Abstract
Autosomal dominant deafness-15 which is caused by mutation in the POU4F3 gene, has been reported with a wide degree of clinical heterogeneity, even between intrafamilial members. However, the reason is still elusive. In this study, A four-generation Chinese family with 11 patients manifesting late-onset progressive non-syndromic hearing loss was recruited. The phenotype of hearing loss in this family showed a large variability in terms of onset age and progression speed. A novel mutation (c.706 C > T, p.L236F) was identified by the whole exome sequencing, and its pathogenicity was confirmed by altering the subcellular localization of POU4F3. In addition, we found that two individuals with earlier age of onset and more rapid progression of hearing loss carry additional pathogenic variants in other deafness genes (III-7, STRC:c.4057 C > T; IV-1, GJB2:c.109G > A; CDC14A:c.935G > A). By using the real time quantitative PCR, western blot, luciferase assays and electrophoretic mobility-shift assay, POU4F3 was proved to directly regulate the expression of STRC, GJB2 and CDC14A respectively. ChIP-seq further revealed that POU4F3 can also bind to a series of deafness genes. In summary we expanded the mutation spectrum of POU4F3 by identifying a novel mutation and its pathogenicity. Meanwhile, three genes STRC, GJB2 and CDC14A were validated as POU4F3 new targets, implicating that the variants in the three genes may play a role of genetic modifier to generate a synergistic and enhancement effect on the progression of DFNA15.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-025-85881-8.
Keywords: Autosomal dominant deafness, POU4F3, Novel mutation, Oligogenic effect, Clinical heterogeneity
Subject terms: Clinical genetics, Disease genetics
Introduction
Autosomal dominant deafness-15 (DFNA15, OMIM #602459) is a form of progressive non-syndromic sensorineural hearing loss with postlingual onset between the second and sixth decades of life, which is caused by heterozygous mutation in the POU4F3 gene1. It is one of the most common forms of autosomal dominant non-syndromic hearing loss (ADNSHL) in Chinese Hans population2,3. Lots of studies have revealed that audiometric profiles among the DFNA15 patients, even from one family with the same mutation show a large variability in terms of onset age, level of progression, and shape of the audiogram4,5, however, the mechanisms underlying this phenomenon remains unclear.
POU4F3 has two exons and encodes a protein of 338 amino acids. It is a member of the POU family of transcription factors, which comprises two conserved DNA-binding domains (the POU-specific domain and the POU homo domain), and exclusively distributes in the nucleus. POU4F3 mainly expresses in cochlear, playing an important role in the development of hair cells in the inner ear sensory epithelia6. Haploinsufficiency of POU4F3 is the main underlying cause of DFNA15 deafness7.
More and more clinical and genetic evidence shows that even for classically monogenic disorders, the mutation of a single gene is not enough to explain all of the biological phenomena8. In recent years, with the progress of phenotype-genotype research, some hereditary disorders have been proved to be related to oligogenic or polygenic inheritance, such as inheritable cardiac disorders9 and hypercholesterolemia10. In oligogenic disorders, a variant in one gene is sufficient to produce the phenotype, but an additional variant in a second gene like genetic modifiers (usually related to the same pathway/organ system) impacts the disease phenotype or alters the age of onset8,11. Although several syndromic deafness types are caused by the co-existence of variants in two different genes, which defined as a digenic inheritance, such as Usher syndrome12, the oligogenic or polygenic effect in deafness is still elusive.
In this study, we identified a novel mutation of POU4F3 from a four-generation Chinese family with late-onset progressive non-syndromic hearing loss. Moreover, to explore the causes of clinical heterogeneity of deafness in this family, we proved that STRC, GJB2 and CDC14A were the target genes of transcription factors POU4F3. Mutations of STRC, GJB2 and CDC14A may impact the disease phenotype and alters the age of onset of DFNA15. This provides a new insight into the clinical heterogeneity of DFNA15 deafness.
Materials and methods
Subjects
A four-generation Chinese family named ADNSHL have been recruited through Hunan Jiahui Genetics Hospital. All the patients in this family represented bilateral late-onset progressive hearing loss. There were sixteen members (II-2, II-4, II-8, III-2, III-3, III-5, III-7, III-8, IV-1 with hearing loss and II-7, II-9, IV-2, IV-3, IV-4, IV-5, IV-6 with normal hearing) in this study. And 1 ml of peripheral blood was taken from all members for genetic examination. Only seven members (II-8, II-9, III-2, III-7, III-8, IV-1, IV-2) got detailed hearing information. Informed consent was obtained from all subjects. This study was approved by the Ethics Committee of Center for Medical Genetics, Central South University, Hunan, China.We confirmed that all experiments were performed in accordance with relevant guidelines and regulations.
Whole exome sequencing and sanger sequencing
All the subjects’ genomic DNA was extracted from peripheral blood samples using the Quickgene DNA Whole Blood Kit L (FUJIFILM, Tokyo, Japan) according to standard extraction methods. And 1 ml peripheral blood of each patient (III-2, III-7, IV-1) was performed the whole exome sequencing (WES, Berry Genomics Inc., Beijing, China). Sanger sequencing was performed in all the subjects in our study. The primers were designed using Primer Premier 5 and used to amplify the mutations filtrated from the WES data by polymerase chain reaction. Sequencing reactions were performed by Tsingke (Tsingke Biotechnology Co., Ltd). Data was analyzed using Lasergene-SeqMan software. GRCh38/hg38 assembly was used as the reference sequences. Guideline and standard of American College of Medical Genetics and Genomics (ACMG)/Association for Molecular Pathology (AMP)13, recommendations for ACMG/AMP mutation criterion14 and the expert specification of the ACMG/AMP mutation interpretation guidelines for genetic hearing loss15 were used as the reference of data interpretation.
Transient transfection and immunofluorescence analysis
The wild type and mutant sequences of POU4F3 cDNA and HA-tag were synthesized and cloned into the pcDNA3.1(+) vector respectively by Sangon (Sangon Biotech Co., Ltd. Shanghai, China). 293T cell lines obtained from the Center for Medical Genetics & Hunan Key Laboratory of Medical Genetics, School of Life Sciences, Central South University (Changsha, China). 293T cells were inoculated in 24-well plates with 5 × 104 cells per well, when cells were cultured to 60% confluence, the wild type or mutated POU4F3 vectors were transfected into 293T cell lines using Lipofectamine™ 3000 Transfection Reagent (Thermo Fisher Scientific, Shanghai, China). After 48 h, cells were fixed in 4% paraformaldehyde for 30 min, permeabilized in PBS containing 0.5% Triton X-100 for 20 min and then blocked in 5%BSA for 30 min. The cells were then incubated with primary anti-HA antibody for 12 h at 4°C, secondary Dylight 488 Rabbit Anti-Goat antibody for 60 min and DAPI for 10 min. The immunofluorescence test was performed using TCS SP5 laser confocal microscopy (Leica).
Real time fluorescence quantitative polymerase chain reaction (RT-qPCR)
Three small interfering RNAs (siRNA) were synthesized by GenePharma (Suzhou Genepharma Co.,Ltd, Suzhou, China). The siRNA, wild-type, mutated POU4F3 vectors and empty vector were transfected into 293T cell. Total RNA was extracted by Trizol (Invitrogen, Carlsbad, CA, USA) from 293T cells after 48 h transfection. RNA was reverse transcribed into cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, CA, USA). RT- qPCR was performed with an ASA-9600 Real-Time PCR System (Suzhou Baiyuan Gene Technology Co.,Ltd, Suzhou, China) using SYBR Premix Ex Taq (Thermo Fisher, CA, USA) with primers against POU4F3, STRC, GJB2, CDC14A and GAPDH (supplement Table 1).Three replicate wells were tested for each measurement experiment, and more than three replicate experiments were completed at different time periods.
Western blot
293T cells were lysed in the mixture containing SDS buffer and protease inhibitor PMSF (Beyotime Biotechnology Co.,Ltd, Shanghai, China ) after 48 h transfection and the harvested protein was quantitated using the BCA Protein Assay Kit (Thermo Fisher Scientific, USA). Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis with 8% acrylamide, then the transferred and blocked protein was incubated with rabbit polyclonal antibody to STRC (Biorbyt Ltd., Cambridge, UK), mouse monoclonal antibody to GJB2 (Invitrogen, Carlsbad, CA, USA), rabbit polyclonal antibody to CDC14A (Invitrogen, Carlsbad, CA, USA) and rabbit polyclonal antibody to Vinculin(GeneTex, Irvine, CA, USA ) at 1:1000 dilution, followed by peroxidase-conjugated affinipure goat anti-mouse/rabbit IgG (Jackson ImmunoResearch, USA) at a 1:10000 dilution. Enhanced chemiluminescence (Thermo Fisher Scientific, Rockford, USA) was used and then scanned to visualize the specific protein band.Three replicate wells were tested for each measurement experiment, and more than three replicate experiments were completed at different time periods.
Dual luciferase reporter gene assay
The 2.8 kb upstream regulatory sequences of STRC (STRC-P2.8, chr15: 43618801–43621600), GJB2 (GJB2-P2.8, chr13:20192939–20195738) and CDC14A (CDC14A-P2.8, chr1:100349706–100352505) were synthesized and cloned into the dual luciferase reporter gene vector pDuaLuc-Basic (GenScript (Nanjing) Co.Ltd., Nanjing, China) as promotor vectors. The promotor vectors STRC-P2.8, GJB2-P2.8 and CDC14A-P2.8 were co-transfected with wild type, mutated POU4F3 vectors and empty vector into 293T cell, after 48 h, Cells were harvested and assayed using the Dual Luciferase Reporter Gene Assay Kit (Beyotime Biotechnology Co.,Ltd, Shanghai, China).Three replicate wells were tested for each measurement experiment, and more than three replicate experiments were completed at different time periods.
Electrophoretic mobility shift assay (EMSA)
Six possible POU4F3-binding sites of the STRC, GJB2 and CDC14A upstream regulatory sequence were predicted by using the JASPAR CORE (http://jaspar.genereg.net/). The sequences were extended by 10 bases at the upstream and downstream of the biding sites, and then labeled by biotin at the 5’ end and synthesized by Sangon (Sangon Biotech Co., Ltd. Shanghai, China) as EMSA detection probes (supplement Table 1). The nuclear protein and cytoplasmic protein from the 293T cell which were transfected with wild type, mutated POU4F3 vectors and empty vector were extracted by Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime Biotechnology Co.,Ltd, Shanghai, China). The assay reactions of the detection probes and proteins were detected by Chemiluminescent EMSA Kit (Beyotime Biotechnology Co.,Ltd, Shanghai, China). Three replicate wells were tested for each measurement experiment, and more than three replicate experiments were completed at different time periods.
Chromatin immunoprecipitation followed by sequencing (ChIP-seq) analysis
293T cells were lysed after 48 h transfected with wild type POU4F3 vector. DNA were broken into 100–500 bp fragments by ultrasound. The DNA fragments that bind to the protein were pulled down by the HA antibody. After washing and purification, these fragments were sequenced by Novogene (Novogene Co., Ltd., Beijing, China). The interactions of POU4F3 and genes identified from ChIP-seq data were analysed by the search tool for the retrieval of interacting genes/proteins (STRING, http://string-db.org/).
Data statistics
The data differentiation analysis were conducted using GraphPad Prism software. significance was determined using Tukey’s Multiple Comparison Test and Paired t test.
Results
Clinical features
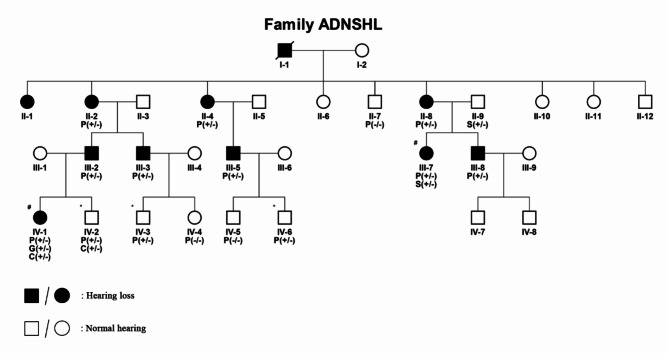
The pedigree and disease statements of all individuals were shown in Fig. 1. There were thirty-one members in this family. Sixteen members (II-2, II-4, II-8, III-2, III-3, III-5, III-7, III-8, IV-1 with hearing loss and II-7, II-9, IV-2, IV-3, IV-4, IV-5, IV-6 with normal hearing) were recruited to perform WES or sanger sequencing. Based on the results of audiometric examination and description of proband (III-7), the onset age among the affected individuals in this family ranges from 8 to 31years. However, unlike other relative patients, III-7 and IV-1 manifested earlier age of onset and faster progression of hearing loss. The age of onset of proband (III-7) was 18 years old and developed moderately severe hearing loss at 28 years old. IV-1 had a much earlier age of onset (8 years old) and developed moderately severe hearing loss at the age of 12 (Table S2).
Fig. 1.
The pedigree and segregation pattern of the ADNSHL family. P(+/−): The heterozygous mutation of POU4F3. P(−/−): Wild type of POU4F3. S(+/−): The heterozygous mutation of STRC. G(+/−): The heterozygous mutation of GJB2. C(+/−):The heterozygous mutation of CDC14A. #Individuals (III-7, IV-1) with a faster progression of hearing loss and an earlier age of onset in this family. *The individuals who carried the mutation of POU4F3 but have not yet experienced hearing loss.
A novel pathogenic variant in POU4F3 accounts for the familial hearing loss
To uncover the genetic cause for the hearing loss in the family, WES was conducted based on the proband. After the bioinformatics analysis for the data, a novel heterozygous missense mutation in POU4F3 gene (NM_002700.3: c.706 C > T, p.L236F) was identified in the proband, which was inherited from her affected mother. The variant (c.706 C > T) was then tested in all members of this family by Sanger sequencing. The results showed that the variant was co-segregated among the patients (the affected individuals including II-2, II-4, II-8, III-2, III-3, III-5, III-7, III-8 and IV-1 all carried the mutation), while the unaffected members II-7, IV-4 and IV-5 were wild type (Fig. 1; Fig. S1). It should be noted that although the individuals IV-2, IV-3 and IV-6 also carried the variant, they did not present hearing loss, which may because they were too young to reach the age of onset.
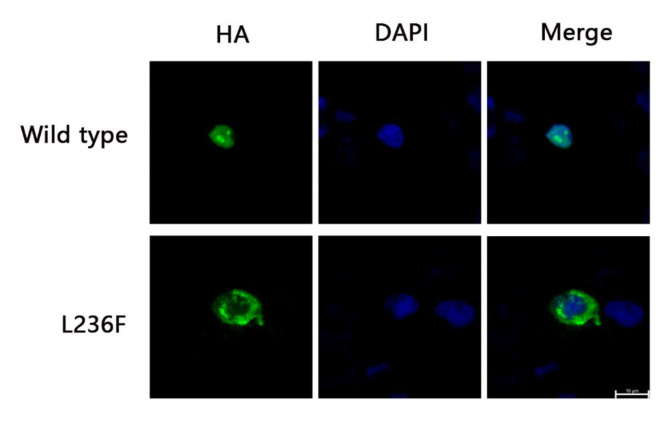
Given that POU4F3 is a transcription factor and mainly locate in nucleus, we further performed functional tests for the subcellular localization of the mutant proteins to confirm the pathogenicity of the variant (c.706 C > T). Expression cassette encoding either the wild type or the mutant POU4F3 protein was constructed to fuse with an HA-tag C-terminally. After transient transfection into the 293T cell lines, the subcellular localization of POU4F3 was detected by using anti-HA immunofluorescence analysis. The results showed that almost all of the expression of wild-type POU4F3 is located in the nucleus, while the mutants mostly distributed in the cytoplasm (Fig. 2). This indicates that the variant has altered the subcellular localization of the POU4F3 protein, thus resulted in great loss of its transcriptional function. Together with the functional change, co-segregation results, the low frequency in population and potential damaging effect predicted by various softwares, the variant c.706 C > T in POU4F3 can be classified as likely pathogenic, according to the standards and guidelines of ACMG/AMP (Table S3, Fig. S1), which would be the primary genetic cause for this familial hearing loss.
Fig. 2.
The variation L236F compromised transportation of POU4F3 from the cytoplasm to the nucleus. Immunofluorescence staining was performed after transient transfection in 293 T cells. Images display HA-tagged protein in green, DAPI in blue and merged pictures. Mutant protein located mostly in cytoplasm while the wild type protein was exclusively located in nucleus. Scale bars = 10 μm. This experiment is repeated three times or more in biology.
Identification of additional pathogenic variants in STRC, GJB2 and CDC14A in two patients
Considering a much faster progression of hearing loss and an earlier age of onset in III-7 and IV-1, we speculated that additional modifiers may contribute to such a high clinical variability other than the c.706 C > T in POU4F3. By analyzing the WES data of III-7 and IV-1, an extra novel heterozygous variant of STRC gene (NM_153700.2: c.4057 C > T, p.Q1353*) was identified in III-7, which is inherited from her unaffected father. In IV-1, two additional heterozygous variants (GJB2: NM_004004.6: c.109G > A, p.V37I; CDC14A: NM_003672.4: c.935G > A, p.R312Q) were detected, while her affected father (III-2) did not carry either of the variants. The variants STRC: c.4057 C > T and CDC14A: c.935G > A have not been reported in previous studies. According to the standards and guidelines of ACMG/AMP, the variants of STRC (c.4057 C > T), GJB2 (c.109G > A) and CDC14A (c.935G > A) can be classified as pathogenic, pathogenic16 and likely pathogenic respectively (Table S3, Fig. S1). The bi-allelic mutations in the STRC, GJB2 and CDC14A can cause autosomal recessive deafness 16, 1 A and 15 respectively17,18.
POU4F3 regulates STRC, GJB2 and CDC14A expression
To investigate whether the POU4F3 could regulate the STRC, GJB2 and CDC14A expression, we firstly performed transient transfection assays to overexpress wild type and mutated POU4F3 (L236F). By using the RT-qPCR, the results revealed that compared with the blank vector group, overexpression of wild type POU4F3 in 293T cell significantly increased the mRNA transcription of endogenous STRC (1.83 fold, p < 0.001), GJB2 (1.41 fold, p < 0.001) and CDC14A (1.31 fold, p < 0.05), while overexpression of the mutant POU4F3 did not change the mRNA levels of three genes (Fig. 3A).
Fig. 3.
POU4F3 promoted the expression by activating the promotors of STRC, GJB2 and CDC14A. (A) mRNA expression analyses of STRC, GJB2 and CDC14A by RT-qPCR after POU4F3 overexpression. The results revealed that compared with the blank vector group, overexpression of wild type POU4F3 in 293T cell significantly increased the mRNA transcription of endogenous STRC (1.83 fold, P-values = 0.0007, F = 31.55, df = 8), GJB2 (1.41 fold, P-values < 0.0001, F = 64.47, df = 8) and CDC14A (1.31 fold, P-values = 0.018, F = 8.44, df = 8), while overexpression of the mutant POU4F3 did not change the mRNA levels of three genes. (B) mRNA expression analyses of STRC, GJB2 and CDC14A by RT-qPCR after POU4F3 knockdown by transfected with siRNA2. After knock-down of the POU4F3, the mRNA transcription of STRC, GJB2 and CDC14A was accordingly reduced to 63% (P-values = 0.0048, t = 7.560, df = 3), 79% (P-values = 0.025, t = 4.178, df = 3) and 80% (P-values = 0.0004, t = 18.37, df = 3) of untreated 293T cells respectively. (C) Protein expression analyses of CDC14A by western blot after POU4F3 overexpression. The results revealed that compared with the blank vector group, overexpression of wild type POU4F3 in 293T cell significantly increased the protein expression of endogenous CDC14A (2.02 fold, P-values = 0.0009, F = 28.63, df = 8). D. Protein expression analyses of CDC14A by western blot after POU4F3 knockdown by transfected with siRNA2. After knock-down of the POU4F3, the protein expression of CDC14A was accordingly reduced to 90% (P-values = 0.0061, t = 12.79, df = 2). NC negative control, transfection of empty vector, WT transfection of wild type POU4F3 vector, siRNA2 transfection of wild type siRNA2, L236F transfection of mutant POU4F3 vector. Values represent mean ± SD, significance was determined using 1way ANOVA Tukey’s Multiple Comparison Test and Paired t test. *P < 0.05, **P < 0.01, ***P < 0.001. This experiment is repeated three times or more in biology.
In addition, the RNA interference was implemented to knock down the endogenous POU4F3 and the sequence of siRNA2 demonstrated the highest interference efficiency (Fig.S2). After knock-down of the POU4F3, the mRNA transcription of STRC, GJB2 and CDC14A was accordingly reduced to 63%, 79% and 80% of untreated 293T cells respectively (all p < 0.05) (Fig. 3B). These results illustrated a positive correlation between the POU4F3 amount and the transcription of STRC, GJB2 and CDC14A.
Protein expression of endogenous STRC, GJB2 and CDC14A were also tested by Western blot under the overexpression or knock down of POU4F3, but only the CDC14A could be detected and exhibited a similar tendency as the mRNA change (Fig. 3C and D). STRC, GJB2 proteins failed to present may due to their low endogenous expression in 293T cells.
POU4F3 activated the promotors of STRC, GJB2 and CDC14A
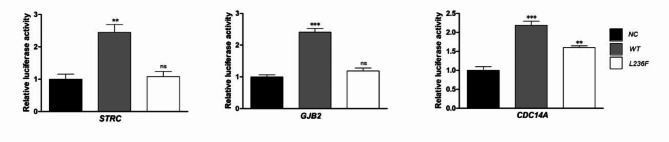
In order to verify whether the regulatory effect of POU4f3 is related to the STRC, GJB2 and CDC14A promoter, about 2.8 kb promotor regions flanked upstream of the coding sequence of each gene were cloned into dual luciferase plasmid. Each luciferase vector was co-transfected with wild-type, mutated POU4F3 expression vectors and empty vector into 293T cell separately. The results suggested that wild type POU4F3 significantly increased the promotor vectors’ luciferase activity of STRC (2.45 fold, P < 0.01), GJB2 (2.40 fold, P < 0.001) and CDC14A (2.18 fold, P < 0.001). The mutant POU4F3 did not change the luciferase activity under the promotor of STRC and GJB2, except an increase in the CDC14A group (Fig. 4). These data suggest that POU4F3 regulates the expression of STRC, GJB2 and CDC14A by activating the promotor of them.
Fig. 4.
Dual luciferase reporter gene assay of STRC, GJB2 and CDC14A promotors. The results suggested that wild type POU4F3 significantly increased the promotor vectors’ luciferase activity of STRC (2.45 fold, P-values = 0.0029, F = 18.03, df = 8), GJB2 (2.40 fold, P-values < 0.0001, F = 66.55, df = 8) and CDC14A ( P-values = 0.0002, F = 46.37, df = 8) NC: negative control, cotransfection of empty vector and the promotor vector; WT: transfection of wild type POU4F3 vectorand the promotor vector; L236F: transfection of mutant POU4F3 vector and the promotor vector. This experiment is repeated three times or more in biology.
Six sites located on the promotors of STRC, GJB2 and CDC14A were identified to bind to POU4F3
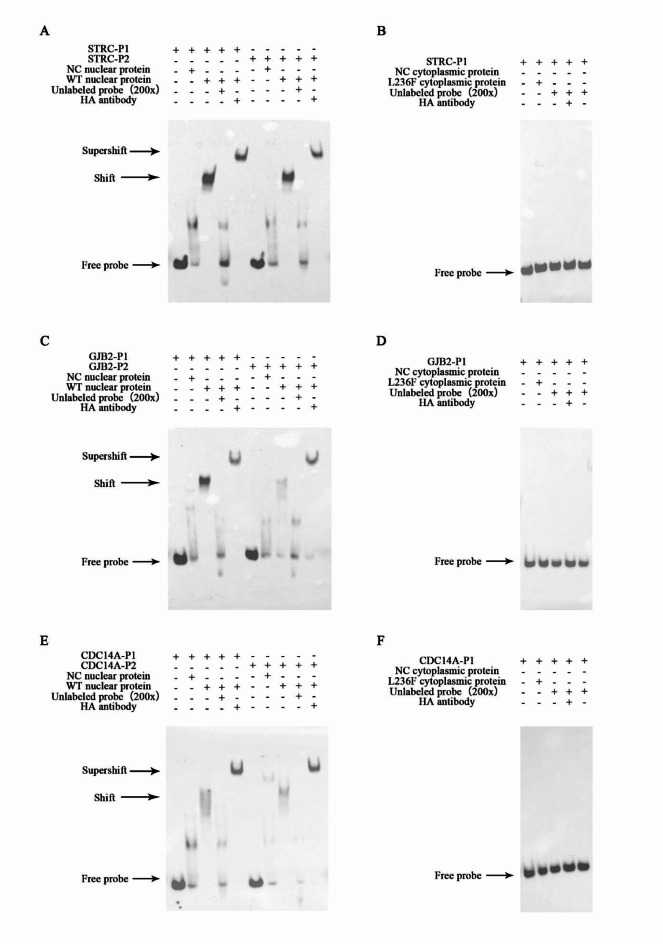
To further find the POU4F3 direct binding sites in STRC, GJB2 and CDC14A, the putative binding sites were predicted by JASPAR CORE. Two of predicted sites in the promotor region of each gene with relative high score were selected and validated via EMSA. Compared with the only biotin-labeled probe group of each gene (STRC-P1, STRC-P2, GJB2-P1, GJB2-P2, CDC14A-P1, CDC14A-P2), a shifted band could be observed after co-incubation with nuclear proteins containing overexpression of WT POU4F3(fused with HA), which would disappear when an overdosage of unlabeled probe competitively bound to POU4F3, while a super shifted band could also consequently display after adding HA antibody (Fig. 5A, C and E). These results demonstrated that POU4F3 indeed directly bind to STRC, GJB2 and CDC14A, thus regulate the expression of three genes. Nevertheless, the mutant POU4F3 which located in the cytoplasm seemed to lose the DNA-binding ability (Fig. 5B, D and F).
Fig. 5.
EMSA results of direct interaction between POU4F3 and the putative binding sites. STRC-P1, STRC-P2, GJB2-P1, GJB2-P2, CDC14A-P1 and CDC14A-P2: the detection probe which were labeled by biotin at the 5’ end. NC nuclear/cytoplasmic protein: nuclear/cytoplasmic protein of empty vector transfected cell. WT nuclear/cytoplasmic protein: nuclear/cytoplasmic protein of wild type POU4F3 vector transfected cell. L236F nuclear/cytoplasmic protein: nuclear/cytoplasmic protein of mutant POU4F3 vector transfected cell. This experiment is repeated three times or more in biology.
POU4F3 may regulate more deafness genes
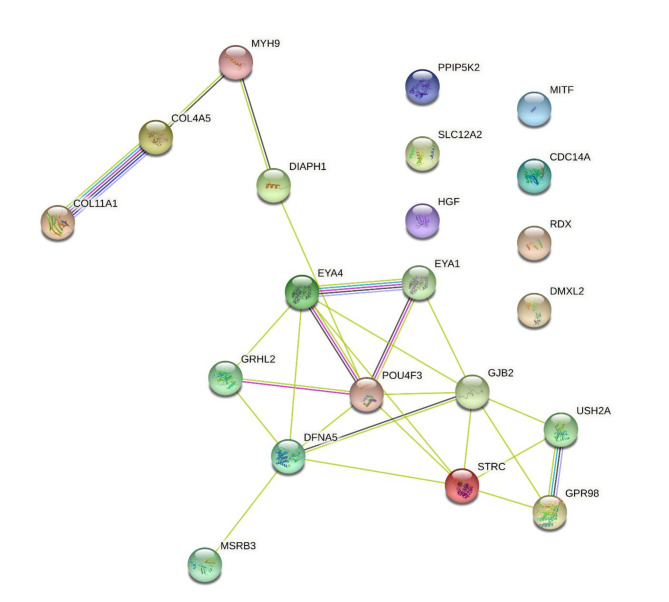
Since DFNA15 has a deep degree of clinical heterogeneity, except the STRC, GJB2 and CDC14A, we inferred that POU4F3 as a transcription factor may regulate the expression of more extensive deafness genes. Hence, ChIP-seq was carried out to identify other POU4F3 binding genes. After transfection of the wild type POU4F3 vector into 293T cell, the fragmental DNA were pulled down by HA antibody and sequenced. According to the sequencing results, those genes which have been reported to be associated with deafness from the Hereditary Hearing Loss Homepage were analyzed. As expected, the results showed that POU4F3 could bind to the promoter regions of additional 17 deafness genes, including MITF, USH2A, ADGRV1 and so on. The results of STRING analysis showed that 13 of them interacted with POU4F3 (Fig. 6). In addition, POU4F3 can also bind to the intron or exon regions of 33 deafness genes and 3’ UTR regions of 4 deafness genes (supplement Table 4). These results suggest that these deafness genes may also be the modifiers for DFNA15.
Fig. 6.
Interaction network of deafness genes that bind to POU4F3 in the promotor region from ChIP-seq data.
Discussion
Heterozygous mutation in the POU4F3 can cause DFNA-15 and it is a form of progressive non-syndromic sensorineural hearing loss with postlingual onset between the second and sixth decades of life. In this study, we recruited a four-generation Chinese family with eleven individuals affected by hearing loss. By whole exome sequencing in the proband III-7 and sanger sequencing in her families, we found a novel mutation of POU4F3 gene (c.706 C > T) co-segregating with hearing loss symptoms in this family. The resulted amino acids variation L236F was located on the evolutionarily conserved POU-specific domain. The POU-specific domain is required for high affinity sequence-specific DNA binding and confers DNA bending by interaction with the DNA19. Nuclear proteins usually have a common sequence NLS which is important for active protein transport into the nucleus. POU4F3 contains two NSLs, the monopartite (amino acids 274 to 278 [RKRKR]) and the bipartite NLS (amino acids 314 to 331 [KKNVVRVWFCNLQRQKQKR])20. Previous studies have demonstrated that variation in bipartite NLS could compromise nuclear localization ability of POU4F321. In this study, the results of immunofluorescence analysis showed that the mutant POU4F3 proteins L236F were located in the cytoplasm. However, the variation L236F) was not located on the monopartite or the bipartite NLS of POU4F3. Moreover, the results of the other reports showed that mutations which were not located on the monopartite or the bipartite NLS of POU4F3 could also impair the nuclear localization function5,22. The potential mechanism of this mutation compromised transportation of POU4F3 from the cytoplasm to the nucleus needs further exploration.
In deafness, digenic inheritance is a well know pattern, such as GJB2, CDH23 and SLC26A4 had been reported to cause hearing loss under digenic inheritance23–26. In these reports, variants at both genes are required for disease, but if only one of the two gene carrying the variants would not result in phenotype. However, the oligogenic inheritance in deafness is rarely reported, especially in those autosomal dominant non-syndromic hearing loss (ADNSHL) with the synergistic effect of other recessive pathogenic variants. In 2010, Yan X et al. once reported a Chinese family with autosomal dominant Waardenburg syndrome type 2 caused by a heterozygous mutation in MITF. The affected individuals in the family had moderate and mild sensorineural deafness, except for the proband, who presented with profound hearing loss as a newborn and had both MITF c.(742_743delAAinsT;746_747delCA) and GJB2 mutations (c.109G > A and c.235delC)27. The author thought that digenic effect of MITF and GJB2 genes may contribute to deafness of the proband, although they did not provide any further functional verification. To our knowledge, there is no report about digenic or oligogenic inheritance among POU4F3 and other genes leading to more severe hearing loss. In our study, except for the mutation of POU4F3, the proband (III-7) and IV-1 carried extra mutations of STRC, GJB2 and CDC14A. The results of RT-qPCR and western blot showed that POU4F3 promoted mRNA expression of STRC, GJB2 and CDC14A and protein expression of CDC14A. POU4F3 bound to six sites which located on the promoter regions and activated the promotors of STRC, GJB2 and CDC14A. All the above results suggested that STRC, GJB2 and CDC14A were the target genes of POU4F3. However, the mutant POU4F3 significantly lose the regulatory effect. This is the first time that the target gene of POU4F3 has been identified in humans. In the inner ear of mature mammals, POU4F3 was expressed in hair cells28 and GJB2 was expressed in supporting cells29. However, during embryonic development, hair cells and supporting cells are generated from a common pool of postmitotic pro-sensory progenitors30. This suggests that the regulatory effects of POU4F3 and GJB2 may occur in the early embryonic development. The oligogenic effect of POU4F3 and STRC/GJB2/CDC14A may contribute to the onset age and progression level of hearing loss in III-7 and IV-1.
POU4F3 is the only POU4 transcription factor present in the sensory receptor hair cells of the cochlea. The regulation of POU4F3 on the expression of its target genes (Caprin-1, Gfi-1, Lhx3 and Nr2f2) is essential for the development and survival of hair cells31–33. And most deafness genes specifically expressed in the hair cell, suggesting that POU4F3 may be the transcription factor of them. The results of ChIP-seq showed that POU4F3 could bind to the promotor regions of 17 deafness genes, intron or exon regions of 33 deafness genes and 3’ UTR regions of 4 deafness genes.
More and more studies have shown that the clinical heterogeneity of genetic diseases is related to oligogenetic effects, and the genes that form oligogenetic effects often have interactive relationships or are located on the same signaling pathway. The results of this study indicate that POU4F3, as a transcription factor, regulates the expression of STRC, CDC14A, and GJB2 genes. When STRC, CDC14A, and GJB2 undergo heterozygous mutations, although they do not produce phenotypes alone, their functions may be more affected due to mutations in their transcription factor POU4F3, leading to premature or aggravated phenotypes. This study only validated the relevant functions in cell lines, lacking further confirmation from animal models, and the research results may have certain biases. Due to factors such as distance or inability of participants to cooperate, we did not obtain detailed hearing test results for IV-2, IV-3, and IV-6. Their hearing problems were reported orally by their parents or cousins, which is also a limitation of this study.
Conclusions
In summary, in this study, we identified a novel mutation of POU4F3 gene in Chinese patients with ADNSHL. Function analysis demonstrated that the mutation impaired nuclear localization of POU4F3 and compromised protein function. We proved that STRC, GJB2 and CDC14A were the new target gene of POU4F3. This indicates that oligogenic effect of POU4F3 and STRC/GJB2/CDC14A may contribute to the clinical heterogeneity of DFNA15.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors greatly appreciate the support and corporation of all patients and their families.
Author contributions
JP, ZL, LW designed the study and collected the data. JP, FL, HT, SC, YT performed the experiments and analyzed the data. YL performed the clinical evaluation of the patients. JP drafted the manuscript. ZL, DL, and LW provided critical editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from the National Key R&D Program of China (2022YFC2703700), the National Natural Science Foundation of China (82171711, 82371724, 82402172), the Hunan Provincial Natural Science Foundation of China (2023JJ30725), the Open Research Funds of the State Key Laboratory of Ophthalmology (303060202400382), and the Fundamental Research Funds for the Central Universities of Central South University (1053320212016, 1053320215502).
Data availability
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2024), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA009740) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhuo Li, Email: lizhuo@sklmg.edu.cn.
Lingqian Wu, Email: wulingqian@sklmg.edu.cn.
References
- 1.Kim, H. J. et al. SNP linkage analysis and whole exome sequencing identify a novel POU4F3 mutation in autosomal dominant late-onset nonsyndromic hearing loss (DFNA15). PLoS One8, e79063. 10.1371/journal.pone.0079063 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vahava, O. et al. Mutation in transcription factor POU4F3 associated with inherited progressive hearing loss in humans. Science279, 1950–1954. 10.1126/science.279.5358.1950 (1998). [DOI] [PubMed] [Google Scholar]
- 3.He, L., Pang, X., Chen, P., Wu, H. & Yang, T. Mutation in the Hair Cell Specific Gene POU4F3 is a Common cause for autosomal Dominant Nonsyndromic hearing loss in Chinese Hans. Neural Plast.2016 (9890827). 10.1155/2016/9890827 (2016). [DOI] [PMC free article] [PubMed]
- 4.Pauw, R. J. et al. Audiometric characteristics of a Dutch family linked to DFNA15 with a novel mutation (p.L289F) in POU4F3. Arch. Otolaryngol. Head Neck Surg.134, 294–300. 10.1001/archotol.134.3.294 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Collin, R. W. et al. Missense mutations in POU4F3 cause autosomal dominant hearing impairment DFNA15 and affect subcellular localization and DNA binding. Hum. Mutat.29, 545–554. 10.1002/humu.20693 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu, F., Yan, W. & Cheng, Y. Pou4f3 gene mutation promotes autophagy and apoptosis of cochlear hair cells in cisplatin-induced deafness mice. Arch. Biochem. Biophys.680, 108224. 10.1016/j.abb.2019.108224 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Zhu, G. J. et al. Aldh inhibitor restores auditory function in a mouse model of human deafness. PLoS Genet.16, e1009040. 10.1371/journal.pgen.1009040 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kousi, M. & Katsanis, N. Genetic modifiers and oligogenic inheritance. Cold Spring Harb Perspect. Med.5. 10.1101/cshperspect.a017145 (2015). [DOI] [PMC free article] [PubMed]
- 9.Cerrone, M., Remme, C. A., Tadros, R., Bezzina, C. R. & Delmar, M. Beyond the one gene-one disease paradigm: Complex Genetics and Pleiotropy in Inheritable Cardiac disorders. Circulation140, 595–610. 10.1161/CIRCULATIONAHA.118.035954 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tada, H. et al. Oligogenic familial hypercholesterolemia, LDL cholesterol, and coronary artery disease. J. Clin. Lipidol.12, 1436–1444. 10.1016/j.jacl.2018.08.006 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Gazzo, A. M. et al. A curated and annotated digenic diseases database. Nucleic Acids Res.44, D900–907. 10.1093/nar/gkv1068 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebermann, I. et al. PDZD7 is a modifier of retinal disease and a contributor to digenic Usher syndrome. J. Clin. Invest.120, 1812–1823. 10.1172/JCI39715 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med.17, 405–424. 10.1038/gim.2015.30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abou Tayoun, A. N. et al. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum. Mutat.39, 1517–1524. 10.1002/humu.23626 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oza, A. M. et al. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum. Mutat.39, 1593–1613. 10.1002/humu.23630 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen, J. et al. Consensus interpretation of the p.Met34Thr and p.Val37Ile variants in GJB2 by the ClinGen hearing loss Expert Panel. Genet. Med.21, 2442–2452. 10.1038/s41436-019-0535-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francey, L. J. et al. Genome-wide SNP genotyping identifies the stereocilin (STRC) gene as a major contributor to pediatric bilateral sensorineural hearing impairment. Am. J. Med. Genet. A158A, 298–308. 10.1002/ajmg.a.34391 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delmaghani, S. et al. Mutations in CDC14A, encoding a protein phosphatase involved in hair cell ciliogenesis, cause autosomal-recessive severe to Profound Deafness. Am. J. Hum. Genet.98, 1266–1270. 10.1016/j.ajhg.2016.04.015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verrijzer, C. P., van Oosterhout, J. A., van Weperen, W. W. & Vliet, P. C. POU proteins bend DNA via the POU-specific domain. EMBO J.10, 3007–3014 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss, S. et al. The DFNA15 deafness mutation affects POU4F3 protein stability, localization, and transcriptional activity. Mol. Cell. Biol.23, 7957–7964. 10.1128/mcb.23.22.7957-7964.2003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, Y. H. et al. A novel missense variant in the nuclear localization signal of POU4F3 causes autosomal dominant non-syndromic hearing loss. Sci. Rep.7, 7551. 10.1038/s41598-017-08236-y (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai, X. et al. Identification of two novel mutations in POU4F3 gene associated with autosomal dominant hearing loss in Chinese families. J. Cell. Mol. Med.24, 6978–6987. 10.1111/jcmm.15359 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.del Castillo, F. J. et al. A novel deletion involving the connexin-30 gene, Del(GJB6-d13s1854), found in trans with mutations in the GJB2 gene (connexin-26) in subjects with DFNB1 non-syndromic hearing impairment. J. Med. Genet.42, 588–594. 10.1136/jmg.2004.028324 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, X. Z. et al. Digenic inheritance of non-syndromic deafness caused by mutations at the gap junction proteins Cx26 and Cx31. Hum. Genet.125, 53–62. 10.1007/s00439-008-0602-9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng, Q. Y. et al. Digenic inheritance of deafness caused by mutations in genes encoding cadherin 23 and protocadherin 15 in mice and humans. Hum. Mol. Genet.14, 103–111. 10.1093/hmg/ddi010 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, M. et al. Digenic inheritance of mutations in EPHA2 and SLC26A4 in Pendred syndrome. Nat. Commun.11, 1343. 10.1038/s41467-020-15198-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan, X. et al. A novel mutation in the MITF may be digenic with GJB2 mutations in a large Chinese family of Waardenburg syndrome type II. J. Genet. Genomics38, 585–591. 10.1016/j.jgg.2011.11.003 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Xiang, M., Maklad, A., Pirvola, U. & Fritzsch, B. Brn3c null mutant mice show long-term, incomplete retention of some afferent inner ear innervation. BMC Neurosci.4. 10.1186/1471-2202-4-2 (2003). [DOI] [PMC free article] [PubMed]
- 29.Del Castillo, F. J. & Del Castillo, I. DFNB1 non-syndromic hearing impairment: diversity of mutations and Associated Phenotypes. Front. Mol. Neurosci.10, 428. 10.3389/fnmol.2017.00428 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korrapati, S., Roux, I., Glowatzki, E. & Doetzlhofer, A. Notch signaling limits supporting cell plasticity in the hair cell-damaged early postnatal murine cochlea. PLoS One8, e73276. 10.1371/journal.pone.0073276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Towers, E. R., Kelly, J. J., Sud, R., Gale, J. E. & Dawson, S. J. Caprin-1 is a target of the deafness gene Pou4f3 and is recruited to stress granules in cochlear hair cells in response to ototoxic damage. J. Cell. Sci.124, 1145–1155. 10.1242/jcs.076141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hertzano, R. et al. Lhx3, a LIM domain transcription factor, is regulated by Pou4f3 in the auditory but not in the vestibular system. Eur. J. Neurosci.25, 999–1005. 10.1111/j.1460-9568.2007.05332.x (2007). [DOI] [PubMed] [Google Scholar]
- 33.Tornari, C., Towers, E. R., Gale, J. E. & Dawson, S. J. Regulation of the orphan nuclear receptor Nr2f2 by the DFNA15 deafness gene Pou4f3. PLoS One9, e112247. 10.1371/journal.pone.0112247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2024), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA009740) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human.