Abstract
Yeast strains can reversibly interconvert between [PSI+] and [psi-] states. The [PSI+] state is caused by a prion form of the translation termination factor eRF3. The [PSI+] state causes read-through at stop codons and can lead to phenotypic variation, although the molecular mechanisms causing those phenotypic changes remain unknown. We identify an interaction between [PSI+]-induced phenotypic variation and defects in nonstop mRNA decay. Nonstop mRNA decay is triggered when a ribosome reaches the 3′ end of the transcript. In contrast, we observed little interaction between [PSI+]-induced phenotypic variation and defects in nonsense-mediated decay, which lead to suppression of premature stop codons. These results suggest that at least some of the phenotypic effects of [PSI+] may be due to read-through of “normal” stop codons, thereby producing extended proteins. Moreover, these observations suggest that nonstop mRNA decay may limit [PSI+]-induced phenotypic variation. Such a process would allow periodic sampling of the 3′ UTR, which can diverge rapidly, for novel and beneficial protein extensions.
Keywords: capacitor, evolution, prion
The [PSI+] state is a heritable characteristic of yeast that shares key characteristics with prions (reviewed in refs. 1-4). eRF3 is one of two key factors that function in the termination of translation at stop codons (5, 6). In [psi-] cells the eRF3 protein is in a conformation that is functional in translation termination, which occurs efficiently. In contrast, in [PSI+] cells the majority of eRF3 is in an altered conformation, and [PSI+] cells have a reduced efficiency of translation termination (3, 4). Both the [PSI+] and [psi-] states are relatively stable, because the preexisting prion form of eRF3 is needed to efficiently convert eRF3 to the prion form. However, the stability is not absolute, and [PSI+] cells convert to [psi-] and vice versa at a low rate (1, 2).
The functions of the eRF3 protein in translation termination and prion propagation are easily separated. The N-terminal domain of eRF3 is required for prion propagation (7, 8). Point mutations in, or deletion of, the N-terminal domain disrupt the prion propagation function of yeast eRF3, and the N-terminal domain by itself is sufficient for prion propagation (3, 7, 9). The translation termination function of eRF3 requires the well conserved C-terminal domain. Deletion of the C-terminal domain is lethal, presumably because of defects in translation termination, and point mutations in the C-terminal domain reduce translation termination efficiency (9). Thus, the prion property of eRF3 is a property of the N-terminal domain, whereas the C-terminal domain is required for translation termination.
The ability of eRF3 to exist in [PSI+] and [psi-] states has been conserved during evolution and is present in the eRF3 proteins of several species and genera of yeast (10-14). The fact that [PSI+] has been conserved during evolution but that mutations that disrupt it can easily be isolated suggests that there is some evolutionary advantage to [PSI+].
True and Lindquist (15) showed that [PSI+] and [psi-] strains of the same genotype have many different growth phenotypes, and that the specific phenotypic effects of [PSI+] depend on the genetic background of the strain used. Most, if not all, of these phenotypic effects are caused by differences in translation termination efficiency, rather than some other effect of PSI state (16). Based on these observations, it has been suggested that the ability to convert from [PSI+]to[psi-] and vice versa may aid the evolution of new traits (15). It is currently not known what kinds of target genes are differentially expressed in [PSI+] versus [psi-] cells and what the underlying molecular mechanisms are by which [PSI+] affects expression of these genes.
One mechanism by which [PSI+] state might affect phenotypes is by causing translational read-through of premature stop codons. Sequence analysis has led to the identification of possible target genes that contain premature stop codons in the one sequenced strain (17, 18). If [PSI+] causes phenotypes mostly by suppressing premature stop codons, then the strain specific consequences would be caused by different premature stop codons in laboratory strains of yeast. Consistent with the hypothesis that [PSI+] induces phenotypic variation by suppressing premature stop codons, it has been reported that in seven cases the effect of [PSI+] was similar to the effect of upf1Δ, which also suppresses premature stop codons (see below) (16). However, it was also reported that in four other cases the effect of [PSI+] was not mimicked by upf1Δ, suggesting that there may be additional mechanisms by which [PSI+] affects phenotypes (16).
Two lines of evidence suggest that [PSI+] affects translation termination not only at premature stop codons, but also at wild-type stop codons naturally found at the 3′ end of coding regions. First, the frequency of extra stop codons just 3′ of the normal stop codon of yeast genes is higher than expected, suggesting that read-through of the normal stop codons occurs at a frequency that is a significant factor over evolutionary time (19). Second, Namy et al. (20) identified eight genes that had a poor stop codon context. In an artificial reporter gene, translation termination at these stop codon contexts was affected by [PSI+]. Thus, [PSI+] might affect phenotypic variation by promoting read-through of normal stop codons (16-18, 20).
In addition to PSI directly affecting translation termination of premature and/or normal stop codons, there may be indirect effects on mRNA stability because translation termination and mRNA stability are intimately linked (16). Two aspects of this link are nonsense-mediated mRNA decay and nonstop mRNA decay. Nonsense-mediated mRNA decay is the process of rapidly degrading mRNAs that contain a premature stop codon (reviewed in ref. 21). It is thought that nonsense-containing mRNAs are recognized as a consequence of premature translation termination. Because [PSI+] affects the efficiency of translation termination, it may also affect nonsense-mediated mRNA decay. Nonstop mRNA decay is the process of degrading mRNAs that do not contain stop codons (22, 23). Nonstop mRNAs are thought to be recognized when a ribosome reaches the 3′ end of an mRNA. In the [PSI+] state, ribosomes may reach the 3′ end of a normal mRNA after reading through one or more stop codons and may trigger nonstop mRNA decay of such normal mRNAs. Importantly, even a single ribosome that reaches the 3′ end of an mRNA is predicted to trigger nonstop decay and reduce the level of mRNA available for translation. Thus, the [PSI+] state may affect both nonsense-mediated and nonstop decay of some mRNAs, which in turn could affect phenotypic variation.
To examine how PSI status interacts with nonsense-mediated decay and nonstop decay, we examined [PSI+]-induced phenotypic variation in strains defective in either of these mRNA decay pathways. Strikingly, we identified a clear effect of defects in nonstop mRNA decay on [PSI+]-induced phenotypic variation. These results suggest that read-through of “normal” stop codons, which produces extended proteins, may be an additional mechanism that contributes to the phenotypic effects of [PSI+] and that, at least in some cases, nonstop mRNA decay limits [PSI+]-induced phenotypic variation. These results are consistent with the hypothesis that the [PSI+] state allows yeast to periodically explore 3′ UTR sequences for novel adaptations (15, 16).
Materials and Methods
Strains. The genotypes of the used strains are listed in Table 1. By sequencing we demonstrated that ade2-1 is a premature UAA codon at codon 64, and can1-100 contains a premature UAA stop codon at codon 47 (24).
Table 1. Strains used.
| Strain | Genotype | Source |
|---|---|---|
| 5V-H19A | mata, ade2–1, can1–100, leu2–3,112, ura3–52, SUP16, SUP35, [PSI+] | Ref. 9 |
| yAV617 | mata, ade2–1, can1–100, leu2–3,112, ura3–52, SUP16, SUP35, upf1Δ::KanMX4, [PSI+] | This study |
| yAV621 | mata, ade2–1, can1–100, leu2–3,112, ura3–52, SUP16, SUP35, ski7Δ::KanMX4, [PSI+] | This study |
| yAV703 | mata, ade2–1, can1–100, leu2–3,112, ura3–52, SUP16, sup35-ΔN, [psi–] | This study |
| yAV705 | mata, ade2–1, can1–100, leu2–3,112, ura3–52, SUP16, sup35-ΔN, upf1Δ::KanMX4, [psi–] | This study |
| yAV707 | mata, ade2–1, can1–100, leu2–3,112, ura3–52, SUP16, sup35-ΔN, ski7Δ::KanMX4, [psi–] | This study |
| yAV734 | mata, ade2–1, can1–100, leu2–3,112, ura3–52, SUP16, SUP35, ski2Δ::KanMX4, [PSI+] | This study |
| yAV736 | mata, ade2–1, can1–100, leu2–3,112, ura3–52, SUP16, SUP35, ski3Δ::KanMX4, [PSI+] | This study |
| yAV738 | mata, ade2–1, can1–100, leu2–3,112, ura3–52, SUP16, SUP35, ski8Δ::KanMX4, [PSI+] | This study |
| yAV740 | mata, ade2–1, can1–100, leu2–3,112, ura3–52, SUP16, SUP35, ski7-ΔC::KanMX4, [PSI+] | This study |
| yAV831 | mata, ade2–1, can1–100, leu2–3,112, ura3–52, SUP16, SUP35, [psi–] | This study |
| yAV833 | mata, ade2–1, can1–100, leu2–3,112, ura3–52, SUP16, SUP35, ski7Δ::KanMX4, [psi–] | This study |
To ensure that the various strains were completely isogenic, we created various deletions in this [PSI+] strain by transformation with gene knockout cassettes. The ski7-ΔC gene was introduced similarly by amplifying the ski7-ΔC:KanMX4 gene (23). All deletions were confirmed by PCR.
Unless otherwise indicated, the [psi-] strains used were created by precisely deleting the prion-forming domain (amino acids 1-123) of eRF3. Other methods of creating [psi-] variants have the disadvantage that they also cure other yeast prions {e.g., [URE3] and [PIN+] (1, 18)}. In addition, successful conversion to [psi-] is usually monitored by failure to grow on synthetic complete medium (SC) lacking adenine (ADE), but this phenotype is modified by the upf1Δ and ski7Δ mutations used in this study (see below). The prion-forming N domain was precisely deleted, using the pop-in, pop-out method with HindIII-cut pAv208, which is a pRS306 derivative that contains the promoter and M domain of the eRF3 gene of strain 5V-H19A (25, 26).
Alternatively, yAV603 and yAV621 were converted to [psi-] strains yAV831 and yAV833 by growing them on yeast extract/peptone/dextrose (YPD) plus 5 mM guanidium hydrochloride, and identifying [psi-] variants by their slower growth on SC lacking ADE and faster growth on complete supplement mixture medium lacking arginine and containing canavanine.
Media and Growth. Media were prepared as described in refs. 15 and 27. To test growth on various media, strains were grown in YPD at 30°C, serially diluted in 5-fold steps, plated on various media, and incubated at 30°C with growth monitored daily for up to 14 days.
Northern Blot Analysis. Strains were grown in YPD at 30°C to early to middle log phase. ADE2 RNA was analyzed by using Northern blot, probe oAV92 (tcactggcttgttccacagggacactttgg), and a Storm imager (Molecular Dynamics). mRNA levels were corrected for loading by using oRP100 (gtctagccgcgaggaagg), which detect the signal recognition particle RNA. PGK1pG mRNA was detected by using probe oRP140 after transformation with plasmid pRP469 and growth in SC-URA containing 2% galactose and 2% sucrose (28).
Results
[PSI+] and Premature Stop Codons. Previous results suggest that one mechanism by which [PSI+] affects phenotype is by allowing read-through of premature stop codons (16). To further test this hypothesis, we first compared the phenotype of a [PSI+] strain to the phenotype caused by another method of suppressing premature stop codons. Many mutations can suppress premature stop codons (e.g., sup35, sup45, and suppressor tRNAs), but most of these are also expected to cause read-through at normal stop codons and are not useful for distinguishing the contribution of read-through at premature versus normal stop codons. In contrast, Upf1p appears to specifically recognize mRNAs containing premature stop codons. Upf1p recognition of an mRNA containing a premature stop codon has been reported to have a variety of effects in vivo. One major effect is to specifically stabilize such mRNAs (29). Upf1p may also specifically alter translation termination or translation initiation on these mRNAs (24, 30-33). The net effect of upf1Δ is a suppression of the phenotype of premature stop codons. These in vivo data are supported by the recent demonstration that in in vitro translation extracts the fate of the ribosome is different at premature stop codons as compared with normal stop codons, and that this difference depends on Upf1p (33). Thus, all available data suggest that upf1Δ specifically suppresses the phenotype caused by premature stop codons. To analyze the significance of suppression of premature stop codons to PSI variable phenotypes, we compared the effect of [PSI+] with the effect of loss of Upf1p.
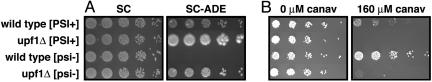
We first confirmed the suppression of the phenotype of known premature stop codons by [PSI+] and upf1Δ. As expected, premature stop codons in ADE2 and CAN1 are partially suppressed in both the UPF1 [PSI+] strain and in the upf1Δ [psi-] strain when compared with the UPF1 [psi-] strain (Fig. 1). The ade2-1 allele contains a premature stop codon in a gene required for synthesis of adenine, and ade2-1 strains grow slowly in the absence of added adenine. Both [PSI+] and upf1Δ caused increased growth on plates lacking adenine, indicating that both suppressed ade2-1. The can1-100 allele contains a premature stop codon in a gene for an arginine transporter, and can1-100 strains are resistant to the arginine analog canavanine. Both [PSI+] and upf1Δ caused decreased growth on plates containing canavanine, indicating that both suppressed can1-100. From these results, we conclude that [PSI+] and upf1Δ are approximately equally effective at phenotypic suppression of premature stop codons. Thus, if the phenotype of a [PSI+] strain is only caused by suppression of premature stop codons, then upf1Δ [psi-] and UPF1 [PSI+] strains should have a largely overlapping phenotype.
Fig. 1.
upf1Δ and [PSI+] have qualitatively similar effects on the suppression of premature stop codons in ade2-1 and can1-100. (A) Strains were grown at 30°C after spotting on media containing (SC) or lacking (SC-ADE) adenine. (B) Strains were spotted on media containing 0, 20, 40, 160, or 320 μM canavanine. Other concentrations of canavanine give essentially the same results (data not shown).
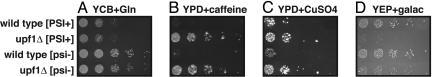
Next, we compared the effect of upf1Δ on phenotypes previously reported to be affected by [PSI+]. In this analysis, we observed phenotypic effects of deleting upf1Δ from a [psi-] strain, but these effects were generally not similar to those observed for [PSI+]. For example, Fig. 3A shows that the [PSI+] strain grows slower when using glutamine as a nitrogen source (compare first and third rows), but upf1Δ has no effect on growth under this condition (compare third and fourth rows). Similar results were seen for growth using serine, threonine, proline, ornithine, or glutamate as nitrogen sources and growth in the presence of ZnCl2 (shown in Fig. 5, which is published as supporting information on the PNAS web site). In each of these cases, [PSI+] causes a clear growth phenotype, but upf1Δ does not have the same effect. Under a second set of conditions, upf1Δ affects the phenotype, and [PSI+] does not have the same effect. Specifically, we observed that upf1Δ leads to increased growth in the presence of caffeine and CuSO4 and decreased growth on galactose, and that the [PSI+] state did not cause the same phenotype (Figs. 2B and 5).
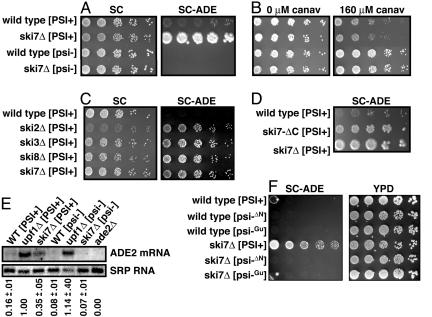
Fig. 3.
Defects in nonstop mRNA decay suppress the premature stop codon in ade2-1 ina[PSI+]-dependent manner. (A) ski7Δ only suppresses ade2-1 ina[PSI+] background. (B) ski7Δ does not affect can1-100 suppression. (C) ski2Δ, ski3Δ, ski7Δ, and ski8Δ each suppress ade2-1 ina[PSI+] strain. (D) A mutation in SKI7 that inactivates nonstop decay but does not interfere with general exosome-mediated mRNA decay (ski7-ΔC) suppresses ade2-1 ina[PSI+] strain. (E) The ade2-1 mRNA level is increased by ski7Δ ina[PSI+]-dependent manner. The numbers in E are the ade2-1 mRNA levels from two independent experiments, relative to the level in the upf1Δ [PSI+] strain.
Fig. 2.
Comparison of upf1Δ and [PSI+] effects on growth. (A)[PSI+] slows growth on yeast carbon base (YCB) plus 1 g/liter glutamine, but upf1Δ has no effect (3 days of growth). Many other conditions where [PSI+] affects growth but upf1Δ does not affect growth are shown in Fig. 5. (B) upf1Δ increases growth on YPD plus 10 mM caffeine, in both the [PSI+] and [psi-] strain (4 days of growth). (C) upf1Δ increases growth on YPD plus 2.5 mM CuSO4, in both the [PSI+] and [psi-] strain (6 days of growth). (D) upf1Δ decreases growth on yeast extract/peptone (YEP) plus 2% galactose, in both the [PSI+] and [psi-] strain (4 days of growth).
The experiment described above also indicates that indirect effects on nonsense-mediated mRNA decay are unlikely to be the only mechanism for [PSI+]-induced phenotypic variation. This conclusion is based on the observation that all of the phenotypic differences we detected between a [PSI+] and a [psi-] strain are maintained when comparing a upf1Δ [PSI+] strain with a upf1Δ [psi-] strain. Thus, [PSI+] affects many phenotypes even in a strain background incapable of nonsense-mediated mRNA decay. We also note that the dissimilar effects of upf1Δ and [PSI+] are not what would be expected if both simply suppressed premature stop codons. Thus, in addition to affecting the phenotype by suppressing premature stop codons, both upf1Δ and/or [PSI+] may also affect the phenotype by other mechanisms (see Discussion) (16).
[PSI+] Genetically Interacts with Mutations Disrupting Nonstop mRNA Degradation. Read-through at normal stop codons may cause the ribosome to reach the 3′ end of an mRNA. Eukaryotic mRNAs are rapidly degraded when a ribosome reaches the 3′ end of an mRNA [referred to as nonstop mRNA decay (22, 23)]. Moreover, because nonstop decay is likely triggered by a single ribosome, even inefficient translation read-through caused by could significantly decrease the abundance of some normal mRNAs. Thus, [PSI+] could reduce the abundance of the normal protein encoded by those mRNAs. Nonstop mRNA decay requires Ski7p, in part to recognize the stalled ribosome (23). Thus, ski7Δ results in stabilization of mRNAs that lack a stop codon but generally has no effect on the stability of normal mRNAs. Therefore, if [PSI+] triggers nonstop mRNA decay, this effect will be blocked in a ski7Δ [PSI+] strain.
We first examined suppression of known premature stop codons, because [PSI+] is thought to suppress premature stop codons independently of triggering nonstop decay, and, thus, suppression should not be blocked by ski7Δ. As expected, ski7Δ did not block [PSI+]-mediated suppression of premature stop codons (Fig. 3 A and B). Instead, we observed that ski7Δ increased ade2-1 expression in the [PSI+] state (Fig. 3A, first and second rows) but not in the [psi-] state (Fig. 3A, third and fourth rows). One possible explanation is that [PSI+] causes read-through of all in-frame stop codons in ade2-1, which triggers nonstop mRNA decay. The ski7Δ would block nonstop decay and would result in increased ade2-1 mRNA levels. Conversely, in a [psi-] strain the ribosome would not reach the 3′ end of the ade2-1 mRNA, and ski7Δ would have no effect. We did not see a similar effect on can1-100 expression (as measured by canavanine sensitivity; Fig. 3B). However, the genetic interaction between [PSI+] and ski7Δ for the expression of ade2-1 suggests that alterations in nonstop mRNA decay could be important for [PSI+]-induced phenotypic variation.
To further test for a genetic interaction between [PSI+] and nonstop decay, we analyzed the effect of other mutations that inhibit nonstop decay. Ski2p, Ski3p, and Ski8p form a heterotrimeric complex that interacts with Ski7p and is required for nonstop decay (23, 34, 35). We therefore deleted the genes for each of these proteins from the [PSI+] strain and analyzed their effect on ade2-1 expression. Fig. 3C shows that each of these deletions increases ade2-1 expression. Finally, we deleted just the C-terminal domain of Ski7p. This domain is specifically required for nonstop decay and likely functions in the recognition of ribosomes that have reached the 3′ end of an mRNA (23). Importantly, this ski7-ΔC mutation also results in increased expression of ade2-1 (Fig. 3D). Thus, analyses of growth phenotype indicate that [PSI+] genetically interacts with the nonstop mRNA decay pathway to alter ade2-1 expression.
To confirm that the observed growth phenotypes on SC-ADE indeed reflected ade2-1 expression, we analyzed the ade2-1 mRNA by Northern blotting. Fig. 3E shows that the ade2-1 mRNA level is increased by the upf1Δ in either the [PSI+] or [psi-] background. This finding is consistent with the increase in growth seen on SC-ADE in Fig. 2 A. As expected from the growth phenotype, ski7Δ resulted in an increase in ade2-1 mRNA levels in the [PSI+] state but had no detectable effect in the [psi-] state (Fig. 3E). This result is consistent with our interpretation that the [PSI+] state triggers nonstop mRNA degradation of the ade2-1 mRNA.
The results above were obtained by using a [psi-] strain that is lacking the prion-forming N terminus of eRF3. Our data therefore indicate that the N terminus of eRF3 genetically interacts with nonstop decay. The most likely explanation is that the [PSI+] prion form of the N terminus of eRF3 triggers nonstop decay. Alternatively, the N terminus of eRF3 can physically interact with several other proteins, and one of these physical interactions may be responsible for the genetic interaction with the nonstop mRNA decay pathway (36, 37). To test whether [PSI+] is responsible for the genetic interaction, we converted [PSI+] strains to [psi-] by growing them in the presence of guanidium HCl (1, 2). Fig. 3F shows that strains that contain the N terminus of eRF3 in the [psi-] conformation have the same phenotype as strains missing the prion-forming domain of eRF3. Therefore, we conclude that [PSI+], and not some other feature of the N terminus of eRF3, genetically interacts with nonstop mRNA decay.
The results above suggest that the [PSI+] state could trigger read-through of all stop codons in some other mRNAs and thereby reduce their levels in a manner dependent on nonstop decay. To test this prediction, we examined the steady state levels of several other mRNAs in [PSI+] and [psi-] strains, with or without a functional nonstop decay system. We found no significant differences in the levels of the RPS15 or RPL25 mRNAs. Strikingly, we observed that the [PSI+] state showed 2-fold reduced levels of the PGK1pG mRNA and that, in a [PSI+] ski7Δ strain, the mRNA level was increased (Table 2). This result provides additional evidence that the [PSI+] state can trigger nonstop decay of mRNAs that contain a stop codon. Most importantly, the conclusion that [PSI+] can trigger decay of mRNAs that contain stop codons in a process that is indistinguishable from nonstop mRNA decay raises the possibility that some of the phenotypic variation caused by [PSI+] is caused or modulated by nonstop mRNA decay.
Table 2. Effect of [PSI+] and ski7Δ on PGK1pG mRNA.
| Strain | PGK1pG mRNA level, % |
|---|---|
| [psi-] | 100 |
| ski7Δ [psi-] | 83 ± 15 |
| [PSI+] | 55 ± 8 |
| ski7Δ[PSI+] | 244 ± 10 |
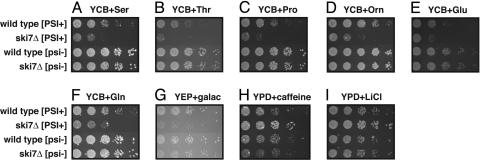
Nonstop mRNA Decay Modulates [PSI+] Phenotypes. To examine whether the genetic interaction between [PSI+] and ski7Δ was specific to ade2-1 or also affected other phenotypes, we compared the phenotype of wild-type [PSI+] and [psi-] strains to isogenic ski7Δ strains under 16 different conditions where growth has previously been shown to be affected by [PSI+] (15). This experiment showed that nonstop decay modulates the phenotypic effect of [PSI+]. In six conditions, we found results similar to those for ade2-1: The phenotypic effect of [PSI+] is further increased by ski7Δ (growth on YCB plus serine, threonine, proline, ornithine, glutamate, or glutamine; Fig. 4 A-F). Under these conditions, [PSI+] isolates grow slower than [psi-] isolates (compare first and third rows). Deletion of SKI7 further decreases growth in the [PSI+] state (compare first and second rows) but has no effect in the [psi-] state (compare third and fourth rows). Thus, in some cases the capacity of [PSI+] to reveal new phenotypes is limited by nonstop decay. Additional examples of this limiting effect can be seen on YEP plus galactose, YPD plus caffeine, and YPD plus LiCl. In these three cases, wild-type [PSI+] and [psi-] strains have (almost) the same phenotype (Fig. 4 G-I, first and third rows), but a clear difference can be seen between ski7Δ [PSI+] and ski7Δ [psi-] strains (second and fourth rows). Under the remaining conditions, we did not see a clear reproducible difference between the phenotypes of [PSI+] and [psi-] strains, or the difference was not affected by ski7Δ (Fig. 5 and data not shown). Thus, in 9 of 16 conditions examined, ski7Δ affects the phenotype in the [PSI+] state but has no effect in the [psi-] state. We conclude that the genetic interaction between nonstop mRNA decay and [PSI+] is not limited to ade2-1 and that nonstop mRNA decay may, in some cases, limit the capacity of [PSI+] to reveal new phenotypes.
Fig. 4.
ski7Δ has various phenotypic consequences in a [PSI+] strain but not in an isogenic [psi-] strain. (A) YCB plus 1 g/liter serine, 3 days of growth. (B) YCB plus 1 g/liter threonine, 3 days of growth. (C) YCB plus 1 g/liter proline, 3 days of growth. (D) YCB plus 1 g/liter ornithine, 3 days of growth. (E) YCB plus 1 g/liter glutamate, 2 days of growth. (F) YCB plus 1 g/liter glutamine, 3 days of growth. (G) YEP plus 2% galactose, 5 days of growth. (H) YPD plus 10 mM caffeine, 6 days of growth. (I) YPD plus 50 mM LiCl, 3 days of growth.
Discussion
[PSI+] Genetically Interacts with Nonstop mRNA Decay. We have shown a genetic interaction between [PSI+] and mutations that inactivate the nonstop mRNA decay pathway. The central observation is that strains defective in nonstop mRNA decay show a greater variation in phenotype between [psi-] and [PSI+] states than corresponding wild-type [psi-] and [PSI+] pairs. This genetic interaction affects the expression of ade2-1 and growth under 9 of 16 conditions tested. It is unclear whether these nine phenotypes reflect read-through events on different mRNAs, or whether several of these phenotypes are caused by the same read-through event of the same mRNA. In either case, a simple explanation for this interaction is that [PSI+] causes read-through of normal stop codons and any other stop codons in the 3′ UTR. Such read-through would cause a ribosome to reach the very 3′ end of the mRNA and would trigger nonstop mRNA decay (22, 23). This possibility is supported by the observation that [PSI+] leads to a Ski7p-dependent reduction in the levels of the PGK1 and ade2-1 mRNAs (Fig. 3 and Table 2). Because yeast 3′ UTRs are relatively short, with many shorter than 100 nt, the ribosome would only have to read through one or two additional stop codons in the 3′ UTR to reach the 3′ end of such mRNAs (38). Thus, [PSI+] may trigger some degree of nonstop mRNA decay on a variety of different mRNAs.
The ability of [PSI+] to induce nonstop mRNA decay of specific mRNAs is affected by several factors. One likely factor is the identity and sequence context of the stop codons, which can affect the degree of suppression by [PSI+] (16). Another likely contributor is the number and type of in-frame stop codons within the 3′ UTR. These additional factors may explain why the ade2-1 allele is affected strongly by [PSI+] ski7Δ, whereas the can1-100 allele is less affected. In the ade2-1 mRNA, the premature stop codon, the normal stop codon, and the next stop codon in the 3′ UTR are all UAA codons. In contrast, the can1-100 mRNA contains a UAA premature stop codon, a UAG at the end of the natural ORF, and a UGA in the 3′ UTR. Although the read-through frequency of these stop codons within ade2-1 and can1-100 is unknown, the read-through frequency of UAA, UAG, and UGA codons in the [PSI+] 5V-H19A strain has been measured (16). Using these frequencies, the combination of the normal UAG codon and UGA at +2 in can1-100 is expected to be read through 33 times less efficiently than the combination of the normal UAA codon and the UAA codon at +12 in ade2-1. This analysis suggests that one possible reason for the difference in effects of ski7Δ may be that the combination of stop codons in ade2-1 is read through more efficiently than that in can1-100, and thus the ribosome is more likely to reach the 3′ end of the ade2-1 mRNA. This interpretation implies that the ability of [PSI+] to read through normal stop codons and/or trigger nonstop mRNA decay on individual mRNAs will vary significantly.
Significance of Read-Through of Normal Stops and Nonstop mRNA Decay for [PSI+]-Induced Phenotypes. We can envision two possible mechanisms by which translation past the normal stop codon and into the 3′ UTR may affect the phenotype. First, translation all of the way through the 3′UTR should trigger nonstop mRNA decay and thereby reduce the level of protein that can be made from that mRNA by subsequent rounds of translation. This mechanism is inconsistent with our findings, because it predicts that [PSI+] and [psi-] phenotypes are the same in a strain that is incapable of nonstop mRNA decay (e.g., ski7Δ). Thus, triggering nonstop decay per se is not a main mechanism that explains the phenotypic consequences of [PSI+] that we observe. In an alternative model, translation into the 3′ UTR produces C-terminally extended proteins. Our observation that ski7Δ enhances [PSI+] phenotypes is consistent with this hypothesis, because ski7Δ would increase the level of target mRNAs and, in turn, increase production of the C-terminally extended proteins encoded by that mRNA. A corollary of this interpretation is that the C-terminally extended proteins are functional and actively contribute to the phenotype of [PSI+] strains.
A final understanding of all of the molecular mechanisms behind each of the [PSI+]-induced phenotypes requires identification of individual target genes. Our results suggest that one way to find some of these target genes is by microarray analyses on [PSI+] versus ski7Δ [PSI+]. Our results also indicate that a bioinformatic approach to finding target genes may be more successful if it takes into account the presence of additional stop codons within the 3′ UTR. It should also be noted that we examined phenotypic variation in strain 5V-H19A, because in this strain [PSI+] is especially effective at modifying growth phenotypes. This strain contains a weak suppressor tRNA (SUP16). Thus, we cannot rule out the formal possibility that the genetic interaction we observe requires the suppressor tRNA. However, this seems unlikely because [PSI+] induces efficient read-through of stop codons and phenotypic variation even in the absence of suppressor tRNAs (15, 39).
Multiple Mechanisms of [PSI+]-Induced Phenotypic Variation. Earlier work by True et al. (16) showed that in 7 of 11 cases examined, [PSI+]-induced phenotypic variation could be mimicked by upf1Δ, which affects nonsense-mediated decay and allows suppression of premature stop codons. This finding suggests that, in some cases, [PSI+]-induced phenotypic variation is caused by the suppression of premature stop codons. However, True et al. (16) also observed that some phenotypes were not mimicked by upf1Δ, suggesting there are additional manners by which [PSI+] affects phenotypic variation. Consistent with this latter conclusion, we also observed different phenotypic consequences for [PSI+] and upf1Δ in our experiments. Thus, the data presented by True et al. and us suggest that [PSI+] affects gene expression by at least two distinct mechanisms. First, [PSI+] causes read-through of premature stop codons, thereby restoring protein production. Second, [PSI+] causes read-through of normal stop codons, thereby possibly producing C-terminally extended proteins. Database searches for premature stop codons and normal stop codons in poor context have identified a number of putative [PSI+] target genes (17, 18, 20). However, True et al. have shown that most of the phenotypic consequences of [PSI+] are multigenic. This finding suggests that [PSI+]-induced phenotypic variation will involve changes in the expression of multiple mRNAs. Thus, a single [PSI+]-induced phenotypic difference could be due to a combination of both the read-through of premature and normal stop codons.
Implications for a Role for [PSI+] in Evolution. The possible exploration of 3′ UTRs for advantageous encoded protein sequences may have advantages as an evolutionary mechanism (15, 16). In the [psi-] state the 3′ UTR could accumulate sequence variation with little selective pressure. Indeed, comparison of genome sequences of closely related yeast species shows that 3′ UTR sequences are less conserved than coding regions or promoters (40, 41). An occasional switch to the [PSI+] state would allow the strain to explore the accumulated diversity for advantageous proteins. If any of the newly translated sequences offered an advantage, this advantage could, over time, become independent of [PSI+], by mutation of a stop codon to a sense codon. Subsequently, the cell could revert to [psi-] without losing the novel adaptation, while losing other C-terminally extended proteins that may be deleterious. The advantage of this mechanism may be further increased by preferentially switching to [PSI+] under severely adverse conditions, which can affect the conversion of [psi-] to [PSI+] (18, 42).
Supplementary Material
Acknowledgments
We thank Susan Lindquist, Heather True, Tricia Serio, and David Bedwell for fruitful discussions and strains. This work was supported by the Howard Hughes Medical Institute (R.P.) and a PEW Scholarship in the Biomedical Sciences (to A.v.H.)
Abbreviations: ADE, adenine; SC, synthetic complete medium; YCB, yeast carbon base; YEP, yeast extract/peptone; YPD, yeast extract/peptone/dextrose.
References
- 1.Uptain, S. M. & Lindquist, S. (2002) Annu. Rev. Microbiol. 56, 703-741. [DOI] [PubMed] [Google Scholar]
- 2.Wickner, R. B., Taylor, K. L., Edskes, H. K., Maddelein, M. L., Moriyama, H. & Roberts, B. T. (1999) Microbiol. Mol. Biol. Rev. 63, 844-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patino, M. M., Liu, J. J., Glover, J. R. & Lindquist, S. (1996) Science 273, 622-626. [DOI] [PubMed] [Google Scholar]
- 4.Paushkin, S. V., Kushnirov, V. V., Smirnov, V. N. & Ter-Avanesyan, M. D. (1996) EMBO J. 15, 3127-3134. [PMC free article] [PubMed] [Google Scholar]
- 5.Stansfield, I., Jones, K. M., Kushnirov, V. V., Dagkesamanskaya, A. R., Poznyakovski, A. I., Paushkin, S. V., Nierras, C. R., Cox, B. S., Ter-Avanesyan, M. D. & Tuite, M. F. (1995) EMBO J. 14, 4365-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhouravleva, G., Frolova, L., Le Goff, X., Le Guellec, R., Inge-Vechtomov, S., Kisselev, L. & Philippe, M. (1995) EMBO J. 14, 4065-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chernoff, Y. O., Derkach, I. L. & Inge-Vechtomov, S. G. (1993) Curr. Genet. 24, 268-270. [DOI] [PubMed] [Google Scholar]
- 8.Ter-Avanesyan, M. D., Dagkesamanskaya, A. R., Kushnirov, V. V. & Smirnov, V. N. (1994) Genetics 137, 671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ter-Avanesyan, M. D., Kushnirov, V. V., Dagkesamanskaya, A. R., Didichenko, S. A., Chernoff, Y. O., Inge-Vechtomov, S. G. & Smirnov, V. N. (1993) Mol. Microbiol. 7, 683-692. [DOI] [PubMed] [Google Scholar]
- 10.Chernoff, Y. O., Galkin, A. P., Lewitin, E., Chernova, T. A., Newnam, G. P. & Belenkiy, S. M. (2000) Mol. Microbiol. 35, 865-876. [DOI] [PubMed] [Google Scholar]
- 11.Zadorskii, S. P., Sopova Iu, V. & Inge-Vechtomov, S. G. (2000) Genetika 36, 1322-1329. [PubMed] [Google Scholar]
- 12.Santoso, A., Chien, P., Osherovich, L. Z. & Weissman, J. S. (2000) Cell 100, 277-288. [DOI] [PubMed] [Google Scholar]
- 13.Kushnirov, V. V., Kochneva-Pervukhova, N. V., Chechenova, M. B., Frolova, N. S. & Ter-Avanesyan, M. D. (2000) EMBO J. 19, 324-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakayashiki, T., Ebihara, K., Bannai, H. & Nakamura, Y. (2001) Mol. Cell 7, 1121-1130. [DOI] [PubMed] [Google Scholar]
- 15.True, H. L. & Lindquist, S. L. (2000) Nature 407, 477-483. [DOI] [PubMed] [Google Scholar]
- 16.True, H. L., Berlin, I. & Lindquist, S. L. (2004) Nature 431, 184-187. [DOI] [PubMed] [Google Scholar]
- 17.Harrison, P., Kumar, A., Lan, N., Echols, N., Snyder, M. & Gerstein, M. (2002) J. Mol. Biol. 316, 409-419. [DOI] [PubMed] [Google Scholar]
- 18.Chernoff, Y. O. (2001) Mutat. Res. 488, 39-64. [DOI] [PubMed] [Google Scholar]
- 19.Williams, I., Richardson, J., Starkey, A. & Stansfield, I. (2004) Nucleic Acids Res. 32, 6605-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Namy, O., Duchateau-Nguyen, G. & Rousset, J. P. (2002) Mol. Microbiol. 43, 641-652. [DOI] [PubMed] [Google Scholar]
- 21.Hilleren, P. & Parker, R. (1999) Annu. Rev. Genet. 33, 229-260. [DOI] [PubMed] [Google Scholar]
- 22.Frischmeyer, P. A., van Hoof, A., O'Donnell, K., Guerrerio, A. L., Parker, R. & Dietz, H. C. (2002) Science 295, 2258-2261. [DOI] [PubMed] [Google Scholar]
- 23.van Hoof, A., Frischmeyer, P. A., Dietz, H. C. & Parker, R. (2002) Science 295, 2262-2264. [DOI] [PubMed] [Google Scholar]
- 24.Maderazo, A. B., He, F., Mangus, D. A. & Jacobson, A. (2000) Mol. Cell Biol. 20, 4591-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothstein, R. (1991) Methods Enzymol. 194, 281-301. [DOI] [PubMed] [Google Scholar]
- 26.Sikorski, R. S. & Hieter, P. (1989) Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burke, D., Dawson, D. & Stearns, T. (2000) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual (Cold Spring Harbor Lab. Press, Cold Spring Harbor, NY).
- 28.Decker, C. J. & Parker, R. (1993) Genes Dev. 7, 1632-1643. [DOI] [PubMed] [Google Scholar]
- 29.Leeds, P., Peltz, S. W., Jacobson, A. & Culbertson, M. R. (1991) Genes Dev. 5, 2303-2314. [DOI] [PubMed] [Google Scholar]
- 30.Muhlrad, D. & Parker, R. (1999) Mol. Biol. Cell 10, 3971-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weng, Y., Czaplinski, K. & Peltz, S. W. (1996) Mol. Cell. Biol. 16, 5477-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bidou, L., Stahl, G., Hatin, I., Namy, O., Rousset, J. P. & Farabaugh, P. J. (2000) RNA 6, 952-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amrani, N., Ganesan, R., Kervestin, S., Mangus, D. A., Ghosh, S. & Jacobson, A. (2004) Nature 432, 112-118. [DOI] [PubMed] [Google Scholar]
- 34.Brown, J. T., Bai, X. & Johnson, A. W. (2000) RNA 6, 449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Araki, Y., Takahashi, S., Kobayashi, T., Kajiho, H., Hoshino, S. & Katada, T. (2001) EMBO J. 20, 4684-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailleul, P. A., Newnam, G. P., Steenbergen, J. N. & Chernoff, Y. O. (1999) Genetics 153, 81-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosoda, N., Kobayashi, T., Uchida, N., Funakoshi, Y., Kikuchi, Y., Hoshino, S. & Katada, T. (2003) J. Biol. Chem. 278, 38287-38291. [DOI] [PubMed] [Google Scholar]
- 38.Graber, J. H., Cantor, C. R., Mohr, S. C. & Smith, T. F. (1999) Proc. Natl. Acad. Sci. USA 96, 14055-14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keeling, K. M., Lanier, J., Du, M., Salas-Marco, J., Gao, L., Kaenjak-Angeletti, A. & Bedwell, D. M. (2004) RNA 10, 691-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. (2003) Nature 423, 241-254. [DOI] [PubMed] [Google Scholar]
- 41.Cliften, P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterston, R., Cohen, B. A. & Johnston, M. (2003) Science 301, 71-76. [DOI] [PubMed] [Google Scholar]
- 42.Derkatch, I. L., Bradley, M. E., Masse, S. V., Zadorsky, S. P., Polozkov, G. V., Inge-Vechtomov, S. G. & Liebman, S. W. (2000) EMBO J. 19, 1942-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.