Abstract
Heparan sulfate glycosaminoglycans, present at the cell surface and in the extracellular matrix that surrounds cells, are important mediators of complex biological processes. Furthermore, it is now apparent that cells dynamically regulate the structure of their heparan sulfate “coat” to differentially regulate extracellular signals. In the present study, the importance of sequence information contained within tumor cell-surface heparan sulfate was investigated. Herein, we demonstrate that the heparan sulfate glycosaminoglycan coat present on tumor cells contains bioactive sequences that impinge on tumor-cell growth and metastasis. Importantly, we find that growth promoting as well as growth inhibiting sequences are contained within the polysaccharide coat. Furthermore, we find that the dynamic balance between these distinct polysaccharide populations regulates specific intracellular signal-transduction pathways. This study not only provides a framework for the development of polysaccharide-based anti-cancer molecules but also underscores the importance of understanding a cell's polysaccharide array in addition to its protein complement, to understand how genotype translates to phenotype in this postgenomic age.
Many, if not most, of the molecular events associated with tumor growth, neovascularization, and metastasis are influenced by interactions between cells and their extracellular matrix (ECM). Heparan sulfate-like glycosaminoglycans (HSGAGs), along with structural proteins, are key components of the cell surface–ECM interface. Whereas collagen-like proteins provide the necessary scaffold for cell attachment and tissue formation, the HSGAG complex polysaccharides fill the scaffold and act as a molecular sponge by specifically binding to and regulating the activities of numerous signaling molecules such as growth factors and cytokines (1, 2). Important progress has been made in understanding the diverse roles of collagen (and related proteins) and enzymes (namely, collagenases) that degrade the proteinaceous component of the ECM in regulating tumor growth and metastasis (3, 4). However, the chemical heterogeneity of HSGAGs, coupled with the lack of effective tools to study these polysaccharides, has seriously limited investigations into the roles of HSGAGs in tumor growth and metastasis. Despite extensive observations made regarding both the level of expression as well as the changes in the fine structure of tumor cell surface HSGAGs, it remains to be seen whether these changes are merely a result of tumor progression or whether they actually play a more active role in tumor invasion and metastasis (5). Currently, it is unclear whether tumor cell-surface HSGAGs contain biologically relevant information that can serve to regulate tumor progression. The recent cloning of tumor heparanase genes has led to the proposition that the expression of HSGAG-degrading enzymes represents a switch from a primary tumor to a metastatic disease state (6, 7). However, the diversified structural characteristics and information density of HSGAGs (5, 8) might allow them to regulate tumor pathophysiology in multiple ways. Thus, what is required at the present time is direct evidence of the roles of HSGAGs in tumor growth, neovascularization, and metastasis as well as an understanding of the biological information encoded in the HSGAGs at the tumor-cell surface.
Herein, we used heparinases I (Hep I) and III (Hep III) (9, 10), which have very distinct HSGAG substrate specificities (11), as tools to investigate the role of HSGAGs in tumor growth, neovascularization, and metastasis. Hep I cleaves at the highly sulfated regions of HSGAGs, whereas Hep III only cleaves at the undersulfated regions of the polysaccharide chain. Because these enzymes cleave divergent regions of HSGAGs, leaving behind intact structurally distinct saccharide fragments, they have become powerful tools to investigate the in vivo and in vitro roles of HSGAGs in processes such as development (12) and neovascularization (13).
Materials and Methods
Materials.
Recombinant Hep I and III were expressed and purified to homogeneity, as described (9, 10). The enzymes were incubated with endotoxin removal resin (Associates of Cape Cod) to ensure its removal. HSGAG fragments were collected by incubating 90–100% confluent B16BL6 cells with 1.5 ml of PBS containing 200 nM of Hep I (9 μg) or Hep III (15 μg) at 37°C on a shaker for 1 h. Thus, supernatant was pooled into a tube, centrifuged for 8 min at 4500 × g, boiled for 15 min, and filtered. This procedure yielded reproducible amounts of HSGAG fragments for in vivo analysis. Rabbit polyclonal IgG antibodies specific to Erk-1, 2 (#9102), phospho-p44/42 Erk 1, 2 (#9101), Akt (#9272), and phospho-Akt (Ser-473) (#9271) were purchased from New England Biolabs. Mouse anti-FAK monoclonal antibody and RC20 (phosphotyrosine-specific antibody conjugated with horseradish peroxidase) were purchased from Transduction Laboratories (Lexington, KY). Polyclonal anti-fibroblast growth factor (FGF) receptor (FGFR)-1 antibody was obtained from Santa Cruz Biotechnology. SuperSignal West Pico Chemiluminescent Substrate was purchased from Pierce. vWF and Ki-67 antigen-staining kits for immunohistochemical study were purchased from Dako. Fluorescein apoptosis detection kit was obtained from Promega. DNase, RNase, and Pronase were purchased from Roche Molecular Biochemicals.
Tumor Implantation and Lung Metastasis.
For primary tumor implantation, 4 × 105 log growth-phase B16BL6 melanoma cells or Lewis lung carcinoma cells in 0.1 ml PBS were injected s.c. to the right flank of C57BL/6 mice on day 1 (n = 5). In mice treated with Hep, osmotic pumps (Alza) delivering 0.5 μl of Hep solution per hour for 7 days were implanted s.c. on day 7, while an additional daily injection of enzyme at a selected site distant from the tumor started on day 4 and continued throughout the experiment. In mice treated with HSGAG fragments, osmotic pumps delivering 0.5 μl of an HSGAG fragment solution per hour for 14 days were implanted on day 2, and additional daily injections of fragment solutions were started on day 5 and continued throughout the experiment. Tumor volume was measured daily after day 7 with a caliper and calculated using the formula [volume (in mm3) = 0.52 × (width)2 × (length)]. For the lung metastasis model, 0.2 ml of cell suspensions (2 × 105 tumor cells) were injected slowly by means of the tail vein (n = 8) on day 1. For the enzyme experiments, cells were treated with either PBS, Hep I (9 μg/ml, 200 nM), or Hep III (15 μg/ml, 200 nM) for 30 min at 37°C before injection. Microscopic analysis of hep-treated cells revealed no morphological differences from PBS-treated cells. In HSGAG fragment experiments, cells were resuspended in Hep I- or III-derived HSGAG fragment solution before injection. Mice were killed on day 14 and lungs were harvested. The number of nodules on the lung surface was counted with the assistance of a dissection microscope.
Saccharide Isolation and Structural Analysis of HSGAG Fragments.
To complete the structural analysis, HSGAG fragments were bound to an Ultrafree-DEAE membrane that had been equilibrated with pH 6.0 sodium phosphate and 0.15 M NaCl. The fragments were washed with the same buffer and eluted with 0.1 M sodium phosphate buffer pH 6.0 that contained 1.0 M NaCl. The fragments then were concentrated and buffer-exchanged into ultra-pure water by application to a Microcon filter (molecular weight cutoff = 3,000 Da). The sample was digested overnight with a mixture of Hep I-III (1 milliunit each) in 25 mM sodium acetate and 1 mM calcium acetate, pH 7.0. Analysis was completed by capillary electrophoresis using a high-sensitivity flow cell under reverse polarity with a running buffer of 50 mM Tris/phosphate, pH 2.5 (14). Disaccharide identification was made by comigration with known standards.
Immunohistochemistry.
Tumor tissues were fixed and embedded in paraffin according to standard histological procedures. Immunohistochemical staining of von Willebrand factor (vWF) and Ki-67 antigens as well as terminal deoxynucleotidyltransferase (TdT) labeling were performed according to manufacturer's instructions, with minor modifications. Capillary density was determined by counting the number of vWF-positive capillaries per high power field (×200). The proliferative and apoptotic indices of tumor cells within areas of viable tumor were estimated from the percentage of cells scored under a microscope at 400× magnification and are shown in Table 1 (15). A minimum of 2,000 cells was counted in each animal.
Table 1.
Immunohistochemical analysis of tumor samples
| Vessels/HPF | Apoptotic index | Proliferative index | |
|---|---|---|---|
| Treatment | |||
| PBS | 16.8 ± 0.81* | 2.5 ± 0.33 | 13.9 ± 0.45 |
| Heparinase III | 10.8 ± 0.77 | 6.4 ± 0.35 | 6.5 ± 0.35 |
| Heparinase I fragments | 23.7 ± 1.27 | 1.7 ± 0.25 | 23.3 ± 0.71 |
Indicates mean and SE.
Analysis of Signaling Intermediates in Tumor-Signaling Pathways.
Primary B16BL6 tumors were grown and treated as described; at day 15, the tumor was harvested in cold modified RIPA buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) containing enzyme inhibitors and homogenized. The homogenates were passed through a 25-gauge needle three times and centrifuged at 12,000 × g. The supernatant was adjusted for protein concentration and subjected to immunoprecipitation with 15 μl of either polyclonal anti-FGFR-1 or mouse anti-FAK monoclonal antibody overnight at 4°C. Immunoprecipitated samples were electrophoresed on a 7.5% polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with RC20. Erk-1, 2 and Akt protein in cell lysates were probed with antibodies specific to either phosphorylated or total protein.
Results and Discussion
Substrate Specificity of the Heps Dictates Whether They Promote or Inhibit Tumor Growth and Metastasis.
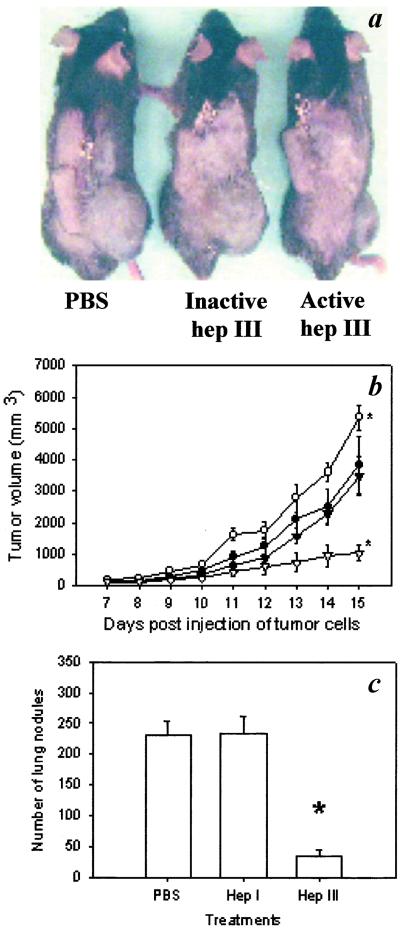
We used B16BL6 melanoma as a model system and treated tumor-bearing mice with either Hep I or III (9, 10) to investigate their effect on tumor progression (Fig. 1). Hep I treatment accelerated tumor growth in treated mice (Fig. 1b). This result is consistent with the current model of heparanase expression being associated with tumor progression (6, 7). Conversely, Hep III treatment significantly inhibited primary tumor growth (Fig. 1 a and b). The opposing effects on tumor growth observed by these two enzymes suggest that differences in their substrate specificity play a specific role in modulating tumor growth, and that in vivo HSGAGs are more than a passive barrier to tumor metastasis (6, 7, 16). Exploring further Hep III's anti-tumor activity, we completed more extensive studies to understand how the action of Hep III inhibits tumor growth, using Hep I treatment to provide a frame of reference for these studies.
Figure 1.
Effect of Hep treatment on tumor growth and metastasis. (a) Representative pictures of tumors in mice treated with PBS (Left), inactive Hep III (Center), and active Hep III (Right) 15 days after tumor implantation. No signs of pathological changes or inflammatory responses were found at nontumor bearing organs or tissues at the time of necropsy. (b) Tumor growth curves for mice treated with PBS (●), Hep I (○), inactive Hep III (▾), and active Hep III (▿). Tumor-bearing mice were treated with Hep III by both s.c. injection and osmotic pump delivery at a total dose of 12 mg/kg/day. The Hep I group received only osmotic pump delivery at 0.5 mg/kg/day. As stated in the text, the route of administration was found not to play a significant role in the enzyme activity. (c) B16BL6 lung metastases, 13 days after tail vein injection of melanoma cells (n = 8). *, P < 0.05 (Mann–Whitney test). Error bars represent SE.
At a dosage of 12 mg/kg per day of Hep III, about 75% inhibition of tumor growth was observed; the inhibition of tumor growth by Hep III was found to be dose-dependent, with initial inhibition observed at 2 mg/kg/day (about 30% inhibition). The route of administration was found not to play a significant role in the activity of Heps, as repeated s.c. or i.p. injections had similar effects as delivery by osmotic pump. Control mice treated with heat- or chemically inactivated Hep III exhibited comparable growth curves to those of mice treated with PBS (Fig. 1 a and b), indicating that the catalytic activity of Hep III alone was responsible for its ability to inhibit primary tumor growth. In addition, Hep III inhibited B16BL6 growth in nude mice at a level comparable to that seen in immunocompetent mice (see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org.), suggesting the inhibition was not immune cell-mediated. Furthermore, histological examination of enzyme-injection sites and internal organs revealed no inflammatory responses in any of the enzyme-treated mice. To ensure that these observations were not limited to the tumor model chosen, Hep III was used to treat mice bearing Lewis lung carcinoma tumors. Similar to what was observed in the B16BL6 tumor model, Hep III showed marked inhibition of tumor growth at a dose of 12 mg/kg per day (see Fig. 7, which is published as supporting information on the PNAS web site). The inhibitory effect of Hep III on B16 tumor growth also was observed in cell-proliferation assays in vitro, where treatment of B16 cells with Hep III at 200 nM resulted in about 30% inhibition of cell proliferation (see Fig. 8, which is published as supporting information on the PNAS web site). Thus, the in vivo studies, along with in vitro experiments, point to the enzymatic action of Hep III reducing the tumorigenicity of a variety of tumor cell types.
In addition, spontaneous tumor metastasis was examined at the end of primary tumor experiments. Specifically, mice were thoroughly examined for metastasis to internal organs and tissues away from the site of primary tumor at the time of necropsy. We found that in the PBS control group there was an average of 2 (0–4) metastatic foci identified in various organs and tissues such as lung, i.p. cavity, and mesenteric membrane. Conversely, no metastasis was found in mice treated with Hep III, whereas the extent of metastasis in mice treated with Hep I was comparable to that of the PBS control group. To test the influence of Hep treatment more directly, we studied tumor metastasis by using a lung colonization model.
Hep Treatment of Tumor Cells Modulates Lung Colonization.
To investigate the role of HSGAGs in tumor metastasis, B16BL6 cells were treated with either Hep I or III, injected into the tail vein of syngeneic mice, and assessed for their ability to colonize to the lungs. Whereas the potential of Hep I-treated cells to colonize the lung was comparable to that of PBS-treated cells, Hep III-treated B16BL6 cells were significantly less able to colonize to the lungs (Fig. 1c). Similarly, treatment of the HSGAG coat on the surface of Lewis lung carcinoma cells by Hep III inhibited their ability to metastasize and colonize the lungs (see Fig. 7). In vitro invasion assays showed similar results, where treatment of B16 cells with Hep III significantly inhibited, by 3-fold, the ability of cells to invade and migrate through a Matrigel membrane. Conversely, Hep I treatment showed the opposite effect; i.e., it increased over 2-fold the ability of cells to migrate through a Matrigel membrane (see Fig. 9, which is published as supporting information on the PNAS web site).
Whereas systemic delivery of Hep may be predicted to have wide-ranging effects beyond targeting the tumor compartment, it was found, through histological examination and dosing experiments completed in nude mice, that no immune-mediated or coagulation-mediated effects upon Hep treatment could be observed. Taken together with the in vivo and in vitro data on B16 melanoma, these results suggest two possible explanations for the opposing effects of Hep I and Hep III treatment on tumor cells. First, by digesting the HSGAG coat on the tumor cell surface, it is possible that Hep treatment directly impinges on the growth and metastatic ability of tumor cells. Alternatively, Hep treatment might indirectly influence tumor-cell behavior through the release of bioactive HSGAG fragments from the cell surface. To differentiate between these two possibilities, we tested whether the in vivo activity of Hep treatment could be recapitulated by directly injecting HSGAG fragments released from tumor cells upon either Hep I or III treatment.
Tumor-Cell HSGAG Fragments Modulate Tumor Growth and Lung Colonization.
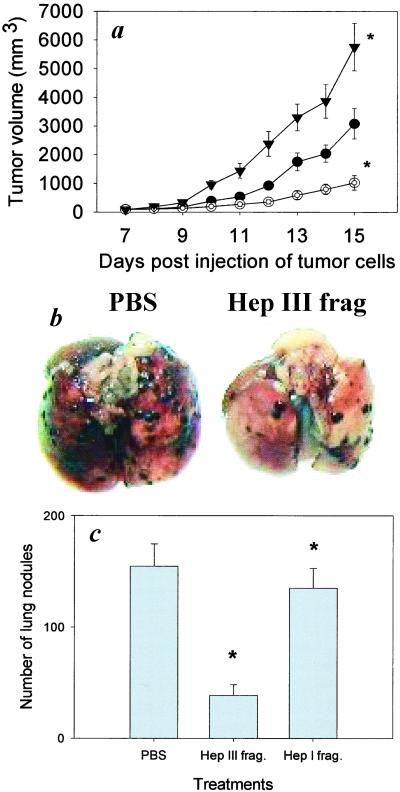
To investigate directly the role of HSGAG fragments in modulating tumor progression, B16 cells were treated in vitro with either Hep I or Hep III, and then the Hep I- and Hep III-generated B16 cell HSGAG fragments were isolated and tested in vivo. Treatment of mice with Hep I-generated HSGAG fragments promoted primary tumor growth (Fig. 2a), consistent with Hep I's enzymatic effect. On the other hand, Hep III-derived HSGAG fragments at 10 μg/kg/day showed significant inhibition of primary tumor growth by about 70% (Fig. 2a), which is comparable with Hep III's enzymatic effect on primary tumor growth. As with enzyme treatment, the inhibition of tumor growth by HSGAG fragments was found to be dose-dependent, with initial inhibition observed at 2 μg/kg/day. Notably, the pronounced biological potency of both Hep I and Hep III treatment could be recapitulated with injection of the fragments alone, suggesting that the effects of enzyme treatment are caused by the release of bioactive HSGAG fragments from the cell surface of B16BL6 cells. To ensure that HSGAG fragments present in the solution were the primary mediators of the observed effects, the HSGAG solution was first boiled and filtered before DEAE purification in an effort to inactivate proteins that are potentially released by Hep treatment. We expected that any proteins or other biological material attached to HSGAGs would be inactivated and removed by using this procedure. To confirm that the effect was HSGAG-specific, treatment of the purified HSGAG solution with either Pronase, DNase/RNase, Chondroitinase ABC, or a combination of these three enzymes did not affect the observed activities of either the Hep I- or Hep III-derived fragment solution.
Figure 2.
Effect of tumor-cell-derived HSGAG fragments on tumor growth and metastasis. (a) Tumor growth curves of mice treated with either Hep III- (○, 10 μg/kg/day) or Hep I (▾, 2 μg/kg/day)-derived B16BL6 saccharide fragments or a PBS control (●, n =5). (b) Representative lungs are shown from mice treated with either PBS or Hep III-derived fragments. (c) Measurement of lung colonization by B16BL6 cells subsequent to treatment with HSGAG fragments. B16BL6 cells were resuspended in either a PBS solution (Left) or a solution containing either Hep III (Center) or Hep I-derived B16BL6 saccharide fragments (Right) and then injected into the tail veins of mice (n = 7 or 8). No signs of morbidity such as weight loss are associated with any of the treatments. *, P < 0.05 (Mann–Whitney test). Error bars represent SE.
To extend the above findings, when B16BL6 cells were suspended or ‘doped’ in a PBS solution containing Hep III-generated B16 HSGAG fragments (2.0 μg/ml) prior to injection through the tail veins of mice, it was found that these cells were 75% less able to colonize the lung, suggesting that the HSGAG fragments were extremely potent inhibitors of tumor metastasis (Fig. 2c). Taken together, these results are consistent with the following model. Hep I treatment releases bioactive HSGAG fragments from the surface of the tumor cell that are able to promote primary tumor growth. Hep III, with an orthogonal substrate specificity, releases HSGAG fragments from the tumor-cell surface that inhibit both primary tumor growth and tumor metastasis.
HSGAG Fragments with Distinct Composition Are Potent Inhibitors of Tumor Growth and Metastasis.
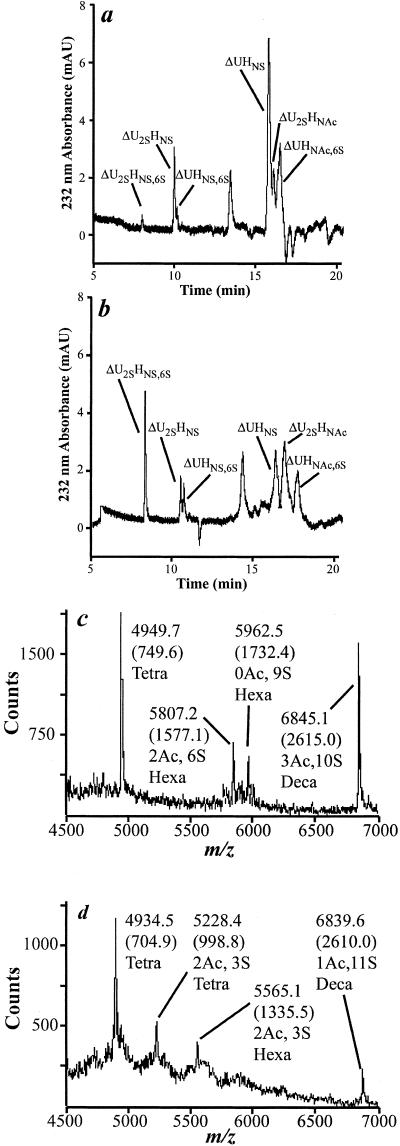
Compositional studies of the HSGAG saccharide fragments generated upon Hep treatment confirmed that the HSGAG fragments released from B16BL6 cells by Hep I or Hep III are compositionally different and structurally distinct (Fig. 3). Capillary electrophoresis, in combination with exhaustive enzymatic digest, was used to derive compositional information on the saccharide fragments (Fig. 3; ref. 14). The saccharide fragments derived from Hep III treatment had more of tri- and di- sulfated disaccharides, whereas the Hep I-treated HSGAGs had more mono- and un-sulfated disaccharides (Fig. 3). This result is consistent with the known substrate specificities of the Heps. Mass spectrometric investigation of Hep I- and III-derived HSGAGs using described techniques (14) yielded an oligosaccharide “fingerprint” proving that the saccharide fragments generated from the different treatments are structurally distinct (Fig. 3). Specifically, the Hep I-derived fragments yielded tetra-, hexa-, and deca-saccharide fragments with sulfation and acetylation patterns that are distinct from the Hep III-derived pool (Fig. 3).
Figure 3.
Structural analysis of HSGAG fragments released from the cell surface of B16BL6 cells. B16BL6 cells were treated with either Hep I or Hep III, and the harvested fragments were compositionally defined and structurally mapped. Representative capillary electrophoretograms of Hep I-generated and Hep III-generated fragments are shown in a and b, respectively. Compositional analysis was completed as described (14). The identity of peaks, as determined by comigration with authentic standards, is enumerated in the figures. (a and b) ▵U, ▵4,5 uronic acid; H, glucosamine; 2S, NS, and 6S refer to 2-O, 6-O, and N-sulfation, respectively; NAc refers to N-acetylation. (c and d) Mass spectrometric oligosaccharide mapping (14) of the fragments also was completed. As expected from their orthogonal substrate specificities, Hep I-derived fragments (c) possess a sulfation pattern distinct from that of Hep III-derived fragments (d).
HSGAG Fragments Target both the Tumor Cell and the Endothelial Cell Compartments.
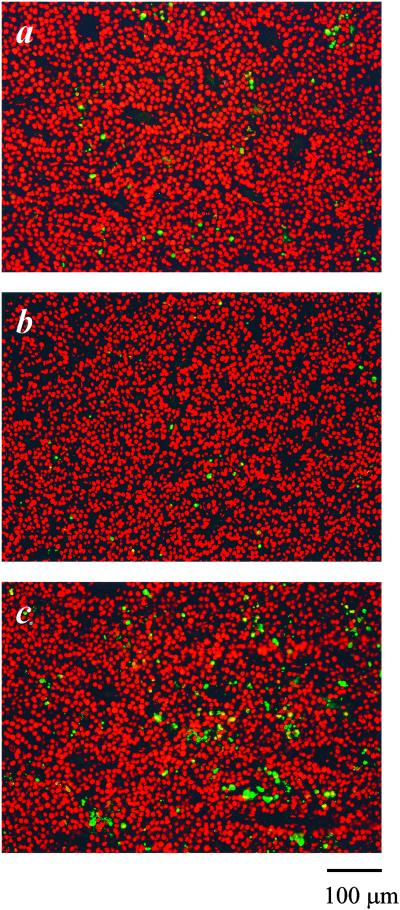
To understand how Hep I- or III-generated HSGAG fragments elicited their function, immunohistochemical studies were completed (Table 1; Fig. 4). For Hep III-treated tumors, immunohistological examination (17, 18) of tumor samples revealed reduced neovascularization (19), increased cellular apoptosis, and decreased cellular proliferation compared with that of PBS-treated animals (Table 1; Fig. 4). Similar results were observed upon immunohistological examination of animals treated with Hep III-generated HSGAG fragments, consistent with the notion that bioactive HSGAG fragments are indeed the mediators of tumor growth and neovascularization (data not shown). Immunohistological examination of animals treated with Hep I-generated HSGAG fragments revealed the opposite, namely increased proliferation, decreased apoptosis, and increased neovascularization (Table 1; Fig. 4). Surprisingly, it seems as if the release of bioactive HSGAG fragments through Hep treatment affects tumor physiology both by modulating the survival and proliferation of the tumor-cell compartment as well as by influencing the ability of the endothelial-cell compartment to form new blood vessels.
Figure 4.
TdT labeling of tumor samples derived from mice treated with PBS (a), Hep I (b), and Hep III (c). Apoptotic cells shown in green were detected by labeling fragmented DNA with Fluorescein-12-dUTP. Propidium iodide was used for background staining of live cells. The apoptotic index of tumor cells within areas of viable tumor was estimated from the percentage of cells scored under a microscope at 400× magnification and are shown in Table 1. A minimum of 2,000 cells were counted in each animal. (Bar = 100 μm.)
Mechanism of Action: HSGAGs Impinge on the Biological Activity of Specific Signaling Molecules.
Having observed the marked and opposite effects that distinct HSGAG fragments have on both the tumor and vascular compartments, we sought to elucidate the underlying molecular mechanism of HSGAGs in tumor progression. Because many HSGAG-binding proteins are growth factors and cytokines, we explored HSGAG-binding growth factors that play key roles in tumor pathobiology to identify an immediate target of the HSGAG fragments generated from the surface of tumor cells. FGF2 signaling has been shown to be a prerequisite for melanoma progression by promoting tumor growth in an autocrine fashion, and the interruption of FGF2 autocrine loops by interfering with either FGF2 or FGFR activity results in inhibition of melanoma progression (20–23). Conversely, up-regulation of the expression of FGF2 in normal melanocytes results in their malignant transformation (24). Furthermore, FGF2 is a potent and essential angiogenic factor regulating melanoma neovascularization (25, 26). Most importantly, specific HSGAG structures are known to bind and modulate FGF2 activity, and there is increasing evidence that HSGAG sequences, depending on their structure, can either promote or inhibit FGF2 activity (27–29). Given the multiple lines of evidence implicating FGF2 as a key switch in melanoma progression and taken together with FGF's strict dependence on HSGAGs for its activity (30, 31), we sought to determine whether the immediate target of tumor-derived HSGAG fragments is indeed FGF2.
To test whether Hep I- and Hep III-derived fragments bind to FGF2 and affect its activity, we first established that Hep III treatment inhibits FGF-induced proliferation of B16BL6 cells in vitro; additionally, we directly confirmed this finding by examining FGF-mediated downstream signaling pathways, namely, the MAP kinase pathway (i.e., Erk-1, 2), the principle signal-transduction pathway of FGF2 leading to cell proliferation and differentiation (32). Hep III treatment of cells prevented activation of Erk-1, 2 by FGF2 stimulation, whereas Hep I treatment resulted in the opposite effect (see Fig. 10, which is published as supporting information on the PNAS web site). The in vitro data were confirmed further by using F32 cells, a prelymphocyte cell line that has been transfected with FGFR and that often has been used as a model system to study FGF-mediated signaling in cell culture unfettered by complications associated with signaling events initiated by other growth factors and/or receptors (33). Similar to what was observed in B16BL6 cells, Hep I-derived fragments promote, and Hep III-derived fragments inhibit, FGF2-mediated cellular proliferation in these cells (see Fig. 10). Together, the in vitro findings point to the fact that HSGAG fragments derived from the cell surface can substantially and specifically affect FGF2 signaling.
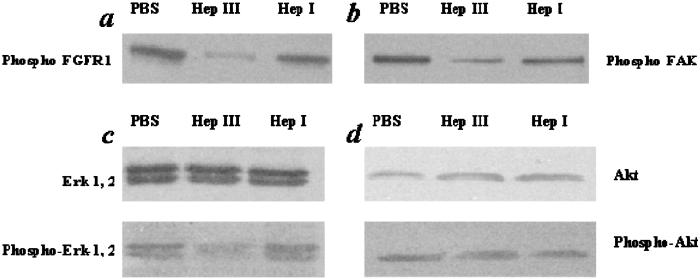
Consistent with the in vitro observations, we find that Hep I- and Hep III-derived B16BL6 fragments significantly affect FGF-signaling pathways in vivo. Within the tumor in the animals, we examined both FGFR phosphorylation in Hep I- and Hep III-treated animals as well as Erk-1 and 2 signaling. Treatment of the primary tumor with Hep III (or its generated fragments) inhibited phosphorylation of FGFR1, whereas Hep I treatment had a minimal effect on the phosphorylation of FGFR1 (Fig. 5a). In addition, treatment of the primary tumor with Hep III resulted in a lower level of activated Erk-1, 2 (Fig. 5c). Additional intracellular signaling events such as focal adhesion kinase (FAK) activity, which is implicated in cell adhesion and migration processes (34, 35), was similarly modulated by Hep I and III treatment of the tumor. Consistent with these findings, Hep III treatment inhibited FAK activation (Fig. 5b). Notably, there was no change in activation of Akt with either Hep I or Hep III treatment (Fig. 5d), indicating that the changes in phosphorylation were specific and resulted from down-regulation of only certain signaling pathways. Together, these results suggest that HSGAG fragments mediate FGF2 signaling; Hep I-generated HSGAG fragments promote FGF2 activity, whereas Hep III-generated HSGAG fragments inhibit it. This effect was observed in key steps of FGF-mediated signaling, from the cell-surface receptor (FGFR) through downstream signaling events.
Figure 5.
Activation of FGFR-1 (a), FAK (b), Erk-1, 2 (c), and Akt (d) in primary B16BL6 melanoma samples treated with Hep. Western blot analysis was completed on isolated in vivo tumor samples to examine the effect of heparanase treatment on intracellular-signaling pathways.
The results presented herein demonstrate that by impinging on the biological activity of specific signaling molecules, HSGAGs play a direct role in tumor growth and metastasis. Most importantly, HSGAGs at the cell surface of tumor cells contain both ‘activatory’ and ‘inhibitory’ HSGAG sequences that are in balance. The specific degradation of one set of sequences (e.g., by Hep I) results in the release of fragments that promote the biological activity of HSGAG-binding signaling molecules and thus act as a switch for tumor growth and metastasis. Conversely, degradation by an enzyme with an orthogonal substrate specificity (e.g., Hep III) tips the balance in the opposite direction, releasing fragments that antagonize HSGAG-binding signaling molecules, leading to the inhibition of tumor growth and metastasis. Thus, we have demonstrated here that chemically complex HSGAGs at the cell surface are “cryptic” promoters or inhibitors of tumor growth and metastasis that become biologically active upon their release from the cell surface by specific HSGAG-degrading enzymes.
Just as collagenases clip the proteinaceous compartment of the ECM, serving either to increase tumor growth (e.g., breakdown of the basement membrane; refs. 36 and 37) or to inhibit tumors (e.g., the formation of endostatin from collagen XVIII; refs. 3 and 4), the polysaccharide compartment exhibits a similar phenomenon. Importantly, like the proteolytically cleaved collagen fragment endostatin, distinct HSGAG oligosaccharides, upon release by enzymatic cleavage from the tumor-cell surface, can serve as potent inhibitors of tumor progression. With the continuing discovery of mammalian HSGAG-degrading enzymes (6, 7, 38), our findings have direct implications for human tumor biology. Thus, the present study not only allows a paradigm of how the polysaccharides modulate tumor growth and metastasis, but it also identifies a therapeutic target by providing a framework toward the development of HSGAG-based anti-cancer molecules.
Our findings have implications beyond tumor growth and metastasis, in that we directly demonstrate a level of fine control of cell kinetics. By changing the composition or ‘signature’ of the polysaccharide coat at their surface, cells provide a systemic mechanism for fine control of the activity of a repertoire of signaling molecules. Together, these studies demonstrate that the display of complex polysaccharides at the cell surface modulate protein function and impinge on cellular phenotype.
Supplementary Material
Acknowledgments
We thank Ms. Kris Holley for help in the production of recombinant Heps and with the immunohistochemistry. We thank Dr. Yiwei Qi for help with animal studies and the Division of Comparative Medicine, Massachusetts Institute of Technology for help with the animal facilities and histological studies. We thank Drs. Bevin Engelward, John Essigmann, and V. Sasisekharan for critical reading of the manuscript. This investigation was supported in part by funds from the Burroughs Wellcome Foundation, the Arnold and Mabel Beckman Foundation, National Institutes of Health Grant GM57073 and HL 59966, the CaPCURE Foundation, and the Whitaker Health Sciences Fund Fellowship (Massachusetts Institute of Technology).
Abbreviations
- ECM
extracellular matrix
- HSGAG
heparan sulfate-like glycosaminoglycans
- Hep
heparinase
- FGF
fibroblast growth factor
- FGFR
FGF receptor
Footnotes
See commentary on page 543.
References
- 1.Sasisekharan R, Venkataraman G. Curr Opin Chem Biol. 2000;4:626–631. doi: 10.1016/s1367-5931(00)00145-9. [DOI] [PubMed] [Google Scholar]
- 2.Esko J D, Lindahl U. J Clin Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehm T, Folkman J, Browder T, O'Reilly M S. Nature (London) 1997;390:404–407. doi: 10.1038/37126. [DOI] [PubMed] [Google Scholar]
- 4.O'Reilly M S, Boehm T, Shing Y, Fukai N, Vasios G, Lane W S, Flynn E, Birkhead J R, Olsen B R, Folkman J. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 5.Sanderson R D. Semin Cell Dev Biol. 2001;12:89–98. doi: 10.1006/scdb.2000.0241. [DOI] [PubMed] [Google Scholar]
- 6.Hulett M D, Freeman C, Hamdorf B J, Baker R T, Harris M J, Parish C R. Nat Med. 1999;5:803–809. doi: 10.1038/10525. [DOI] [PubMed] [Google Scholar]
- 7.Vlodavsky I, Friedmann Y, Elkin M, Aingorn H, Atzmon R, Ishai-Michaeli R, Bitan M, Pappo O, Peretz T, Michal I, et al. Nat Med. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- 8.Borsig L, Wong R, Feramisco J, Nadeau D R, Varki N M, Varki A. Proc Natl Acad Sci USA. 2001;98:3352–3357. doi: 10.1073/pnas.061615598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst S, Venkataraman G, Winkler S, Godavarti R, Langer R, Cooney C L, Sasisekharan R. Biochem J. 1996;315:589–597. doi: 10.1042/bj3150589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pojasek K, Shriver Z, Hu Y, Sasisekharan R. Biochemistry. 2000;39:4012–4019. doi: 10.1021/bi992514k. [DOI] [PubMed] [Google Scholar]
- 11.Ernst S, Langer R, Cooney C L, Sasisekharan R. Crit Rev Biochem Mol Biol. 1995;30:387–444. doi: 10.3109/10409239509083490. [DOI] [PubMed] [Google Scholar]
- 12.Binari R C, Staveley B E, Johnson W A, Godavarti R, Sasisekharan R, Manoukian A S. Development (Cambridge, UK) 1997;124:2623–2632. doi: 10.1242/dev.124.13.2623. [DOI] [PubMed] [Google Scholar]
- 13.Sasisekharan R, Moses M A, Nugent M A, Cooney C L, Langer R. Proc Natl Acad Sci USA. 1994;91:1524–1528. doi: 10.1073/pnas.91.4.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkataraman G, Shriver Z, Raman R, Sasisekharan R. Science. 1999;286:537–542. doi: 10.1126/science.286.5439.537. [DOI] [PubMed] [Google Scholar]
- 15.O'Reilly M S, Holmgren L, Chen C, Folkman J. Nat Med. 1996;2:689–692. doi: 10.1038/nm0696-689. [DOI] [PubMed] [Google Scholar]
- 16.Eccles S A. Nat Med. 1999;5:735–736. doi: 10.1038/10455. [DOI] [PubMed] [Google Scholar]
- 17.Parangi S, O'Reilly M, Christofori G, Holmgren L, Grosfeld J, Folkman J, Hanahan D. Proc Natl Acad Sci USA. 1996;93:2002–2007. doi: 10.1073/pnas.93.5.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Reilly M S, Holmgren L, Shing Y, Chen C, Rosenthal R A, Moses M, Lane W S, Cao Y, Sage E H, Folkman J. Cell. 1994;79:315–328. doi: 10.1016/0092-8674(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 19.Weidner N, Semple J P, Welch W R, Folkman J. N Engl J Med. 1991;324:1–8. doi: 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
- 20.Becker D, Meier C B, Herlyn M. EMBO J. 1989;8:3685–3691. doi: 10.1002/j.1460-2075.1989.tb08543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker D, Lee P L, Rodeck U, Herlyn M. Oncogene. 1992;7:2303–2313. [PubMed] [Google Scholar]
- 22.Rodeck U, Becker D, Herlyn M. Cancer Cells. 1991;3:308–311. [PubMed] [Google Scholar]
- 23.Torcia M, Lucibello M, De Chiara G, Labardi D, Nencioni L, Bonini P, Garaci E, Cozzolino F. Biochem Biophys Res Commun. 1999;262:838–844. doi: 10.1006/bbrc.1999.1292. [DOI] [PubMed] [Google Scholar]
- 24.Nesbit M, Nesbit H K, Bennett J, Andl T, Hsu M Y, Dejesus E, McBrian M, Gupta A R, Eck S L, Herlyn M. Oncogene. 1999;18:6469–6476. doi: 10.1038/sj.onc.1203066. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Becker D. Nat Med. 1997;3:887–893. doi: 10.1038/nm0897-887. [DOI] [PubMed] [Google Scholar]
- 26.Birck A, Kirkin A F, Zeuthen J, Hou-Jensen K. Melanoma Res. 1999;9:375–381. doi: 10.1097/00008390-199908000-00006. [DOI] [PubMed] [Google Scholar]
- 27.Guimond S E, Turnbull J E. Curr Biol. 1999;9:1343–1346. doi: 10.1016/s0960-9822(00)80060-3. [DOI] [PubMed] [Google Scholar]
- 28.Nurcombe V, Smart C E, Chipperfield H, Cool S M, Boilly B, Hondermarck H. J Biol Chem. 2000;275:30009–30018. doi: 10.1074/jbc.M003038200. [DOI] [PubMed] [Google Scholar]
- 29.Pye D A, Vives R R, Hyde P, Gallagher J T. Glycobiology. 2000;10:1183–1192. doi: 10.1093/glycob/10.11.1183. [DOI] [PubMed] [Google Scholar]
- 30.Lin X, Buff E M, Perrimon N, Michelson A M. Development (Cambridge, UK) 1999;126:3715–3723. doi: 10.1242/dev.126.17.3715. [DOI] [PubMed] [Google Scholar]
- 31.Iozzo R V, San Antonio J D. J Clin Invest. 2001;108:349–355. doi: 10.1172/JCI13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seger R, Krebs E G. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- 33.Ornitz D M, Xu J, Colvin J S, McEwen D G, MacArthur C A, Coulier F, Gao G, Goldfarb M. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez-Fernandez J L. BioEssays. 1999;21:1069–1075. doi: 10.1002/(SICI)1521-1878(199912)22:1<1069::AID-BIES13>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 35.Schlaepfer D D, Hauck C R, Sieg D J. Prog Biophys Mol Biol. 1999;71:435–478. doi: 10.1016/s0079-6107(98)00052-2. [DOI] [PubMed] [Google Scholar]
- 36.MacDougall J R, Matrisian L M. Cancer Metastasis Rev. 1995;14:351–362. doi: 10.1007/BF00690603. [DOI] [PubMed] [Google Scholar]
- 37.Wilson C L, Heppner K J, Labosky P A, Hogan B L, Matrisian L M. Proc Natl Acad Sci USA. 1997;94:1402–1407. doi: 10.1073/pnas.94.4.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenzie E, Tyson K, Stamps A, Smith P, Turner P, Barry R, Hircock M, Patel S, Barry E, Stubberfield C, et al. Biochem Biophys Res Commun. 2000;276:1170–1177. doi: 10.1006/bbrc.2000.3586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.