Skoble et al., 2001). Biochemical data support a role for Ena/VASP proteins in actin dynamics (Huttelmaier et al., 1999; Harbeck et al., 2000; Lambrechts et al., 2000) and the proteins localize to cellular structures rich in actin assembly such as protruding lamellipodial and filopodial tips (Reinhard et al., 1992; Gertler et al., 1996; Lanier et al., 1999; Rottner et al., 1999).
Ena/VASP proteins share a conserved domain structure, consisting of an Ena-VASP Homology (EVH) 1 domain at their amino termini and a carboxyl-terminal EVH2 domain. Ena/VASP proteins all contain a more variable central region between the EVH1 and EVH2 domains rich in polyproline clusters. The EVH1 domain has been crystallized and adopts a structure related to PH and PTB domains (Fedorov et al., 1999; Prehoda et al., 1999). The EVH1 domain binds directly to a consensus motif, (D/E)-FPPPP-X(D/E)(D/E) (Niebuhr et al., 1997), and plays an essential role in focal adhesion targeting of Ena/VASP proteins by binding to proteins containing the EVH1 binding motif (Gertler et al., 1996).
Much less is known about the cellular function of the proline-rich region and the EVH2 domain. Various lines of evidence suggest that the EVH2 domain can promote oligomerization of Ena/VASP proteins and can bind directly to F-actin in vitro (Bachmann et al., 1999; Huttelmaier et al., 1999; Harbeck et al., 2000). The proline-rich region can bind to profilins, small actin monomer-binding proteins, as well as proteins containing SH3 and WW domains, protein modules that bind to specific proline-rich motifs (Reinhard et al., 1995; Gertler et al., 1996; Ermekova et al., 1997). Adjacent to the proline-rich region, the three vertebrate Ena/VASP proteins also contain one or more sites for phosphorylation by the cyclic nucleotide-dependent kinases protein kinase (PK) A and G (Butt et al., 1994; Gertler et al., 1996; Lambrechts et al., 2000). Phosphorylation of Ena/VASP proteins at the single PKA site found in all three proteins induce changes in electrophoretic mobility and protein–protein interactions (Lambrechts et al., 2000). Although the in vivo functional significance of this phosphorylation is unknown, VASP knockouts exhibit platelet aggregation defects associated with misregulation of PKA-mediated intracellular signaling, suggesting that VASP may be the major physiological substrate for PKA in that cell type (Aszodi et al., 1999).
To gain insight into the function of Ena/VASP proteins, we have performed a systematic mutagenesis of conserved motifs within this protein family. We have previously isolated from mena;vasp-null embryos a clonal fibroblastic cell line (MVD7) that lacks detectable EVL protein and therefore is deficient in all known Ena/VASP proteins (Bear et al., 2000). MVD7 cells move more rapidly than do MVD7 cells expressing physiological levels of Mena (Bear et al., 2000), and a companion study finds that they do not support normal Listeria intracellular movement (Geese et al., 2002). We used complementation of the hypermotile phenotype of MVD7 cells to conduct a structure–function analysis of Ena/VASP-mediated regulation of whole cell motility. The average speed of motile cell populations was quantitated in a video microscopy-based long-term cell-tracking assay. This allows us to directly measure the effect of Ena/VASP function on cell motility. Our analysis of cells expressing mutant forms of Mena indicates that the proline-rich region is dispensable for function in random cell motility, but identifies two key features for function of this protein family: an F-actin binding motif and Ser/Thr phosphorylation.
MATERIALS AND METHODS
Subcloning of Enhanced Green Fluorescent Protein (EGFP)-Ena/VASP Proteins and Engineering of EGFP-Mena Structural Variants
EGFP-Ena/VASP family members were subcloned into pMSCV-EGFP retroviral plasmid by using standard techniques. EGFP-Mena structural variants were generated using mutagenic polymerase chain reaction (PCR) primers. For small deletions, two rounds of PCR were required. First, mutagenic primers were used to amplify regions upstream and downstream of the intended deletions. Those PCR products were then purified and combined at equimolar ratios to serve as template for a second PCR reaction to amplify an altered mena open reading frame (ORF). For point mutations, mutagenic primers were used to amplify the entire plasmid (pBSII) containing the mena ORF. Double-mutant ORFs were generated by subcloning. Mutations were confirmed by sequencing and restriction fragment length polymorphism analysis. Mutagenic primers were designed in complementary pairs; only the sense orientation is listed: Q, 5′G A C AA A A T T C A C A G C T A C C T G C T C A A C T G C A A G A A C A G C A G C G A C A G A A G G A A C3′; LER, 5′G A A G G C A A C T G C A A G A A C A G C A G C G A C A G G A G C G C A G A A T G T C C A A T G C T G C T G C C C-C3′; PRO, 5′G G G C C T T G T C T T G G G A G C A T C T G G A A T T T T C T C T G G3′; TLM, 5′G A C A A T C G C C C T T T A A C T T C C C G G G T G G A G G A T G3′; FAB, 5′C T G G G C G T G G G A A T G G A C C T C T T C C T C T A G C T G A G A A G G G A T C A A C A A T A G A A A C A G A AC3′; PWE, 5′-C T G C T A A G G C C C C A T C A A C A A G T A C A C C T A T G A A C G G C A G T A A G T C A C C T G T C A T C T C C3′; COCO, 5′G G C T G A A G C A G G A C A T T A G C A A G T C G A A C A C T G C3′; S236A, 5′G A G C G C A G A A T G G C C A A T G C T G C T G C C3′; S326D, G A G C G C A G A A T G G A C A A T G C T G C T G C C; S376A, 5′G C A A A A C T T A G G A A A G T G G C C C G G G T G G A T G G3′; and S376D: 5′ G C A A A A C T T A G G A A A G T G G A C C G G G T G G A T G G3′.
Retroviral Packaging, Infection, Fluorescence-activated Cell Sorting (FACS), and Cell Culture
Culture of Rat2 cells is described in Gertler et al. (1996). Isolation of MVD7 fibroblastic cells is described in Bear et al. (2000). The MVD7 cells were cultured at 32°C in Immorto media (high-glucose Dulbecco's modified Eagle's with 15% fetal calf serum, penicillin/streptomycin, l-glutamine, and 50 U/ml recombinant mouse interferon-γ (Invitrogen, Carlsbad, CA). Retroviral plasmids described above were transiently calcium-phosphate transfected into Bosc23 packaging cells (3.3 μg of retroviral plasmid and 1.7 μg of pCL-Eco helper plasmid), and supernatant was collected after 48 h. MVD7 cells or Rat2 cells (American Type Culture Collection, Manassas, VA) plated at 50% confluence were exposed to infectious supernatant for 24 h in the presence of 4 μg/ml polybrene. Infected cells were cultured to 90% confluence, trypsinized, and FACS sorted in phosphate-buffered saline/5% fetal calf serum. EGFP-positive cells were harvested and cultured for one passage and then resorted for EGFP signal intensity levels that matched EGFP-Mena wild-type controls. To confirm FACS analysis and assess protein stability, radioimmunoprecipitation assay (RIPA) extracts from each cell line were resolved by SDS-PAGE and probed with Anti-EGFP Ig, by using anti-actin Ig as a loading control.
Immunofluorescence and Microscopy
Cells were plated on acid-washed coverslips coated with 10 μg/ml fibronectin (Sigma-Aldrich, St. Louis, MO), and allowed to spread for 6–8 h. They were fixed and stained as described in Gertler et al. (1996). Anti-vinculin Ig (hvin-1; Sigma-Aldrich) was used at 1:400. Anti-N-WASP was used at 1:1000 (gift from R. Rohgati and M. Kirschner, Harvard University, Cambridge, MA). Coumarin-phallicidin (Molecular Probes, Eugene, OR) was used at 1:20. Images were collected on a Deltavision microscope (Carl Zeiss, Thornwood, NY) and digitally deconvolved using Softworx graphics processing software (SGI, Mountain View, CA).
Video Microscopy, Quantitation, and Statistical Analysis of Motility Movies
Cells were first adapted overnight in CO2-independent video microscopy media (high-glucose Dulbecco's modified Eagle's, 350 mg/l NaHCO3, 25 mM HEPES, l-glutamine, penicillin/streptomycin, 15% fetal calf serum, and 50 U/ml interferon-γ). Then, 4,000 MVD7 or 10,000 Rat2 adapted cells were plated on a ΔT dish (Bioptechs, Butler, PA) pretreated with 10 μg/ml fibronectin and blocked for 1 h with 1 mg/ml tissue culture grade bovine serum albumin. MVD7 cells were plated for 8 h before filming, and Rat2 cells were plated for 2 h before filming. Time-lapse images were collected every 5 min for 4 h for MVD7 cells, and every 5 min for 2 h for Rat2 cells. At least two acceptable movies from each genotype were quantitated using DIAS software (Solltech, Oakdale, IA). Individual cells chosen for quantitation 1) were not in contact with other cells for >15 min (i.e., −3 frames), 2) did not undergo mitosis, and 3) stayed within the viewing area for the duration of the movie. The cell periphery was outlined in each frame by using a Wacom digital tablet (Vancouver, WA). DIAS software then computed an area-based centroid for each cell in each frame that subsequently defined a motility path for each cell. Average speed was calculated from paths for at least 20 cells/genotype. Data sets were analyzed by one-way unstacked analysis of variance (ANOVA) (n is number of cells). Statistical significance was determined by one-way unstacked analysis of variance. EGFP-Mena structural variants complemented loss of Ena/VASP function if their mean 95% confidence intervals overlapped with that of wild-type EGFP-Mena.
Biochemical Analysis of Ser/Thr Phosphorylation of EVH2 Domain
Rat2 cells stably expressing EGFP-EVH2 were grown to 90% confluence and treated with forskolin solubilized in dimethyl sulfoxide (10 or 100 μM final forskolin concentration) or dimethyl sulfoxide alone for 30 min. Cells were then lysed in NP-40 buffer (1% NP-40, 150 mM NaCl, and 50 mM Tris, pH 8.0) containing protease inhibitors (Complete tablets; Roche Applied Science, Indianapolis, IN) and phosphatase inhibitors (1 mM sodium vanadate and 1 mM sodium fluoride). Protein extracts were run on 8% SDS-PAGE gels and probed with mouse monoclonal 16C2 antibody (1:100; Vasopharm, Munich, Germany), rabbit polyclonal anti-EGFP (1:100; CLONTECH, Palo Alto, CA), and anti-Mena rabbit polyclonal 2197 (1:5000). Positive and negative controls for PKA Ser/Thr phosphorylation of Mena, VASP, and EGFP-EVH2 were generated by harvesting Rat2::EGFP-EVH2 extracts in NP-40 buffer with protease inhibitors and running either in vitro phosphatase reactions (New England Biolab) or PKA kinase reactions (New England Biolab) for 30 min.
RESULTS
All Three Murine Ena/VASP Proteins Rescue Ena/VASP-dependent Cell Motility Defects
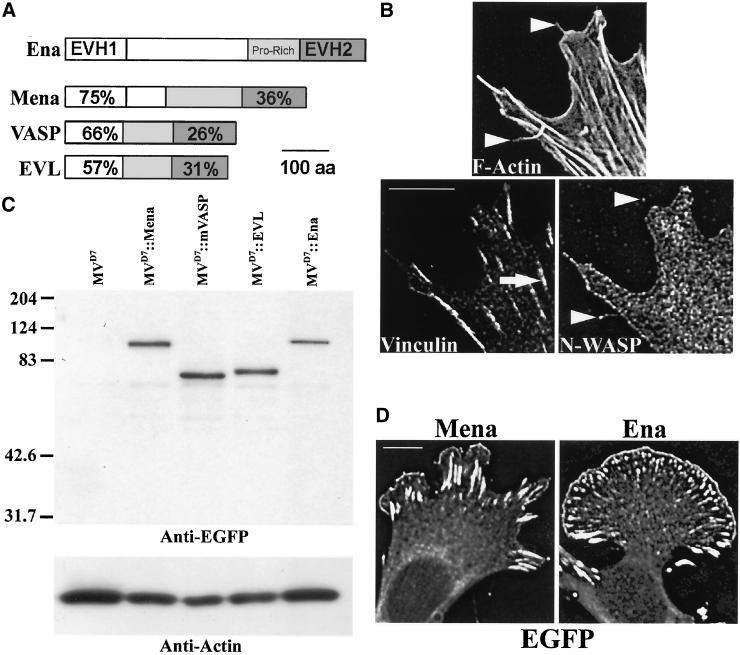
Ena/VASP proteins have a broad, overlapping expression pattern in developing and adult mice, and a variety of genetic and biochemical experiments indicate they likely share overlapping functions within most cells. We tested the ability of different Ena/VASP proteins to complement MVD7 cells in a random motility assay. At subconfluent densities, MVD7 cells have morphological attributes typical to mammalian fibroblasts, including filopodia, focal contacts, and protrusive lamellae (Figure 1B). Previously, we demonstrated that stable expression of EGFP-Mena in MVD7 cells complements loss of Ena/VASP function by decreasing their average speed (Bear et al., 2000). We have extended that analysis to include EGFP-murine VASP (mVASP) and EGFP-EVL. We also tested Drosophila EGFP-Ena because it is structurally similar to mammalian Ena/VASP proteins (Figure 1A) and because mammalian Ena/VASP transgenes complement the loss-of-function phenotype of mutations in Drosophila Ena (Ahern-Djamali et al., 1998; Gertler, unpublished data).
Figure 1.
Expression and subcellular distribution of Ena/VASP proteins in Ena/VASP-null fibroblasts. (A) All Ena/VASP proteins contain two highly conserved domains, EVH1 and EVH2, that flank a less conserved central proline-rich region. Percentages denote degree of amino acid identity between a mammalian Ena/VASP domain and the same domain in Drosophila Ena. (B) Ena/VASP null MVD7 cells make typical fibroblastic actin-dependent structures such as stress fibers terminating at vinculin-positive focal contacts (arrow in Vinculin panel), lamellae with N-WASP–positive leading edges, and F-actin rich filopodia (arrowheads in F-actin panel). Note that filopodia denoted are N-WASP positive (arrowheads in N-WASP panel). Bar, 10 μm. (C) Mammalian EGFP-Mena, EGFP-mVASP, EGFP-EVL, and Drosophila EGFP-Ena are stable in MVD7 cells and accumulate at comparable levels. Ten micrograms of total protein per lane of RIPA extracts was loaded onto an SDS-PAGE gel and analyzed by Western blot probed with anti-EGFP Ig and anti-actin Ig as a loading standard. (D) All four Ena/VASP proteins have identical subcellular distribution properties in MVD7 cells. Two examples are shown. Bar, 10 μm.
Each transgene was stably inserted by retroviral infection into MVD7 cells and sorted by FACS for uniform EGFP signal levels. This approach minimizes genetic drift of the novel transgenic line from the parental cell line and facilitates a direct comparison of activities of different protein variants. All four Ena/VASP proteins were detected by Western blotting at comparable levels (Figure 1C). Therefore, this approach generated cell populations that stably express equivalent levels of EGFP-tagged proteins on a per cell basis within each population, and comparable expression levels of the EGFP-tagged proteins among the different populations.
We previously demonstrated that Ena/VASP activity in cell motility depends on the function of Ena/VASP proteins at the cellular leading edge (Bear et al., 2000). All four family members displayed similar subcellular distributions (Figure 1D; Supplemental Fig S1, panels 8, 13, 18, and 23). Colocalization of EGFP signal with vinculin demonstrated proper localization of all four Ena/VASP proteins to focal adhesions (Supplemental Fig S1, panels 10, 15, 20, and 25), whereas colocalization of EGFP signal with N-WASP, a lamellipodial leading edge marker, demonstrated that all four family members localize to the leading edge (Supplemental Fig S1, panels 10, 15, 20, and 25). All four Ena/VASP proteins were also concentrated at the distal tips of filopodia (our unpublished data). These data indicate that all the Ena/VASP proteins exhibit a subcellular distribution pattern in MVD7 cells similar to those reported for other fibroblastic cell types (reviewed in Bear et al., 2001).
We used time-lapse video and fluorescence microscopy to analyze the ability of the different Ena/VASP proteins to function in MVD7 cells. As we previously observed with EGFP-tagged Mena, expression of EGFP-mVASP, EGFP-EVL, or Drosophila Ena in MVD7 cells did not grossly change the F-actin network of MVD7 cells as judged by phalloidin staining (Supplemental Fig S1, panels 1, 6, 11, 16, and 21). We next analyzed cell behavior. Individual living cells were filmed at high magnification to observe subcellular dynamics and filmed at lower magnification to characterize mean population speeds.
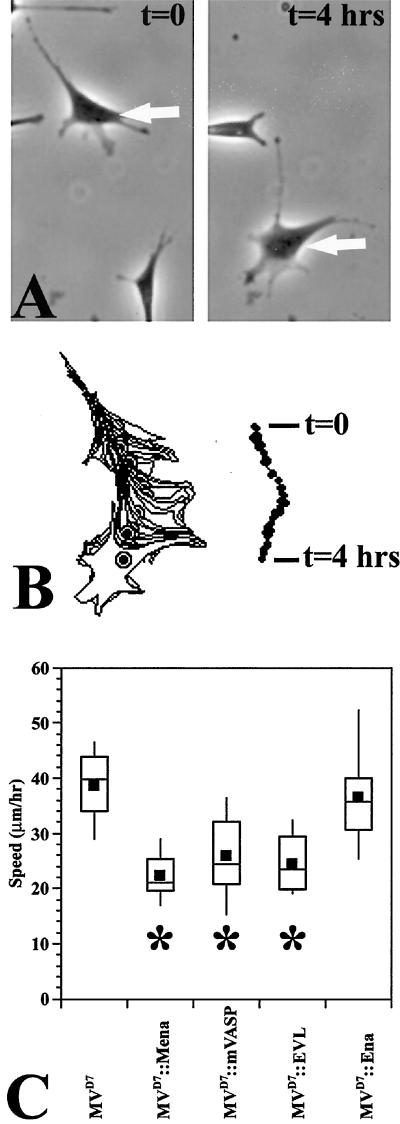
MVD7 cells complemented with any of the family members were able to form filopodia, focal adhesions, and stress fibers that were morphologically indistinguishable from the parental MVD7 cells as judged by fluorescence, phase contrast, and interference reflection microscopy of fixed and living cells (Figure 1B; Supplemental movies M1 and M2; our unpublished data). We then quantitated cell speeds by tracking individual cells over 4 h, calculating a mean speed for each cell, and comparing the population statistics of the experimental group with the parental MVD7 control group (Figure 2, A and B; Supplemental movie M3). We found, as previously shown, that MVD7 cell speeds were reduced by the expression of EGFP-Mena to physiological levels. EGFP-EVL and EGFP-mVASP reduced MVD7 cell speed equivalently, indicating that all three murine Ena/VASP proteins function interchangeably in this assay (Figure 2C). Interestingly, Drosophila EGFP-Ena failed to complement the hypermotility phenotype even though its subcellular distribution and expression levels were indistinguishable from mouse Ena/VASP proteins (Figure 2C), indicating that Drosophila Ena lacks some critical feature for function in cell movement present in all murine Ena/VASP proteins.
Figure 2.
Mammalian Ena/VASP proteins complement MVD7 cells in a random motility assay. (A) Example of MVD7 cell translocation. To quantitate eukaryotic cell speed, time-lapse movies are generated at 5-min intervals for at least 4 h. Arrows point to the same cell at two time points. (B) Left, the cell is then processed digitally by outlining the cell perimeter in each frame. Shown is an overlay of all the time points for one cell. Right, an area-based centroid is calculated from each outline in every frame to generate a path for the cell. Average speed is calculated from this data set; at least 20 cells are quantitated for each data point. Data are displayed in a Box and Whisker format to show the distribution of the entire cell population. In a Box and Whisker format, the top of the upper line marks the 90th percentile. The top of the rectangle marks the 75th percentile. Within the rectangle the black box is the mean and the horizontal line is the median. The bottom of the rectangle is the 25th percentile, and the bottom of the lower line is the 10th percentile. (C) Three mammalian Ena/VASP proteins complement MVD7 cell motility phenotypes (denoted by * beneath box and whisker plot; ANOVA, p < 10−4), but EGFP-Ena fails to complement (ANOVA, p > .05) (MVD7, n = 20; MVD7::EGFP-Mena, n = 20; MVD7::EGFP-mVASP, n = 24; MVD7::EGFP-EVL, n = 25, MVD7::EGFP-Ena, n = 20).
EGFP-Mena Mutant Proteins Are Stable in MVD7 Cells
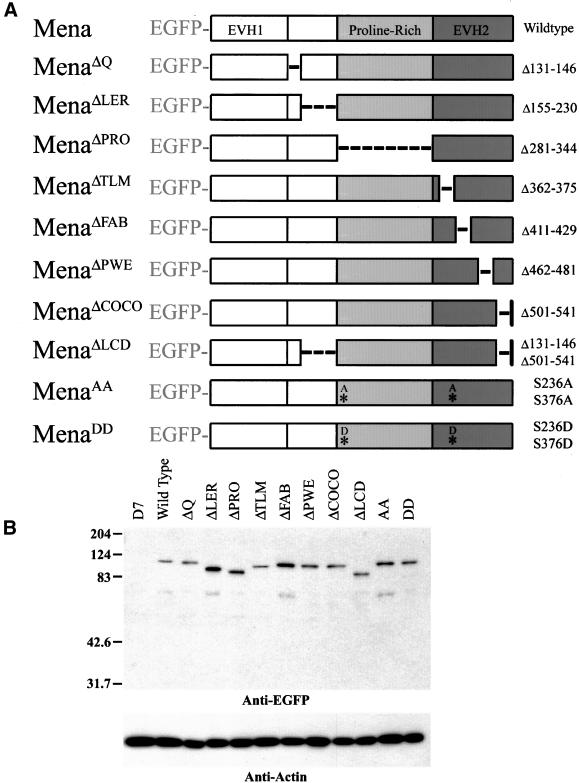
Because all murine Ena/VASP proteins functioned equivalently in the whole cell motility assay, we chose one member, Mena, to use for a structure–function analysis of Ena/VASP proteins. A series of structural variants of EGFP-Mena were generated to define the regions of EGFP-Mena that regulate cell motility properties of MVD7 cells. As noted above, extensive information on the structure and function of the EVH1 domain exists. Therefore, we focused our attention on the remainder of the protein. Small internal deletions and point mutations were chosen on the basis of evolutionary conservation and known biochemical properties (Gertler et al., 1996; Bachmann et al., 1999; Figure 3A). EGFP-Mena mutants were stably expressed in MVD7 cells and sorted for uniform EGFP signal levels as described above. Western blotting confirmed that the mutant EGFP-Mena proteins accumulated to levels comparable with the wild-type protein in MVD7 cells and migrated at their predicted sizes (Figure 3B).
Figure 3.
Small deletions and point mutations do not disrupt protein stability. (A) Series of structural mutants were engineered to determine the structural requirements for Ena/VASP function in cell motility. Small deletions or point mutations were introduced into EGFP-Mena, and those constructs were stably integrated into cell lines by retroviral insertion. Stable cell populations expressing comparable levels of EGFP-Mena transgene were isolated by FACS. (B) SDS-PAGE analysis of RIPA extracts (7.5 μg of total protein/lane) probed with anti-EGFP to detect transgene and anti-actin Ig as a loading standard. EGFP-Mena structural variants are stable and accumulate at comparable levels. Relative mobility shifts of structural variants correlate with their respective deletion sizes.
Central Proline-rich Region Is Dispensable for Ena/VASP Function in Random Whole Cell Motility
Three structural features are present between the EVH1 and the EVH2 domains of Mena. The first is a conserved block of 16 residues distal to the EVH1 domain that is deleted in EGFP-MenaΔQ. Although this block is only conserved among vertebrate Ena/VASP proteins, all Ena/VASP proteins have a high incidence of glutamine residues in their primary structure at this region. In VASP and EVL, the “Q” block is 12 residues from the most conserved Ser/Thr phosphorylation site. However, within Mena a repetitive sequence unique to Mena is inserted between the Q block and the phosphorylation site. The repeat LERER occurs six times in this 77-amino acid stretch, which also contains seven glutamines. These 77 amino acids are deleted in EGFP-MenaΔLER. Finally, all Ena/VASP proteins share a proline-rich region known to bind profilin, Src, and Abl SH3 domains, and the WW domain of FE65. This region is deleted in EGFP-MenaΔPRO.
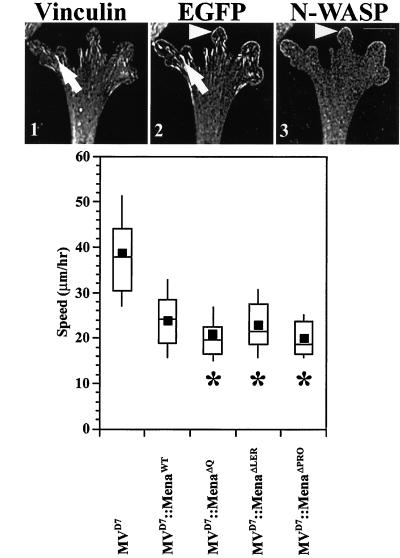
Colocalization with N-WASP and vinculin indicated that the three mutants all localized normally to the leading edge and focal adhesions, respectively (Figure 4A, panels 1–3; our unpublished data). EGFP-MenaΔQ, EGFP-MenaΔLER, and EGFP-MenaΔPRO were each capable of complementing MVD7 cells in the cell motility assay to the same extent as wild-type EGFP-Mena (Figure 4B). These results indicate that the LERER repeat unique to Mena and the conserved Q motifs are dispensable for subcellular targeting and whole cell movement. Surprisingly, the interactions between the polyproline-cluster of Ena/VASP proteins and proteins such as profilin are not required for proper subcellular localization or for function of Ena/VASP proteins in random cell motility.
Figure 4.
Deletion of regions between the EVH1 and EVH2 domain does not affect localization or function in cell motility. (A) EGFP-MenaΔQ, EGFP-MenaΔLER, and EGFP-MenaΔPRO have subcellular distribution properties similar to wild-type EGFP-Mena. EGFP-MenaΔPRO is shown. Anti-vinculin Ig (arrow in panel 1) colocalizes with EGFP-Mena mutants at focal adhesions (compare arrows in panels 1 and 2). Anti-N-WASP Ig (arrowhead in panel 3) colocalizes with EGFP-Mena mutants at the leading edge (compare arrowheads in panels 2 and 3). Bar, 10 μm. (B) Box and Whisker plot of speed distributions indicate that EGFP-MenaΔQ, EGFP-MenaΔLER, and EGFP-MenaΔPRO cell populations have speeds comparable with rescued cell populations (ANOVA, p < 10−4 for each) with speeds comparable with EGFP-MenaWT (MVD7, n = 30; MVD7::EGFP-Mena, n = 24; MVD7::EGFP-MenaΔQ, n = 26; MVD7::EGFP-MenaΔLER, n = 22; MVD7::EGFP-MenaΔPRO, n = 21).
Interaction with F-Actin Network Is Essential for Ena/VASP Function in Cell Motility
Our previous study indicated that the EVH1 domain alone is detected in the cytoplasm, at focal adhesions, weakly along the leading edge, and in the nucleus (a property of EGFP alone) (Bear et al., 2000). Because deletion of the proline-rich region resulted in a subcellular distribution equivalent to that of full-length Mena, we speculated that the EVH2 domain harbors an activity that increases the efficiency of leading edge targeting. We deleted four conserved blocks within the EVH2 domain to test this hypothesis.
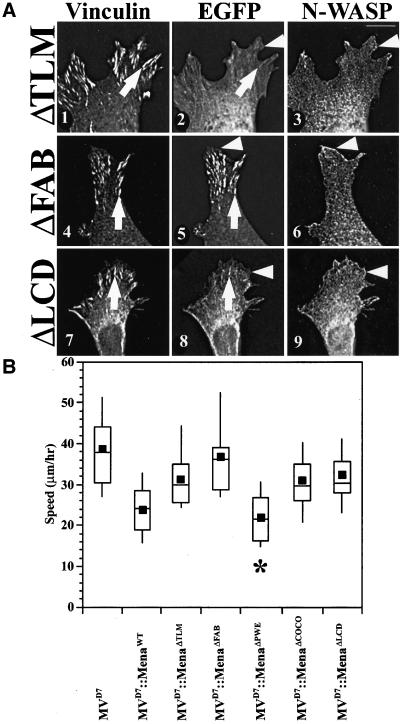
The first conserved region in the EVH2 domain, Thymosin-β4-Like Motif (TLM), is a motif related to the actin-monomer binding site in Thymosin-β4 (Van Troys et al., 1996). Although EGFP-MenaΔTLM was excluded from the nucleus, it was diffusely distributed throughout the cytoplasm (Figure 5A, panels 1–3), weakly detected at focal adhesions, and barely detectable along the leading edge of lamellipodia. EGFP-MenaΔTLM failed to complement the MVD7 motility phenotype (Figure 5B).
Figure 5.
EVH2 domain is necessary for function in cell motility. (A) EGFP-MenaΔTLM and EGFP-MenaΔFAB have altered subcellular distribution properties compared with wild-type EGFP-Mena. EGFP-MenaΔTLM is mostly diffuse throughout the cytoplasm, but is slightly enriched at focal adhesions (arrows in panels 1 and 2). Arrowhead in panel 3 denotes N-WASP–positive leading edge, which is not enriched for EGFP-MenaΔTLM (compare arrowheads in panels 2 and 3). EGFP-MenaΔFAB is enriched at focal adhesions (arrows in panels 4 and 5), but is not enriched at the leading edge compared with wild-type protein (arrowheads in panels 5 and 6). EGFP-MenaΔLCD also had altered subcellular distribution properties compared with wild-type EGFP-Mena. It is mostly diffuse throughout the cytoplasm but is detected at focal adhesions (compare arrows in panels 7 and 8). EGFP-MenaΔLCD is barely detected at the leading edge (compare arrowheads in panels 8 and 9). Bar, 10 μm. (B) Box and Whisker plot of speed distributions indicate that EGFP-MenaΔTLM and EGFP-MenaΔFAB do not complement loss of Ena/VASP function in MVD7 random cell motility assays (ANOVA, p < 10−4 for each), and EGFP-MenaΔPWE cell populations have speeds comparable with rescued cell populations (ANOVA, p < 10−4). Box and Whisker plot of speed distributions also indicate that EGFP-MenaΔCOCO and EGFP-MenaΔLCD do not complement loss of Ena/VASP function in MVD7 random cell motility assays (ANOVA, p < 10−4 for each) (MVD7, n = 30; MVD7::EGFP-Mena, n = 24; MVD7::EGFP-MenaΔTLM, n = 28; MVD7:: EGFP-MenaΔFAB, n = 20; MVD7::EGFP-MenaΔPWE, n = 26; MVD7::EGFP-MenaΔCOCO, n = 27; MVD7::EGFP-MenaΔLCD, n = 30).
The next conserved region within the EVH2 domain binds F-actin in vitro (Bachmann et al., 1999; Lambrechts et al., 2000) and was deleted to create EGFP-MenaΔFAB (F-actin binding). EGFP-MenaΔFAB localized robustly to focal adhesions, but was only barely detectable along the leading edge of lamellipodia, comparable with the signal observed with the EVH1 domain alone, suggesting that the F-actin binding motif plays an important role in concentrating Ena/VASP at lamellipodial tips (Figure 5A, panels 4–6). EGFP-MenaΔFAB failed to complement the motility defects in MVD7 cells, indicating that the ability to interact with F-actin is essential for the function of Ena/VASP proteins in whole cell movement (Figure 5B). Interestingly, EGFP-MenaΔFAB fully supports intracellular Listeria motility (Geese et al., 2002), indicating that this mutant retains some type of biological activity.
The third region is a small set of 12 conserved amino acids defined by the EGFP-MenaΔPWE mutant that is between the FAB and the oligomerization region (COCO; see below). No known function has been ascribed to the PWE region. EGFP-MenaΔPWE localized in a pattern similar to the wild-type protein (our unpublished data), and fully complemented the motility phenotype of MVD7 cells (Figure 5B).
Potential Oligomerization Motifs Are Required for Full Ena/VASP Function in Cell Motility
The C terminus of the EVH2 domain contains a predicted coiled-coil region that can mediate oligomerization of Ena/VASP proteins (Ahern-Djamali et al., 1998; Carl et al., 1999). To assess the requirement for oligomerization in Ena/VASP function, we made two mutants intended to disrupt the formation of EGFP-Mena oligomers. EGFP-MenaΔCOCO harbors an internal deletion that excises the predicted coiled-coil motif in the EVH2 domain. EGFP-MenaΔCOCO localized within the cytosol, was enriched at focal adhesions, but was only weakly detected at lamellipodial leading edges (our unpublished data). In the motility assay, EGFP-MenaΔCOCO provided only partial function compared with the wild-type protein, but it did reduce average cell speeds significantly compared with the parental MVD7 cell line (ANOVA, p < 0.05) (Figure 5B).
We wondered whether the Mena-specific LERER region could contribute to function in the absence of the COCO region. The LERER repeat region is predicted to form a potential extended helix with alternating charges, and therefore could serve to form an additional oligomerization motif. As described above, deletion of the LERER region by itself has no effect on localization or function of EGFP-Mena, we deleted both the LERER region and the coiled-coil region to generate EGFP-MenaΔLCD (LER-COCO double-mutant). Although this mutant had similar subcellular distribution properties to EGFP-MenaΔCOCO, it was more difficult to detect along the leading edge (Figure 5A, panels 7–9). In the motility assay, EGFP-MenaΔLCD did not alter the hypermotile phenotype of MVD7 cells (Figure 5B). These results suggest that oligomerization of Ena/VASP proteins is required for full function of these proteins in cell motility.
Ser/Thr Phosphorylation Regulates Mammalian Ena/VASP Protein Function
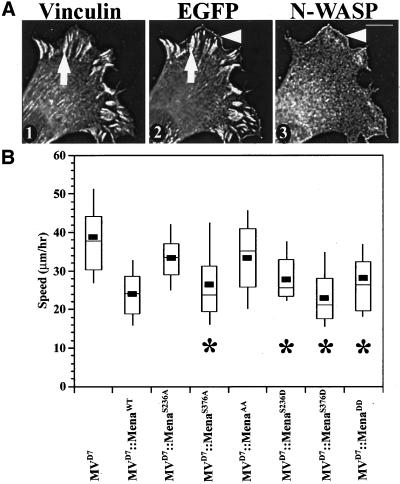
The failure of Drosophila Ena to replace its murine orthologs in the motility assay prompted us to focus on features conserved within the vertebrate proteins that are missing in Ena. Between one and three cyclic nucleotide-dependent kinase phosphorylation sites flank the proline-rich core in all vertebrate Ena/VASP proteins. Drosophila Ena lacks any known PKA/PKG phosphorylation sites. To test whether phosphorylation of the highly conserved PKA/PKG sites is required for Ena/VASP function in mammalian cell motility, six different EGFP-Mena mutants were engineered. Individual phosphorylation sites are mutated from serine to alanine in EGFP-MenaS236A and EGFP-MenaS376A to block phosphorylation of only one site. Conversely, each phosphorylation site is mutated from serine to aspartic acid in EGFP-MenaS236D and EGFP-MenaS376D to mimic constitutive phosphorylation at each site. In EGFP-MenaAA, the two phosphorylation sites were each mutated to alanine (S236A and S376A), whereas in EGFP-MenaDD, both phosphorylation sites were mutated to aspartic acids (S236D and S376D).
All six phosphomutants localized in a subcellular pattern indistinguishable from wild-type EGFP-Mena, colocalizing with vinculin at focal adhesions and with N-WASP at the leading edge (Figure 6A, panels 1–3; our unpublished data). However, EGFP-MenaAA failed to complement loss of Ena/VASP function, whereas EGFP-MenaDD caused a statistically significant reduction of cell speed (Figure 6B). This latter result is consistent with in vitro analysis suggesting that replacing serines with aspartic acid in Ena/VASP proteins mimics phosphorylation (Harbeck et al., 2000). Functional analysis of the single point mutations indicate that Ser236, the only phosphorylation site conserved in all three murine Ena/VASP proteins, is more critical because conversion of that site alone to alanine affects Ena/VASP function. Mutation of Ser376 to alanine alone had no effect on Ena/VASP function. As with EGFP-MenaDD, mutation of either phosphorylation site to aspartic acid did not disrupt function in whole cell motility. These results indicate that although Ena/VASP phosphorylation is essential for function in cell movement, phosphorylation has no obvious role in subcellular targeting of the proteins. Furthermore, blocking phosphorylation at the site common to all three mammalian Ena/VASP proteins is sufficient to block EGFP-Mena function in cell motility.
Figure 6.
Blocking Ser/Thr phosphorylation does not disrupt localization, but affects function. (A) EGFP-MenaAA has subcellular distribution properties indistinguishable from wild-type EGFP-Mena. Anti-vinculin Ig detects focal adhesion complexes (arrow in panel 1) that colocalize with EGFP-Mena phosphomutants (compare arrows in panels 1 and 2). Anti-N-WASP Ig marks the lamellipodial leading edge (arrowhead in panel 3) and colocalizes with EGFP-Mena mutants (compare arrowheads in panels 2 and 3). Bar, 10 μm. (B) Box and Whisker plot of speed distributions indicate that EGFP-MenaS236A and EGFP-MenaAA cells have speeds comparable with parental cell lines (ANOVA, p < 10−4), whereas EGFP-MenaS376A, EGFP-MenaS236D, EGFP-MenaS376D, and EGFP-MenaDD speeds are statistically slower, with speeds comparable with EGFP-MenaWT (ANOVA, p < 10−4; MVD7, n = 30; MVD7::EGFP-Mena, n = 24; MVD7::EGFP-MenaS236A, n = 20; MVD7::EGFP-MenaS376A, n = 20; MVD7::EGFP-MenaAA, n = 31; MVD7::EGFP-MenaS236D, n = 20; MVD7::EGFP-MenaS376D, n = 20; MVD7::EGFP-MenaDD, n = 20).
Some EGFP-Mena Mutants That Fail to Complement MVD7 Cells Induce an Overexpression Phenotype in Rat2 Fibroblasts
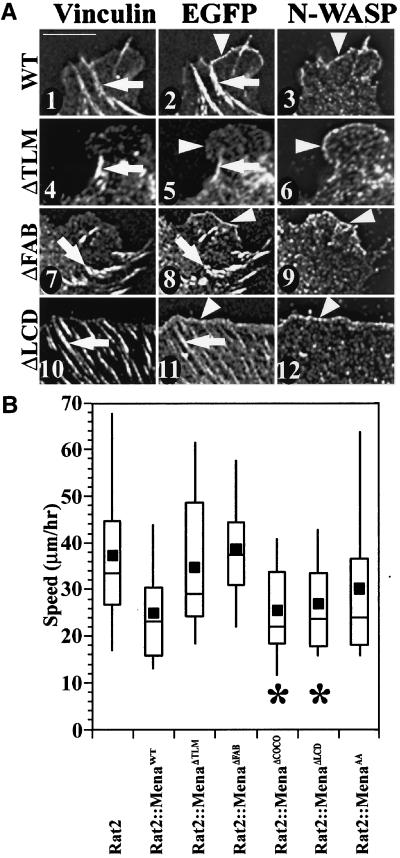
Previously, we reported that overexpression of Mena to 4 times normal levels causes a significant reduction in the speed of Rat2 fibroblasts (Bear et al., 2000). We used this overexpression assay as a second way to test whether any of the mutants that failed to rescue normal motility of MVD7 retained some function in this overexpression assay. We made stable Rat2 cell lines expressing EGFP-Mena mutants and assayed the subcellular distribution (Figure 7A) and functional capacity of those mutants compared with wild-type EGFP-Mena (Figure 7B).
Figure 7.
Some of the mutants that failed to complement MVD7 cells had an overexpression phenotype when expressed in Rat2 cells, which express endogenous Ena/VASP proteins. (A) EGFP-Mena expressed in Rat2 fibroblasts accumulates at focal adhesions (arrows in panels 1 and 2) and along lamellipodial leading edges (arrowheads in panels 2 and 3). EGFP-MenaΔTLM is detected at focal adhesions (arrows in panels 4 and 5), but not at the leading edge (arrowheads in panels 5 and 6). EGFP-MenaΔFAB localizes to both focal adhesions (arrows in panels 7 and 8) and the leading edge (arrowheads in panels 8 and 9). Both EGFP-MenaΔCOCO (our unpublished data) and EGFP-MenaΔLCD have similar subcellular distribution patterns in Rat2 fibroblasts with enrichment at focal adhesions (arrows in panels 10 and 11) and the leading edge (arrowheads in panels 11 and 12). Bar, 10 μm. (B) Box and Whisker plot of speed distributions indicate that EGFP-MenaΔTLM, EGFP-MenaΔFAB, and EGFP-MenaAA do not elicit an overexpression phenotype in Rat2 random cell motility assays (ANOVA, p < 10−4 for each population) and EGFP-MenaΔCOCO and EGFP-MenaΔLCD cell populations have speeds comparable with cell populations overexpressing wild-type EGFP-Mena (ANOVA, p < 10−4 for each) (Rat2, n = 38; Rat2::EGFP-Mena, n = 30; Rat2::EGFP-MenaΔTLM, n = 27; Rat2::EGFP-MenaΔFAB, n = 30; Rat2::EGFP-MenaΔCOCO, n = 31, Rat2::EGFP-MenaΔLCD, n = 22; Rat2::EGFP-MenaAA, n = 33).
We tested whether overexpression of nonphosphorylatable EGFP-Mena affected cell speed. As in MVD7 cells, EGFP-MenaAA localization was indistinguishable from wild-type EGFP-Mena (our unpublished data). Analysis of cell speeds indicated that EGFP-MenaAA failed to induce a statistically significant reduction in the overall speeds of the cell population (Figure 7B). However, visual inspection of the tracking movies suggested that some cells within the EGFP-MenaAA–expressing population did seem to move more slowly than parental Rat2 cells. In fact, the median value of EGFP-MenaAA speed was nearly identical to that of wild-type EGFP-Mena (indicating that at least half the cells within the population were slowed to the same extent as wild-type EGFP-Mena overexpression).
As in MVD7 cells, EGFP-MenaΔTLM again localized diffusely throughout the cytoplasm (Figure 7A, panels 4–6). This was surprising because EGFP-MenaΔTLM should oligomerize with endogenous Ena/VASP proteins, and suggests that deletion of this small conserved region may have broader consequences on the structure of Ena/VASP proteins. Overexpression of EGFP-MenaΔTLM does not reduce Rat2 cell speed (Figure 7B).
The subcellular distribution of EGFP-MenaΔFAB was significantly altered in Rat2 cells compared with MVD7 cells. Whereas EGFP-MenaΔFAB was nearly absent from the leading edge of MVD7 lamellipodia, it was clearly detected along the leading edge of Rat2 lamellipodia, perhaps due to its ability to oligomerize with endogenous Ena/VASP proteins (Figure 7A, panels 7–9). Although EGFP-MenaΔFAB is enriched at the leading edge of Rat2 cells, it does not cause an overexpression phenotype (Figure 7B). This indicates that the F-actin binding motif of the EVH2 domain is essential for the phenotype induced by overexpression of Ena/VASP proteins.
EGFP-MenaΔCOCO and EGFP-MenaΔLCD had the same properties in Rat2 cells. Both proteins were found at focal adhesions, but were mostly diffuse throughout the cytoplasm (Figure 7A, panels 10–12; our unpublished data). However, both EGFP-MenaΔCOCO and EGFP-MenaΔLCD expression in Rat2 cells elicited an overexpression phenotype comparable with overexpression of wild-type EGFP-Mena in Rat2 cells (Figure 7B).
EVH2 Domain Alone Is Sufficient for Ena/VASP Function in Random Whole Cell Motility in Absence of PKA Phosphorylation
Our functional assays confirmed the physiological importance of previous observations indicating that Ena/VASP proteins interact with F-actin and that they can multimerize with each other. Both of these functions map to the conserved EVH2 domain, and mutations within that domain disrupt localization and function. We wondered whether expression of the EVH2 domain alone would complement MVD7 cell motility phenotypes.
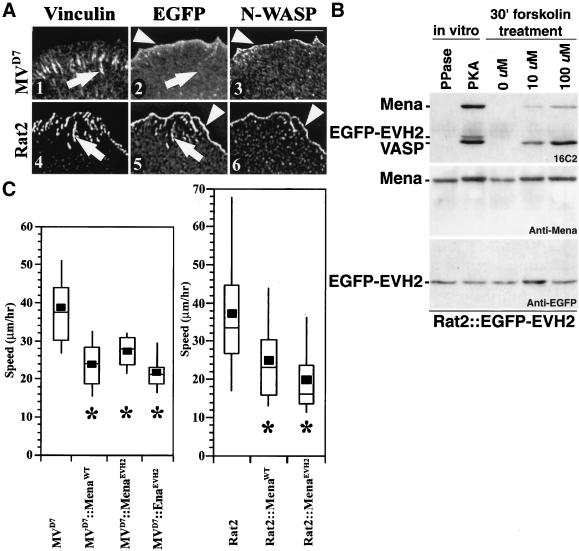
EGFP-EVH2 localized to the lamellipodia of MVD7 cells, although it exhibited a broader distribution within this structure than EGFP-Mena, which concentrates just at the distal edge of lamellipodia (Figure 8A, panels 1–3). EGFP-EVH2 was not enriched at focal adhesions, but did weakly decorate stress fibers. The failure of EGFP-EVH2 to target focal adhesions in MVD7 cells is consistent with previous work indicating the essential role of EVH1-mediated interactions in focal adhesion targeting of Ena/VASP proteins (Gertler et al., 1996; Bear et al., 2000). When EGFP-EVH2 was expressed in Rat2 cells, the EGFP signal was concentrated at the tips of lamellipodia and in focal adhesions in a pattern identical to the endogenous Ena/VASP proteins, likely due to the ability of the EVH2 domain to oligomerize with endogenous Ena/VASP proteins (Figure 8A, panels 4–6). Together, these results indicate that although the EVH2 domain can target to a broad region of the lamellipodia, other parts of Ena/VASP proteins such as the EVH1 domain are required for targeting to the tip of lamellipodia and to focal adhesions.
Figure 8.
EVH2 domain negatively regulates cell motility in both MVD7 and Rat2 fibroblasts. (A) EGFP-EVH2 does not accumulate at focal adhesions in MVD7 cells (compare arrows in panels 1 and 2) but is detected at focal adhesions in Rat2 cells (compare arrows in panels 4 and 5). EGFP-EVH2 is enriched along the leading edge of MVD7 cells (arrowheads in panels 2 and 3), but EGFP-EVH2 has a slightly broader distribution pattern along the edge of MVD7 cells. In contrast, EGFP-EVH2 is tightly colocalized with N-WASP along the leading edge of Rat2 fibroblasts (arrowheads in panels 5 and 6). Bar, 10 μm. (B) EVH2 domain is not phosphorylated in Rat2 cells. Rat2::EGFP-EVH2 cells were treated with 10 or 100 μM forskolin for 30 min and compared with extracts treated in vitro with phosphatase or PKA as negative and positive controls, respectively. Top panel is blotted with 16C2, which recognizes three bands in the PKA-treated positive control lane corresponding from top to bottom with Mena, EGFP-EVH2, and VASP. Although VASP and Mena have a dosage sensitive increase in the phosphorylation of their EVH2 domains, the EVH2 domain by itself is not as good a substrate for forskolin-induced phosphorylation. (C) Box and Whisker plot of speed distributions indicate that EGFP-EVH2 can complement loss of Ena/VASP function in MVD7 cells (left panel; ANOVA, p < 10−4) and can elicit an overexpression phenotype in Rat2 fibroblasts (right panel; ANOVA, p < 10−4). Expression of the EVH2 domain of Ena is also capable of complementing MVD7 hypermotility phenotype. (MVD7, n = 30; MVD7::EGFP-Mena, n = 24; MVD7::EGFP-EVH2, n = 22; MVD7::EGFP-Ena-EVH2, n = 20; Rat2, n = 38; Rat2::EGFP-Mena, n = 30; Rat2::EGFP-EVH2, n = 27).
We next tested EGFP-EVH2 in the cell motility assays. EGFP-EVH2 complemented loss of Ena/VASP function in MVD7 cells to an extent equivalent to the full-length protein (Figure 8C). Similarly, when EGFP-EVH2 was expressed in Rat2 cells, it induced an overexpression phenotype identical to intact Mena (Figure 8C). We also tested whether the EVH2 domain of Ena complements MVD7 cells and found that it, too, reduces average cell speeds (Figure 8C).
We wondered whether the EVH2 domain alone is a substrate for PKA Ser/Thr phosphorylation. To test this we treated Rat2 cells stably expressing EGFP-EVH2 with forskolin, which stimulates adenylate cyclase, thereby increasing the intracellular concentration of cAMP necessary to activate PKA within cells. We found that a mouse monoclonal antibody, 16C2, which was developed for detection of VASP proteins that are phosphorylated on Ser238, cross-reacts in vitro with PKA-phosphorylated Mena and EGFP-EVH2 (Figure 8B). Surprisingly, we found that forskolin-treated Rat2::EGFP-EVH2 cells labeled both VASP and Mena but failed to robustly label EGFP-EVH2. We also found that EGFP-EVH2 is not robustly phosphorylated in forskolin-treated MVD7 cells (our unpublished data). This suggests that phosphorylation of the EVH2 domain is not required for its function in whole cell motility (see below), and it also suggests that corecruitment of EGFP-EVH2 to the leading edge by binding to endogenous Ena/VASP proteins in Rat2 cells is not sufficient to phosphorylate the EVH2 domain. Recall that neither full-length Ena (Figure 2C) nor EGFP-MenaS236A (Figure 6B) complements the MVD7 hypermotility phenotype, suggesting that elements outside the EVH2 domain of full-length Ena/VASP protein regulate the physiological activity of the EVH2 domain. Together, these results indicate that the EVH2 domain contains the core mechanistic elements of Ena/VASP proteins required for their function in random whole cell motility, but that the EVH1 and/or central proline-rich region are required to regulate Ena/VASP function.
DISCUSSION
We have conducted a structure–function analysis to identify features of Ena/VASP proteins required for function in three different assays: subcellular targeting in MVD7 cells, rescue of the hypermotile phenotype of MVD7 cells, and overexpression effects on Rat2 fibroblast motility (Table 1). In a companion study, we analyzed several of these mutants for their ability to support intracellular Listeria motility (Geese et al., 2002; Table 1).
Table 1.
Summary of subcellular localization properties and functional capabilities in three distinct assays
| Localization in MVD7
|
Functional assay
|
||||
|---|---|---|---|---|---|
| Focal adhesions | Leading edge | Complement cell motility in MVD7 | Overexpression phenotype in Rat2 cells | Complement Listeria motility in MVD7 | |
| MenaWT | + | + | + | + | + |
| MenaΔQ | + | + | + | NA | NA |
| MenaΔLER | + | + | + | NA | NA |
| MenaΔPRO | + | + | + | NA | +/− |
| MenaΔTLM | +/− | − | − | − | + |
| MenaΔFAB | + | − | − | − | ++ |
| MenaΔPWE | + | + | + | NA | NA |
| MenaΔCOCO | +/− | +/− | +/− | + | + |
| MenaΔLCD | +/− | − | − | + | NA |
| MenaS236A | + | + | +/− | NA | NA |
| MenaS376A | + | + | + | NA | NA |
| MenaAA | + | + | − | +/− | +/− |
| MenaS236D | + | + | + | NA | NA |
| MenaS376D | + | + | + | NA | NA |
| MenaDD | + | + | + | NA | ++ |
| MenaEVH2 | − | + | + | + | NA |
For subcellular localization: +, denotes localization comparable with wild-type EGFP-Mena; +/−, signifies presence of EGFP signal weaker than wild-type control; and −, signifies no or extremely weak EGFP signal. For functional studies, ++ is more active than wild type, + is comparable to wild type, +/− is less than wild type, and − is no function detected.
NA, not analyzed.
One striking result of these studies is that different experimental assays revealed distinct structural requirements for Ena/VASP function. In the case of whole cell motility, the F-actin binding motif within the EVH2 domain is essential for localization within MVD7 cells and for activity in both MVD7 and Rat2 cells. In contrast, intracellular Listeria motility is unaffected or even enhanced by the absence of the FAB motif. Conversely, the polyproline-rich region of Mena is dispensable for function in random whole cell movement, but seems to play an important role in intracellular Listeria motility (Geese et al., 2002).
We draw three general conclusions from these observations. First, all of the mutants studied displayed partial or complete activity in at least one of the functional assays. This observation combined with the fact that all the mutant proteins were detectable by both FACS analysis and Western blotting suggests that failure of a given mutant to function in one of these assays is unlikely to result from a trivial failure in global protein folding. Second, the role of Ena/VASP proteins in Listeria motility differs from their function in lamellipodia. We postulate that different sets of features within the same molecules are required differentially for each process. Third, on a more general note, the requirements for various Ena/VASP motifs in these assays suggest that Ena/VASP proteins may be used in distinct ways by different actin-driven processes. As a result, it may not be prudent to assess the function of these molecules in other contexts, such as Jurkat T-cell polarization or axonal growth cone guidance (Lanier et al., 1999; Krause et al., 2000), solely by extrapolation from the results obtained in fibroblast motility or in Listeria motility assays. Furthermore, it is possible that other cell types or processes may use the three murine Ena/VASP proteins in ways that are not interchangeable.
Polyproline-rich Region Is Dispensable for Ena/VASP Regulation of Random Cell Motility
The identification of Ena/VASP proteins as profilin ligands, mediated through the proline-rich region, provided a potential mechanism by which this protein family might regulate actin assembly (Reinhard et al., 1995). Profilin binds G-actin, promotes the exchange of ADP for ATP on G-actin, and permits bound ATP-actin to be added onto the barbed ends of actin filaments (reviewed in Pollard et al., 2000). The observations that both Ena/VASP proteins and profilin can increase the rate of Listeria motility within cell-free systems (Loisel et al., 1999) and that profilin recruitment is proportional to intracellular Listeria speed (Geese et al., 2000) supports such a model. In this model, Ena/VASP–profilactin complexes concentrate actin monomer at sites of rapid actin assembly. However, in cell-free systems Ena/VASP proteins can increase the rate of Listeria motility in the absence of profilin (Loisel et al., 1999). Consistent with this, EGFP-MenaΔPRO can partially restore Listeria motility in MVD7 cells (Geese et al., 2002).
Our results indicate that the central proline-rich region is dispensable for normal subcellular targeting and function in fibroblast motility, therefore interactions with profilin, SH3, and WW domains are all dispensable for the function of Ena/VASP proteins in random cell movement. Previous genetic studies suggest that, in the absence of Mena, the process of neurulation is sensitive to the level of profilin I (Lanier et al., 1999). It is possible that the concentration of profilin in MVD7 is high enough such that direct interaction with Ena/VASP proteins is not required for their function, even if the molecules do form complexes within cells. Alternatively, profilin recruitment may not be essential to regulate whole cell motility, but it may be important for other functions of this protein family. Additionally, recent reports have postulated a role for WW-mediated binding of Fe65 to Mena in the regulation of cell motility (Sabo et al., 2001), and a requirement for SH3-mediated binding of IRSp53 to Mena in the promotion of filopodial outgrowth (Krugmann et al., 2001). As noted, Ena/VASP-null cells possess morphologically normal filopodia, indicating that they are not strictly required for filopodial formation. Because expression of EGFP-MenaΔPRO is sufficient to complement cell motility phenotypes, binding of ligands to the proline-rich region is not required for Ena/VASP function in cell motility. Although this discrepancy may reflect cell type differences, it does prompt a reevaluation of models in which profilin or SH3/WW-domain recruitment is critical for the function of Ena/VASP proteins in cell motility.
Role of F-Actin Binding Activity in Ena/VASP Localization and Function
The F-actin binding motif within the EVH2 domain plays an important role in Ena/VASP targeting to the leading edge within MVD7 cells. Interestingly, the EGFP-MenaΔFAB mutant displayed a normal subcellular distribution in Rat2 cells, presumably due to oligomerization with endogenous Ena/VASP proteins. Similarly, subcellular targeting by the isolated EVH2 domain was affected by the presence of endogenous Ena/VASP proteins, a factor not controlled for in two recent studies that proposed a role for the EVH2 domain in subcellular targeting (Price and Brindle, 2000; Nakagawa et al., 2001).
Previously, we reported that the EVH1 domain could direct green fluorescent protein (GFP) to the leading edge and focal adhesions, although the targeting was weak and accompanied by a background nuclear signal. Despite its ability to target GFP, the EVH1 domain alone fails to mediate robust leading edge targeting of full-length Ena/VASP proteins. Deletion of the 18 residue F-actin binding motif within the EVH2 domain resulted in a mutant protein that could target appropriately to focal adhesions, but at best weakly to the leading edge. Consistent with these results, we have recently shown that Ena/VASP proteins are recruited to the leading edge by a direct interaction with the barbed ends of elongating actin filaments (Bear et al., unpublished data).
The EVH2 domain by itself can target GFP to the lamellipodia by a mechanism that depends on the FAB motif (our unpublished data). By itself, EVH2 does not decorate focal adhesions. Close examination of the distribution of EGFP-EVH2 revealed that EVH2 alone targets a broader region of lamellipodia than full-length Ena/VASP proteins, which decorate only the tips of protruding lamellipodia. We speculate that the EVH1 domain refines the EVH2-mediated actin-dependent targeting of Ena/VASP proteins, thereby restricting them to the very tips of lamellipodia by interacting with either unknown protein ligands or perhaps phosphotidylinositol-containing phospholipids.
EVH2 Domain Is Sufficient to Regulate Random Motility
Although the EVH2 domain alone does not fully recapitulate wild-type Ena/VASP localization within lamellipodia, it functions equivalently to full-length Mena protein in fibroblast motility assays. Within MVD7 cells, EVH2 localizes to lamellipodia but not to focal adhesions, confirming our previous observations that the hypermotile phenotypes observed by genetic deletion or neutralization approaches within fibroblasts are a consequence of effects on lamellipodia and do not involve loss of Ena/VASP function at focal adhesions.
The EVH2 domain contains two motifs, FAB and COCO, with established biochemical properties. Although EGFP-MenaΔFAB localizes predominantly to focal adhesions in MVD7 cells, in Rat2 cells it is also detected at the leading edge. Because EGFP-MenaΔFAB fails to elicit an overexpression phenotype in Rat2 cells we conclude that F-actin binding is essential for the function of Ena/VASP proteins within lamellipodia. The capacity of EGFP-MenaΔFAB to support normal, or even enhanced rates of intracellular Listeria motility (Geese et al., 2002) provides conclusive evidence that Ena/VASP proteins are used by this pathogen by a mechanism that is distinct from the ways in which these same molecules function in lamellipodia during whole cell motility.
In Rat2 cells, as in MVD7 cells, oligomerization mutants localize to focal adhesions, but are generally diffuse throughout the cytoplasm, suggesting that oligomerization plays a role in targeting to lamellipodia. Because the EVH2 domain alone is sufficient to support normal motility, and EGFP-MenaΔCOCO is not, we conclude that oligomerization is necessary for full function of the EVH2 domain.
EGFP-MenaΔTLM still contains the coiled-coil region that seems functional in other mutants. TLM is similar to a motif that mediates G-actin binding within molecules such as Thymosin-β4 and Villin (Gertler et al., 1996). The failure of EGFP-MenaΔTLM to localize properly in Rat2 cells suggests that deletion of this 14 amino acid residue region may have a broader impact on Mena structure, although the capacity of EGFP-MenaΔTLM to localize to focal adhesions and to partially support Listeria motility suggests that it retains some function. It will be important to determine whether the TLM region actually binds G-actin and to establish what role this motif plays in the overall function of the EVH2 domain.
Regulation of Ena/VASP Proteins by Phosphorylation
PKA/PKG phosphorylation of Ena/VASP proteins has been correlated with a number of physiological processes that involve cytoskeletal remodeling (Walter et al., 1993). Furthermore, inhibition of platelet aggregation by cyclic nuleotide kinase agonists is dramatically attenuated in VASP-deficient mice (Aszodi et al., 1999; Hauser et al., 1999). There are three PKA/PKG sites in VASP, two are present in Mena, and only the amino terminal site is contained within EVL. We analyzed the functional requirements for the two sites found in Mena. Phosphorylation of the first, highly conserved site is essential for function, whereas we observed no obvious role for the second site in our assays. Because phosphorylation of this first site also induces a shift in the electorphoretic mobility of Mena, EVL, and VASP, we propose that it is the major site for regulation of this protein family in vertebrates.
Surprisingly, EGFP-Ena failed to complement the MVD7 random cell motility phenotype, although it is structurally similar to murine Ena/VASP proteins and localized appropriately. This result is especially striking in light of the ability of vertebrate Ena/VASP proteins to replace Ena function in Drosophila. Although Drosophila Ena is phosphorylated on serine as well as tyrosine (Gertler et al., 1995), it lacks the highly conserved PKA/PKG site found in vertebrates (Gertler et al., 1996). The ability of the isolated Ena EVH2 domain to function in mammalian cells indicates that it contains all of the key properties required for function of the domain. It seems likely that the reason why intact Ena fails to function in mammalian cells is that regulation by PKA/PKG is a feature that has been incorporated into Ena/VASP proteins after the divergence between invertebrates and vertebrates.
The isolated Mena EVH2 domain, which complements MVD7 cells, lacks the key site contained in all the vertebrate proteins. Interestingly, the site within the Mena EVH2 domain is robustly phosphorylated in the intact protein, but not when the EVH2 domain is expressed by itself. It is likely that interactions with the EVH1 or proline-rich region are important for recruiting protein complexes that contain PKA/PKG. One candidate class of proteins may be A-kinase anchoring proteins, which localize PKA to specific regions within cells (reviewed in Diviani and Scott, 2001).
How does phosphorylation regulate Ena/VASP function? Phosphorylation plays no obvious role in subcellular targeting, suggesting that the Ena/VASP proteins are regulated at their sites of function. In addition to causing shifts in electrophoretic mobility, phosphorylation of Ena/VASP proteins is known to alter their affinities for some, but not all, of their binding partners in vitro (Halbrugge et al., 1990; Gertler et al., 1996; Lambrechts et al., 2000). The most conserved phosphorylation site within vertebrate Ena/VASP proteins lies between the EVH1 and EVH2 domains. Substitution of a phosphomimetic aspartic acid for the serine residue at this site permitted Mena function in MVD7 cells, providing further evidence that the phosphorylated form of the protein is active and suggesting that cycling between phospho- and dephospho-forms may not be required for Ena/VASP function. Phosphorylation also seems to increase the ability of Ena/VASP proteins to support Listeria motility. Therefore, phosphorylation likely activates Ena/VASP proteins in the context of a variety of cellular functions. Together, our data lead us to propose that phosphorylation relieves inhibitory interactions that somehow block the activity of the EVH2 domain.
Supplementary Material
ACKNOWLEDGMENTS
This article is dedicated to Irina Libova who died on December 28, 1999, in a mountain climbing accident. Her intellectual and technical contributions to the Gertler laboratory continue to be felt. We thank Katie Fillion and Seth Berman for technical support. We are particularly indebted to the MIT-CCR FACS facility, supervised by Glenn Paradis, for exceptional assistance. We thank R. Rohatgi and M. Kirschner for the N-WASP antibody, and Reinhard Fassler for the VASP knockout mice. We thank members of the Gertler laboratory, particularly Matthias Krause, for helpful suggestions and comments. Work from J.J.L. was supported by the Anna Fuller Molecular Oncology Fund and National Institutes of Health F32 GM-20286. A.V.K. was supported by the Anna Fuller Molecular Oncology Fund. J.E.B. is supported by a Special Fellow award from the Leukemia and Lymphoma Society (3476-02). This work was supported by National Institutes of Health grant GM58801 and by funds from the W.M. Keck Distinguished Young Scholar Award to F.B.G.
Footnotes
Online version of this article contains video material. The online version is available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E01–10–0102. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E01–10–0102.
REFERENCES
- Ahern-Djamali SM, Comer AR, Bachmann C, Kastenmeier AS, Reddy SK, Beckerle MC, Walter U, Hoffmann FM. Mutations in Drosophila enabled and rescue by human vasodilator-stimulated phosphoprotein (VASP) indicate important functional roles for Ena/VASP homology domain 1 (EVH1) and EVH2 domains. Mol Biol Cell. 1998;9:2157–2171. doi: 10.1091/mbc.9.8.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aszodi A, Pfeifer A, Ahmad M, Glauner M, Zhou XH, Ny L, Andersson KE, Kehrel B, Offermanns S, Fassler R. The vasodilator-stimulated phosphoprotein (VASP) is involved in cGMP- and cAMP-mediated inhibition of agonist-induced platelet aggregation, but is dispensable for smooth muscle function. EMBO J. 1999;18:37–48. doi: 10.1093/emboj/18.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann C, Fischer L, Walter U, Reinhard M. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J Biol Chem. 1999;274:23549–23557. doi: 10.1074/jbc.274.33.23549. [DOI] [PubMed] [Google Scholar]
- Bashaw GJ, Kidd T, Murray D, Pawson T, Goodman CS. Repulsive axon guidance: Abelson and Enabled play opposing roles downstream of the roundabout receptor. Cell. 2000;101:703–15. doi: 10.1016/s0092-8674(00)80883-1. [DOI] [PubMed] [Google Scholar]
- Bear JE, Krause M, Gertler FB. Regulating cellular actin assembly. Curr Opin Cell Biol. 2001;13:158–166. doi: 10.1016/s0955-0674(00)00193-9. [DOI] [PubMed] [Google Scholar]
- Bear JE, Loureiro JJ, Libova I, Fassler R, Wehland J, Gertler FB. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 2000;101:717–28. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, Gertler FB. Antagonism between Ena/VASP Proteins and Actin Filament Capping Regulaties Fibroblast Motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Butt E, Abel K, Krieger M, Palm D, Hoppe V, Hoppe J, Walter U. cAMP- and cGMP-dependent protein kinase phosphorylation sites of the focal adhesion vasodilator-stimulated phosphoprotein (VASP) in vitro and in intact human platelets. J Biol Chem. 1994;269:14509–14517. [PubMed] [Google Scholar]
- Carl UD, Pollmann M, Orr E, Gertlere FB, Chakraborty T, Wehland J. Aromatic and basic residues within the EVH1 domain of VASP specify its interaction with proline-rich ligands. Curr Biol. 1999;9:715–718. doi: 10.1016/s0960-9822(99)80315-7. [DOI] [PubMed] [Google Scholar]
- Diviani D, Scott J. AKAP signaling complexes at the cytoskeleton. J Cell Sci. 2001;114:1431–1437. doi: 10.1242/jcs.114.8.1431. [DOI] [PubMed] [Google Scholar]
- Ermekova KS, Zambrano N, Linn H, Minopoli G, Gertler F, Russo T, Sudol M. The WW domain of neural protein FE65 interacts with proline-rich motifs in Mena, the mammalian homolog of Drosophila enabled. J Biol Chem. 1997;272:32869–32877. doi: 10.1074/jbc.272.52.32869. [DOI] [PubMed] [Google Scholar]
- Fedorov AA, Fedorov E, Gertler F, Almo SC. Structure of EVH1, a novel proline-rich ligand-binding module involved in cytoskeletal dynamics and neural function. Nat Struct Biol. 1999;6:661–665. doi: 10.1038/10717. [DOI] [PubMed] [Google Scholar]
- Geese M, Schluter K, Rothkegel M, Jockusch BM, Wehland J, Sechi AS. Accumulation of profilin II at the surface of Listeria is concomitant with the onset of motility and correlates with bacterial speed. J Cell Sci. 2000;113:1415–1426. doi: 10.1242/jcs.113.8.1415. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Comer AR, Juang JL, Ahern SM, Clark MJ, Liebl EC, Hoffmann FM. Enabled, a dosage-sensitive suppressor of mutations in the Drosophila Abl tyrosine kinase, encodes an Abl substrate with SH3 domain-binding properties. Genes Dev. 1995;9:521–533. doi: 10.1101/gad.9.5.521. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- Halbrugge M, Friedrich C, Eigenthaler M, Schanzenbacher P, Walter U. Stoichiometric and reversible phosphorylation of a 46-kDa protein in human platelets in response to cGMP- and cAMP-elevating vasodilators. J Biol Chem. 1990;265:3088–3093. [PubMed] [Google Scholar]
- Harbeck B, Huttelmaier S, Schluter K, Jockusch BM, Illenberger S. Phosphorylation of the vasodilator-stimulated phosphoprotein regulates its interaction with actin. J Biol Chem. 2000;275:30817–3025. doi: 10.1074/jbc.M005066200. [DOI] [PubMed] [Google Scholar]
- Hauser W, Knobeloch KP, Eigenthaler M, Gambaryan S, Krenn V, Geiger J, Glazova M, Rohde E, Horak I, Walter U, Zimmer M. Megakaryocyte hyperplasia and enhanced agonist-induced platelet activation in vasodilator-stimulated phosphoprotein knockout mice. Proc Natl Acad Sci USA. 1999;96:8120–8125. doi: 10.1073/pnas.96.14.8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Harbeck B, Steffens O, Messerschmidt T, Illenberger S, Jockusch BM. Characterization of the actin binding properties of the vasodilator-stimulated phosphoprotein VASP. FEBS Lett. 1999;451:68–74. doi: 10.1016/s0014-5793(99)00546-3. [DOI] [PubMed] [Google Scholar]
- Krause M, Sechi AS, Konradt M, Monner D, Gertler FB, Wehland J. Fyn-binding protein (Fyb)/SLP-76-associated protein (SLAP), Ena/vasodilator-stimulated phosphoprotein (VASP) proteins and the Arp2/3 complex link T cell receptor (TCR) signaling to the actin cytoskeleton. J Cell Biol. 2000;149:181–194. doi: 10.1083/jcb.149.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krugmann S, Jordens I, Gevaert K, Driessens M, Vandekerckhove J, Hall A. Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr Biol. 2001;11:1645–1655. doi: 10.1016/s0960-9822(01)00506-1. [DOI] [PubMed] [Google Scholar]
- Lambrechts A, Kwiatkowski AV, Lanier LM, Bear JE, Vandekerckhove J, Ampe C, Gertler FB. cAMP-dependent protein kinase phosphorylation of EVL, a Mena/VASP relative, regulates its interaction with actin and SH3 domains. J Biol Chem. 2000;275:36143–36151. doi: 10.1074/jbc.M006274200. [DOI] [PubMed] [Google Scholar]
- Lanier LM, Gates MA, Witke W, Menzies AS, Wehman AM, Macklis JD, Kwiatkowski D, Soriano P, Gertler FB. Mena is required for neurulation and commissure formation. Neuron. 1999;22:313–325. doi: 10.1016/s0896-6273(00)81092-2. [DOI] [PubMed] [Google Scholar]
- Lanier LM, Gertler FB. From Abl to actin: Abl tyrosine kinase and associated proteins in growth cone motility. Curr Opin Neurobiol. 2000;10:80–87. doi: 10.1016/s0959-4388(99)00058-6. [DOI] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins [see comments] Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Miki H, Ito M, Ohashi K, Takenawa T, Miyamoto S. N-WASP, WAVE and Mena play different roles in the organization of actin cytoskeleton in lamellipodia. J Cell Sci. 2001;114:1555–1565. doi: 10.1242/jcs.114.8.1555. [DOI] [PubMed] [Google Scholar]
- Niebuhr K, Ebel F, Frank R, Reinhard M, Domann E, Carl UD, Walter U, Gertler FB, Wehland J, Chakraborty T. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J. 1997;16:5433–5344. doi: 10.1093/emboj/16.17.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Prehoda KE, Lee DJ, Lim WA. Structure of the enabled/VASP homology 1 domain-peptide complex: a key component in the spatial control of actin assembly. Cell. 1999;97:471–480. doi: 10.1016/s0092-8674(00)80757-6. [DOI] [PubMed] [Google Scholar]
- Price CJ, Brindle NP. Vasodilator-stimulated phosphoprotein is involved in stress-fiber and membrane ruffle formation in endothelial cells. Arterioscler Thromb Vasc Biol. 2000;20:2051–2056. doi: 10.1161/01.atv.20.9.2051. [DOI] [PubMed] [Google Scholar]
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Halbrugge M, Scheer U, Wiegand C, Jockusch BM, Walter U. The 46/50 kDa phosphoprotein VASP purified from human platelets is a novel protein associated with actin filaments and focal contacts. EMBO J. 1992;11:2063–2070. doi: 10.1002/j.1460-2075.1992.tb05264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K, Behrendt B, Small JV, Wehland J. VASP dynamics during lamellipodia protrusion. Nat Cell Biol. 1999;1:321–322. doi: 10.1038/13040. [DOI] [PubMed] [Google Scholar]
- Sabo SL, Ikin AF, Buxbaum JD, Greengard P. The Alzheimer amyloid precursor protein (APP) and FE65, an APP-binding protein, regulate cell movement. J Cell Biol. 2001;153:1403–1414. doi: 10.1083/jcb.153.7.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoble J, Auerbuch V, Goley ED, Welch MD, Portnoy DA. Pivotal role of VASP in Arp2/3 complex-mediated actin nucleation, actin branch-formation, and Listeria monocytogenes motility. J Cell Biol. 2001;155:89–100. doi: 10.1083/jcb.200106061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GA, Theriot JA, Portnoy DA. The tandem repeat domain in the Listeria monocytogenes ActA protein controls the rate of actin-based motility, the percentage of moving bacteria, and the localization of vasodilator-stimulated phosphoprotein and profilin. J Cell Biol. 1996;135:647–660. doi: 10.1083/jcb.135.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Troys M, Dewitte D, Goethals M, Carlier MF, Vandekerckhove J, Ampe C. The actin binding site of thymosin beta 4 mapped by mutational analysis. EMBO J. 1996;15:201–210. [PMC free article] [PubMed] [Google Scholar]
- Walter U, Eigenthaler M, Geiger J, Reinhard M. Role of cyclic nucleotide-dependent protein kinases and their common substrate VASP in the regulation of human platelets. Adv Exp Med Biol. 1993;344:237–249. doi: 10.1007/978-1-4615-2994-1_19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.