Abstract
BACKGROUND & AIMS:
A genome-wide significant association between anti–Helicobacter pylori (H pylori) IgG titers and Toll-like receptor (TLR1/6/10) locus on 4p14 was demonstrated for individuals of European ancestry, but not uniformly replicated. We re-investigated this association in an updated genome-wide association study (GWAS) meta-analysis for populations with low gastric cancer incidence, address potential causes of cohort heterogeneity, and explore functional implications of genetic variation at the TLR1/6/10 locus.
METHODS:
The dichotomous GWAS (25% individuals exhibiting highest anti–H pylori IgG titers vs remaining 75%) included discovery and replication sampls of, respectively, n = 15,685 and n = 9676, all of European ancestry. Longitudinal analysis of serologic data was performed on H pylori–eradicated subjects (n = 132) and patients under surveillance for premalignant gastric lesions (n = 107). TLR1/6/10 surface expression, TLR1 mRNA, and cytokine levels were measured in leukocyte subsets of healthy subjects (n = 26) genotyped for TLR1/6/10 variants.
RESULTS:
The association of the TLR1/6/10 locus with anti–H pylori IgG titers (rs12233670; b = −0.267 ± SE 0.034; P = 4.42 × 10−15) presented with high heterogeneity and failed replication. Anti–H pylori IgG titers declined within 2–4 years after eradication treatment (P = 0.004), and decreased over time in patients with premalignant gastric lesions (P < 0.001). Variation at the TLR1/6/10 locus affected TLR1-mediated cytokine production and TLR1 surface expression on monocytes (P = 0.016) and neutrophils (P = 0.030), but not mRNA levels.
CONCLUSIONS:
The association between anti–H pylori IgG titers and TLR1/6/10 locus was not replicated across cohorts, possibly owing to dependency of anti–H pylori IgG titers on therapy, clearance, and antibody decay. H pylori–mediated immune cell activation is partly mediated via TLR1 signaling, which in turn is affected by genetic variation.
Keywords: Single-Nucleotide Polymorphism, Serology, Immunity, Bacteria
Graphical Abstract
The discovery of Helicobacter pylori (H pylori) at the epithelial surface of the human stomach as late as 1983 represented a major breakthrough in gastric microbiology.1 This flagellated bacterium has since been implicated in the etiology of atrophic gastritis and gastroduodenal ulcerative disease,2 identified as a class 1 carcinogen for gastric cancer,3–5 and ranked as the most important contributor to infection-attributable cancers in 2018.6 With estimates indicating that more than half of the world’s population is colonized by H pylori, the size of this global health problem is further emphasized.7 Because H pylori gastric presence has been linked to early stages of gastric carcinogenesis according to the Correa model,8 eradication strategies have been implemented to prevent gastric cancer development.9–12 However, global resistance of H pylori to antibiotics is reaching alarming levels,13 which puts further pressure on the H pylori–related health burden and warrants new strategies to prevent colonization and infection-related consequences. It is generally accepted that H pylori infection is acquired during early childhood,14–17 but the overall rate of infection is reported to be much higher in developing countries.18 Although socioeconomic and environmental factors likely explain the wide variation in H pylori prevalence between regions and countries,7 genetic predisposition also needs to be considered. It has been shown that the same rearing environment contributes to a familial tendency to acquire H pylori infection, but higher similarity in monozygotic than dizygotic twin pairs indicates that genetic factors account for a large part of the variation.19 Some individuals are never infected by H pylori, and others are able to clear the infection spontaneously when colonized.14,16 Moreover, only a small proportion of the H pylori–colonized population develop gastric cancer,20 indicating that host-specific factors governing the host-pathogen interactions are involved in disease risk.21 Because the host genetic background is suggested to be involved in the clinical outcome of H pylori infection, 22 a better understanding of the genetic contributions to the interaction between host and H pylori may improve our insight into this complex relationship.
An increasing number of genome-wide association studies (GWASs) have linked single-nucleotide polymorphisms (SNPs) to gastroduodenal ulcer disease,23,24 gastric premalignant lesions,25–29 and gastric cancer.24,27–39 Interestingly, some of the associations found in those studies seem to be influenced by the presence of H pylori infection,25,29–33 suggesting that genomic variants might be involved in H pylori colonization as well. The first and largest GWAS on H pylori combined data of Dutch and German population-based cohorts in a meta-analysis of anti–H pylori IgG titers by means of a dichotomic study design that compared the 25% of individuals exhibiting the highest anti–H pylori IgG titers vs the remaining 75%.40 Two loci, the Toll-like receptor (TLR1/6/10) locus on 4p14 (lead SNP rs10004195) and the Fc gamma receptor 2A (FCGR2A) locus on 1q23.3 (lead SNP rs368433), were identified to be associated with increased anti–H pylori IgG titers.40 A GWAS among Finnish male smokers (n = 1402) confirmed the lead SNP rs10004195 to be associated with the height of IgG titer rather than a seropositive status itself.41 However, no further replication of these findings have been reported so far, and in contrast to those European studies, no genome-wide significant associations of H pylori serology with any loci were found in a Mexican-American population (n = 1931).42 Because the main findings of the first GWAS study have not been uniformly confirmed,40 the present study aimed to update the original meta-analysis with the use of a larger sample size and to investigate the functional relevance of variation at the TLR1/6/10 locus in H pylori colonization.
Materials and Methods
Study Cohorts
The discovery GWAS was conducted in subjects of European ancestry from population-based cohorts in Europe and the United States with low gastric cancer incidence to re-investigate the previous association between TLR1/6/10 and anti–H pylori IgG levels, and to explore the possibility of new genetic associations. A total of 7 cohorts were included and consisted of 15,685 participants (Supplementary Table S1). The replication was conducted in 2 independent European cohorts with a total of 9676 participants with GWAS or de novo genotyping data. In all cohorts, serologic measurements of anti–H pylori IgG were performed by means of either commercial or customized enzyme-linked immunosorbent assay (ELISA) (Supplementary Table S1). As in the initial study, the 25% of subjects with the highest anti–H pylori IgG values were compared with the remaining 75% in a dichotomous study design.40 Informed consents for participation were obtained for all study subjects, and approvals were given by the institutional review boards. More details concerning individual cohorts are described in the Supplementary Methods.
Discovery
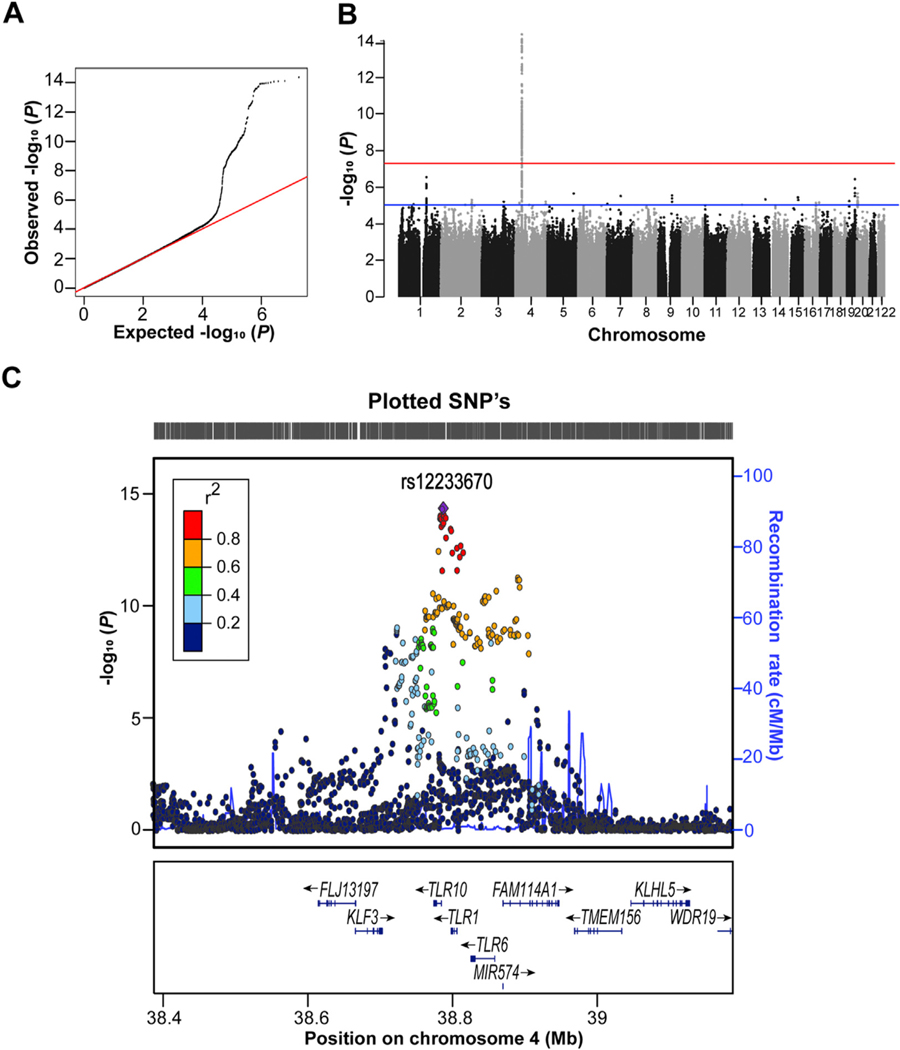
Genome-wide genotyping, imputation to 1kgP1v3, and genome-wide association analyses were conducted separately by the discovery cohorts (Supplementary Methods). EasyQC using standard settings was applied for quality control of individual cohort summary statistics.43 The inverse-variance weighted fixed-effects model approach was used for meta-analysis with the use of METAL.44 A quantile-quantile plot of observed compared with expected —log10 (P value) was computed to investigate genome-wide distribution of P values, and a Manhattan plot to illustrate genome-wide P values. Genome-wide significance was set at a threshold with P value <5.0 × 10−8. A regional plot was generated to show the genomic regions within 100 kb of top hits. In addition, a random-effects model was conducted to explore the association between the TRL locus (rs12233670) with H pylori in more detail.
Replication
Eight top SNPs with the lowest association P values from the discovery phase were selected for replication, particularly rs12233670 within the TLR1/6/10 locus. The ESTHER (Epidemiological Investigations on Chances of Preventing Recognizing Early and Optimally Treating Chronic Diseases in an Elderly Population) cohort achieved in silico replication of 7 out of 8 SNPs (excluding rs147174426), and the Latvian cohort performed de novo genotyping for 4 individual SNPs (rs12233670, rs147174426, rs6107461, rs147900026) (Supplementary Methods). Replication was considered successful with P value <0.05 for individual cohorts and P value <5.0 × 10−8 for the combined analysis.
Longitudinal Analysis of Serologic Data From H pylori–Positive Subsets
To determine whether the timing of anti–H pylori IgG testing may influence serologic outcomes relevant for genetic association studies, 2 different serologic data subsets were analyzed. The first subset consisted of RS (Rotterdam Study) participants with pharmacy records of H pylori eradication treatment before serology (n = 132), allowing analysis of anti–H pylori IgG titers in relation to time following eradication. Anti–H pylori antibodies were measured with the use of the Pyloriset EIA-G III (Orion Diagnostica, Espoo, Finland). The second subset consisted of patients from an ongoing prospective study aimed at the surveillance of atrophic gastritis, intestinal metaplasia, and dysplasia in the Netherlands and Norway.45 Anti–H pylori antibodies were measured as part of the GastroPanel test (Biohit, Helsinki, Finland) using serum samples collected during clinical follow-up. Patients with elevated anti–H pylori IgG levels (>30 enzyme immunoassay units) at baseline in addition to consecutive serum measurements (n = 107) were included to explore fluctuation of the titers over time. All subjects with positive histopathologic/urea breath test findings for H pylori received eradication therapy with efficacy verified by means of fecal antigen testing.
Restriction Fragment Length Polymorphism Polymerase Chain Reaction Assay
Human genomic DNA was isolated from EDTA whole blood with the use of the Kleargene Blood DNA isolation kit (LGC, Teddington, UK) to determine the genotype of subjects included in our functional assays. A restriction fragment length polymorphism polymerase chain reaction (PCR) assay could not be designed for rs10004195, but TLR variant rs28393318 is in complete linkage disequilibrium (LD) (r2 = 1 among Utah residents from north and west Europe [CEUs]) and was therefore used as proxy (Supplementary Table S2). For genotyping of rs28393318, 35 cycles of PCR amplification were performed with custom-designed primers (forward: 5′-TAGCTCAGTGTAGGTGGTCT-3′; reverse: 5′-ATGATTAGT-GACCTTGGGGC-3′) at an annealing temperature of 53◦C. PCR products were confirmed on 2% Tris-borate-EDTA agarose gel and 10 μL of amplicons were subjected to 5 international units of Hin1II restriction enzyme (Thermo Fisher, Waltham, MA) for 2.5 hours at 37◦C. After 20 minutes of enzyme inactivation at 80◦C, digestion products were visualized on agarose gel, showing 1 band of 433 base pairs (bp) for genotype GG, 2 bands of 311 and 122 bp for AA, and 3 bands for GA.
Functional Analysis
Flow cytometry.
To investigate whether genetic variation at the TLR locus on 4p14 affects the expression of the receptor at the cell surface, the presence of TLR1, TLR6, and TLR10 was measured with the use of flow cytometry. Whole-blood samples from non–H pylori–infected individuals without significant comorbidities (n = 26), taken after informed consent, were treated with eBioscience 1-step Fix/Lyse Solution (Thermo Fisher) to lyse red blood cells. Monocytes and polymorphonuclear cells (PMNs) were incubated with antibodies specific for CD14 (APC-cy7, cat. no. A15453), CD66B (APC, cat. no. 17–0666-42), and TLR1 (PE, cat. no. 12–9911-42) (all from Thermo Fisher) or mouse IgG1κ isotype control (PE, cat. no. 554121; BD Biosciences, Franklin Lakes, NJ) for 15 minutes on ice. Because the genes encoding TLR6 (PE, cat. no. MA5–16177) and TLR10 (PE, cat. no. 12–2909-42) reside within the same genetic locus as rs28393318, the surface expression of those proteins was also measured. Flow cytometry was performed on the MACSQuant Flow Cytometer (Miltenyi Biotec, Gladbach, Germany) and analysis conducted with the use of FlowJo v10 (BD Biosciences). Monocytes and PMNs were identified on the basis of the forward/sideward scatter and further gating on CD14 and CD66b, respectively. TLR positivity was measured with gating based on the isotype control of the same sample.
Reverse-transcription quantitative PCR analysis of TLR1 transcript levels.
To explore whether differences in TLR1 surface expression among genotypes were attributable to variation in mRNA expression, quantitative PCR (qPCR) analysis was conducted. Total RNA was isolated from peripheral blood mononuclear cells (PBMCs) of non–H pylori–infected individuals without significant comorbidities (n = 22) by means of the column-based NucleoSpin RNA kit (Macherey-Nagel & Co, Düren, Germany) and reverse transcribed into complementary DNA (cDNA) with the use of PrimeScript RT (Takara, Kusastsu, Shiga, Japan). A qPCR assay of 40 cycles was performed on the StepOnePlus Real-Time PCR system (Thermo Fisher) using SYBR Select Master Mix (Thermo Fisher) and custom-designed TLR1 gene primers (forward: 5′-TGCCAAATGGAACAGACAAGCAG-3′; reverse: 5′-ACA-GATTCCTTTTGTAGGGGTGCC-3′) and RP2 housekeeping gene primers (forward: 5′-AAGCTGAGGATGCTCAAAGG-3′; reverse: 5′-CCCATTAAACTCCAAGGCAA-3′). The annealing temperature was 61◦C for both primer sets. The delta-delta cycle threshold (ΔΔCt) method was applied for data analysis.
ELISA for cytokine analysis on TLR1 stimulation.
To study TLR1 signaling in more detail, PBMCs from non–H pylori–infected individuals (n = 22) were isolated from heparinized blood as described previously.46 In brief, phosphate-buffered saline solution (PBS)–diluted blood was layered onto Ficoll (Amersham, Uppsala, Sweden) and PBMCs harvested after centrifugation, washed in PBS, and plated in Roswell Park Memorial Institute medium (Lonza, Basel, Switzerland) containing 10% fetal bovine serum and penicillin/streptomycin (Lonza). Two million PBMCs were seeded in 12-well plates in a total volume of 2 mL. After 24 hours of incubation, wells were washed and PBMCs stimulated with 1 million colony-forming units of heat-killed H pylori (strain ATCC-43504 [cagA+, vacA(s1/m1), iceA+, babA2+]; Manassas, VA) grown on Trypticase Soy Agar (Oxoid, Hampshire, UK) supplemented with 5% defibrinated sheep blood (VWR, Radnor, PA) and DENT selective medium (Oxoid). Other stimuli used were TLR1 inhibitor CU-CPT-22 (5 μmol/L; Tocris Bioscience, Bristol, UK) and TLR1 agonist Pam3Cys4 (300 ng/mL; InvivoGen, San Diego, CA).47 Supernates were harvested after 8 hours of stimulation for ELISA experiments unless otherwise specified to measure tumor necrosis factor (TNF) α, interleukin-8 (IL8), and IL10 (eBioscience, San Diego, CA) as described previously.48 All samples were tested in duplicate.
Statistical Analysis of Serologic and Functional Data
Statistical differences among 3 groups were determined by means of 1-way analysis of variance or Kruskal-Wallis tests for unpaired data and repeated-measures analysis of variance or Friedman tests for paired data and was followed by post hoc analysis for selected pairs with adjustment for multiple testing. The 2-sample t test or Mann-Whitney test were applied to compare 2 groups with unpaired data. GraphPad Prism software version 5.01 (GraphPad Software, San Diego, CA) was used for calculations and graphic representation.
Results
Genomic Variants Associated With Anti–H pylori IgG Titers in an Updated GWAS
We performed a GWAS meta-analysis based on 7 independent European epidemiologic cohorts with the use of the fixed-effect model. The quantile-quantile plot showed a clear deviation from the null-distribution at the tail (Figure 1A). A genome-wide significant association for the TLR1/6/10 locus on chromosome 4p14 was found with top SNP rs12233670 carrying the lowest P value (β = −0.267 ± SE 0.034 for minor allele T; P = 4.42 × 10−15; minor allele frequency = 25%) (Figure 1B and C), albeit with statistical heterogeneity (Table 1). The association between top SNP rs12233670 and anti–H pylori IgG titers was not significant in either ESTHER (β = 0.041 ± SE 0.050 for the minor allele; P = 0.41) or LATVIA (β = 0.017 ± SE 0.079 for the minor allele; P = 0.83) cohorts, resulting in a failed replication (β = 0.034 ± SE 0.042 for minor allele; P = 0.42) (Table 1). Consequently, the level of genome-wide significance decreased in the combined analysis (β = −0.149 ± SE 0.027 for the minor allele; P = 1.97 × 10−8) (Table 1). Seven other promising SNPs were identified but did not reach genome-wide significance, including the FCGR locus (1q23.3; top-ranked SNP rs147174426; β = 0.480 ± SE 0.094 for major allele A; P = 2.89 × 10−7; minor allele frequency = 7%). Similar results were obtained with the use of a sensitivity model including adult participants only (data not shown). None of these selected SNPs reached genome-wide significance in the combined analysis with discovery and replication cohorts (Supplementary Table S3).
Figure 1.
Results of genome-wide association meta-analysis for anti–Helicobacter pylori IgG titer. (A) Quantile-quantile plot with all single-nucleotide polymorphisms (SNPs) displayed as black dots and the red line corresponding to the null hypothesis of no true association. (B) Manhattan plot of the genome wide association meta-analysis with the chromosome position on the x-axis and —log10 P values on the y-axis. Each dot indicates an SNP, the blue line marks the threshold of P = 1.0 × 10−5, and the red line represents the genome-wide significant threshold of P = 5.0 × 10−8. (C) Regional plot showing locus 4p14 with rs12233670 and other flanking region markers. The P values of SNPs associated with H pylori are plotted against their chromosome position on the x-axis. Each SNP is represented by a colored dot indicating their correlation (linkage disequilibrium) with the top-ranked SNP (purple diamond). The y-axis at the right represents the recombination rates. The bottom part depicts the annotated genes at the locus and their transcriptional direction.
Table 1.
Summary of Single-Nucleotide Polymorphism (SNP) at the TLR1/6/10 Locus in Discovery, Replication, and Combined Meta-analysis
| ID | Chr | Position | Genea | A1/2 b | EAF | Betac | SE | P value | I 2 |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Discovery | |||||||||
| rs12233670 | 4 | 38787216 | TLR10 | T/C | 0.25 | −0.267 | 0.034 | 4.42 × 10−15 | 79.8 |
| Replication | |||||||||
| 0.20 | 0.034 | 0.042 | 0.417 | 0.0 | |||||
| Combined | |||||||||
| Betac | SE | Dird | P value | I 2 | |||||
| −0.149 | 0.027 | −++ | 1.97 × 10−8 | 30.9 | |||||
Chr, chromosome; EAF, effect allele frequency; I2, measure of heterogeneity.
Gene nearest to the SNP.
A1/2, effect allele 1 and other allele 2.
Effect size is relative to the allele 1.
Direction of beta of, respectively, the discovery, ESTHER, and LATVIA cohorts: positive (+) or negative (−).
With high inter-study heterogeneity observed for rs12233670 in the discovery, a random-effects model was applied for this particular SNP. The P value was no longer genome-wide significant (P = 0.0056), but the effect estimate was similar with a odds ratio of 1.26 (95% CI 1.07–1.48) instead of 1.3 (95% CI 1.22–1.39) obtained with the fixed-effect model.
Anti–H pylori Antibody Decay in H pylori–Infected Subjects
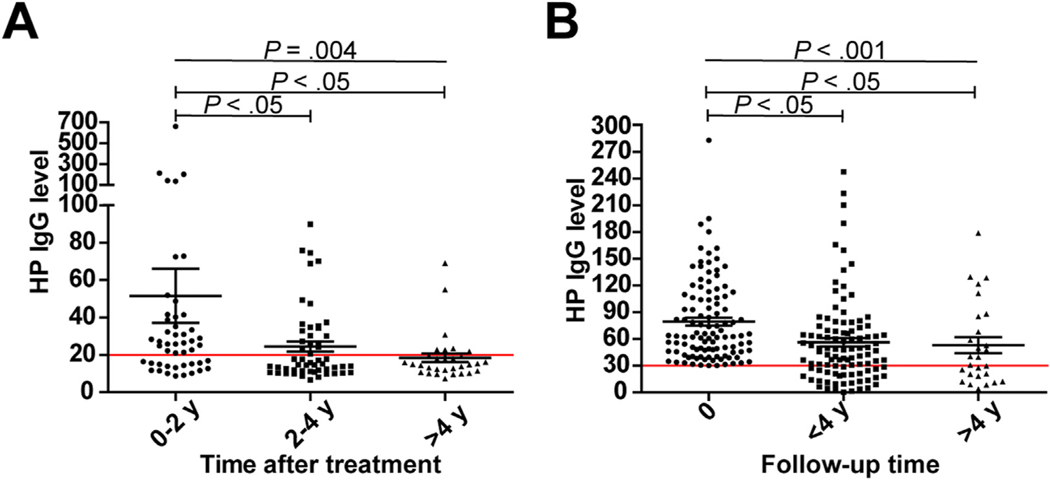
We considered that antibody decay and timing of sampling for H pylori serology may contribute to cohort heterogeneity. To investigate the serologic course of H pylori–infected subjects, anti–H pylori IgG data were studied in 2 settings. In a subset of RS participants that received H pylori eradication treatment at some point before the measurement of IgG antibodies (n = 132), titers were significantly higher in individuals tested within 0–2 years (n = 48) after receiving eradication therapy than in those tested 2–4 (n = 53) or >4 (n = 31) years (P = 0.004) after treatment (Figure 2A). When analyzing sequential anti–H pylori IgG titers from patients with gastric premalignant lesions with positive H pylori serology (n = 107), a significant decline between baseline measurement (time point 0) and retesting at <4 (n = 104) or >4 (n = 25) years of medical follow-up (P < 0.001) was seen (Figure 2B). Together, these data indicate that anti–H pylori antibody decay occurs within 2 years after treatment or clearance of H pylori.
Figure 2.
Anti–Helicobacter pylori (HP) IgG levels over time in 2 subsets. (A) IgG levels of Rotterdam Study subjects (n = 132) who received eradication therapy before serologic testing. The measurements of subjects are divided into 3 groups based on the time between treatment and H pylori serology: 0–2 (n = 48), 2–4 (n = 53), and >4 (n = 31) years. (B) IgG levels of H pylori–positive patients with premalignant gastric lesions (n = 107) at baseline (time point 0 with IgG titers >30 enzyme immunoassay units) and during serologic follow-up at <4 (n = 104) and >4 (n = 25) years. In both plots, the mean ± SEM and the manufacturer’s test cutoff (red horizontal line) are shown.
Higher TLR1 Surface Protein but Not Intracellular mRNA Expression Levels in Leukocytes of G Allele Carriers of rs28393318
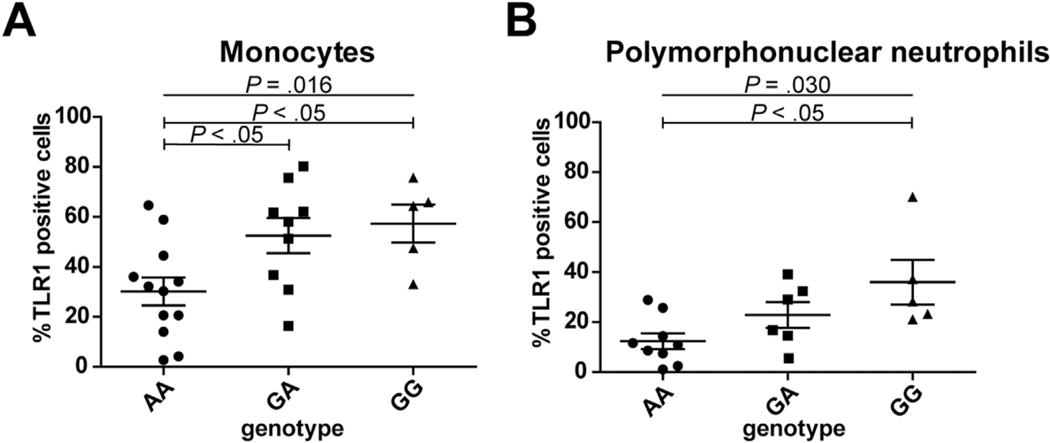
To investigate potential functional consequences of variation at the 4q14 locus, expression of the TLR-encoding genes within this locus was investigated in healthy subjects (n = 26) genotyped for rs28393318. A significant difference in the percentage of TLR1-positive monocytes (P = 0.016) and PMNs (P = 0.030) was observed between AA, GA, and GG genotype carriers (Figure 3A and B). Post hoc analysis revealed significantly higher TLR1 surface expression on monocytes for carriers of the minor rs28393318 allele (G) and on PMNs in subjects homozygous for the G allele compared with A allele carriers. Variation at rs28393318 did not influence TLR6 and TLR10 surface expression on either monocytes or PMNs (Supplementary Figure S1).
Figure 3.
Measurement of Toll-like receptor 1 (TLR1)–positive cells by flow cytometry. Dot plots illustrating the percentage of TLR1 positive cells in healthy subjects genotyped for rs28393318. The mean ± SEM and statistical significance among 3 genotypes are shown. (A) TLR1 positivity of monocytes in AA (n = 12), GA (n = 9), and GG (n = 5) carriers. (B) TLR1 positivity of polymorphonuclear neutrophils in AA (n = 9), GA (n = 6), and GG (n = 5) carriers.
Unlike previous RNA sequencing–based findings of reduced TLR1 mRNA levels for minor rs10004195 A allele carriers,40 our reverse-transcription qPCR findings showed no differences in TLR1 transcript levels in PBMCs between healthy subjects (n = 22) with different genotypes of rs28393318 (Supplementary Figure S2), which is in line with previous reports demonstrating no differences in mRNA and total cellular TLR1 protein levels among other TLR1 variants (in high LD with rs28393318 and rs10004195 among CEUs) tested.49–52
TLR1 rs28393318 Affects Immune Cell Cytokine Production
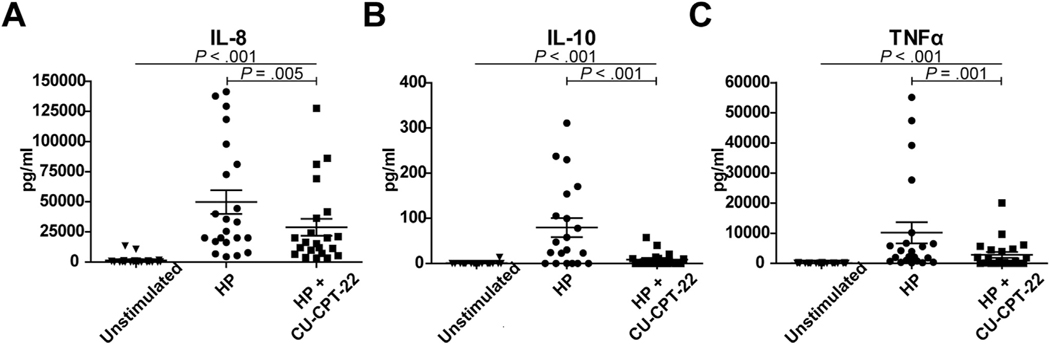
To first confirm TLR1 involvement in H pylori pathogenesis, PBMCs from healthy subjects (n = 22) were treated with H pylori in the presence of selective TLR1 inhibitor CU-CPT-22 or vehicle control. H pylori significantly stimulated IL8, IL10, and TNFα production (all P < 0.001), which was significantly but not fully abrogated on treatment of cells with CU-CPT-22 (P = 0.005 for IL8; P < 0.001 for IL10; P = 0.001 for TNFα) (Figure 4). These findings suggest that H pylori–related cytokine signaling is partly mediated via TLR1.
Figure 4.
Cytokine production by peripheral blood mononuclear cells (PBMCs) on stimulation with Helicobacter pylori. (A) Interleukin-8 (IL8), (B) interleukin-10 (IL10), and (C) tumor necrosis factor α (TNFα) levels in PBMCs of healthy subjects (n = 22). Cytokine levels were measured at baseline without stimulation and after H pylori exposure in the absence and presence of TLR1 inhibitor CU-CPT-22.
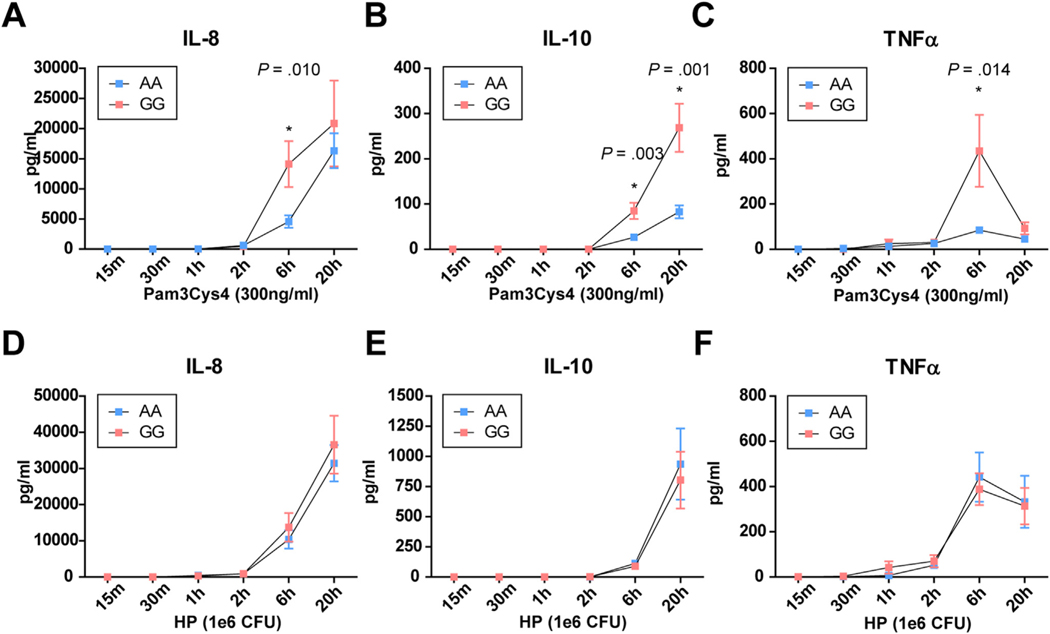
We next explored TLR1-mediated differences in cytokine production between healthy subjects with rs28393318 genotypes AA (n = 12) and GG (n = 8). PBMCs were stimulated with the specific TLR1 agonist Pam3Cys4 or with H pylori, and cytokine levels were measured at different time points afterward (0.25, 0.5, 1, 2, 6, and 20 hours). Pam3Cys4 stimulation resulted in higher IL8 (P = 0.010), IL10 (P = 0.003), and TNFα (P = 0.014) production at 6 hours as well as higher IL10 levels at 20 hours (P = 0.001) in GG carriers compared with AA carriers. Cytokine production on H pylori treatment was considerably higher than on Pam3Cys4 stimulation, but no differences among genotypes were observed (Figure 5).
Figure 5.
Time course of cytokine production by peripheral blood mononuclear cells (PBMCs) on stimulation with Toll-like receptor 1 (TLR1) ligand Pam3Cys4 or Helicobacter pylori in AA and GG carriers of rs28393318. (A, D) Interleukin-8 (IL8), (B, E) IL10, and (C, F) tumor necrosis factor α (TNFα) levels measured at different time points after stimulation with (A–C) TLR1 agonist Pam3Cys4 or (D–F) H pylori (HP). The results are stratified for AA (n = 12) and GG (n = 8) carriers of rs28393318. The mean ± SEM and statistical significance between genotypes are shown. CFU, colony-forming unit.
Discussion
This study aimed to better understand the genome-wide association between the TLR1/6/10 locus and H pylori. We extended the original work of Mayerle et al40 by the inclusion of an additional 4747 subjects of European ancestry in an updated GWAS meta-analysis. An association between anti–H pylori IgG titers and the TLR1/6/10 locus with top SNP rs12233670 was demonstrated in the discovery phase using the fixed-effect model, but replication proved to be challenging. Significant heterogeneity for our top association was observed across cohorts, with the SHIP-TREND and both replication cohorts showing association in the opposite direction. The interpretation of these findings remains complex but might be partially explained by methodologic differences that are inherent in the inclusion of longitudinally population cohorts, including time of recruitment (eg, SHIP vs SHIP-TREND) and use of nonuniform serologic assays. The concession of accounting for false positive assignment of cases by using the 25% highest vs 75% lowest IgG distribution might be another explanation. The allocation of truly H pylori–infected subjects into the control group could have possibly limited the detection and replication of promising SNPs, particularly for high-endemic regions such as Latvia.53 On the other hand, using a dichotomous cutoff based on IgG titers rather than test-defined H pylori positivity precludes bias introduced by the use of different tests with varying sensitivity and specificity across the different cohorts. Heterogeneity in our study may also have been introduced through antibody decay. Studying 2 different serologic datasets, we demonstrated that anti–H pylori IgG antibody decay over time occurs relatively quickly, as was previously observed on H pylori eradication treatment.54 Knowing that this process takes place, it is imperative to know the time of collection in relation to H pylori infection. Different rates of spontaneous clearance, re-infection, and H pylori eradication might have contributed to study heterogeneity, but this information was unfortunately not routinely collected in addition to data regarding H pylori–related disease status (eg, gastric cancer). Finally, various H pylori strains with varying virulence may interact differently with their human host, influencing the clinical outcome.55
Despite technical challenges preventing a clear replication, several studies do point toward a role of the TLR1/6/10 locus in the interaction between H pylori and its human host.40,41 With the data of the discovery phase showing an association with anti–H pylori IgG titers in the same direction as the original report, the relevance of the TLR1/6/10 locus in relation to H pylori pathology was further indicated. Our functional experiments demonstrate that variation at the TLR locus indeed has functional implications, as shown by a higher TLR1 surface expression and higher cytokine production in minor allele G carriers of rs28393318 (which is in complete LD with our top SNP rs12233670 and rs10004195 among CEUs). This might be explained by 2 non-synonymous TLR1 variants affecting intracellular–to–cell surface trafficking (rs5743618; r2 = 0.86 with rs28393318 among CEUs) and transportation of the receptor to the cell membrane (rs4833095; r2 = 0.97 with rs28393318 among CEUs).49–52,56,57 Minor allele carriers of these TLR1 genetic variants displayed higher cytokine responses on targeted TLR1 stimulation (ie, with Pam3Cys4), which was attributed to increased TLR1 surface expression rather than to changes in total protein or mRNA levels measured in cells,49–52 which is in line with our findings. Although H pylori mediates IL8, IL10, and TNFα production at least partially via TLR1 signaling in PBMCs, H pylori–mediated cytokine production was not affected by rs28393318 status of carriers. It is likely that rs28393318 variation effects are masked by other components of the host immune system triggered by this highly virulent H pylori strain.58 Similarly, the effect of rs28393318 variation on serologic titers induced by H pylori infection may be masked by additional H pylori–induced host-specific immune responses.
This study has 2 major limitations that need to be addressed. First, the identification of new genetic variants and replication of promising candidates for anti–H pylori IgG titers may have been hampered by the chosen definition of seropositivity for H pylori antibodies. Future studies might have to reconsider the phenotype definition, because the interpretation of H pylori serologic determination is not straightforward. Anti–H pylori IgG levels are more likely to represent a combination of the host’s ability to mount an immunologic response to infection as well as antibody clearance than actual H pylori incidence. With time from H pylori eradication therapy to serum collection influencing IgG antibody titers, it would be of value to collect those data in future studies. It should also be noted that H pylori infection involves an interplay of factors (host, bacterial, and environmental). Many genetic variants have been identified for H pylori–related conditions, such as ulcerative and (pre)malignant gastric lesions in different ethnic populations,23–25,27–32,38 and therefore it seems plausible that other genetic variants are relevant in H pylori pathogenesis besides the TLR locus. Second, we tested a selective cytokine panel in our functional analysis as a proof of concept. To better understand the immune response with regard to H pylori susceptibility, future experiments with different H pylori strains in different ethnic populations would be of interest.
In summary, the previously observed association between the TLR1/6/10 locus and anti–H pylori antibody titers was not uniformly confirmed across cohorts in this study. The interpretation of H pylori serology is complex and subject to alterations in response to therapy and over time. While variation at the TLR1/6/10 locus regulates surface expression and cytokine production on stimulation, further efforts are required to better understand the clinical relevance of TLR variants and other loci in their complex interaction with H pylori.
Supplementary Material
WHAT YOU NEED TO KNOW.
BACKGROUND
The identification of a genome-wide association between the Toll-like receptor (TLR) locus and anti–Helicobacter pylori IgG titers presented a major breakthrough, but this link is complex and not fully understood.
NEW FINDINGS
Genetic variation at the TLR1/6/10 locus confers immunologic cellular consequences, but high heterogeneity, cohort differences, and antibody decay likely prevent replication of previous associations with anti–H pylori IgG.
LIMITATIONS
Interpretation of study heterogeneity remains difficult owing to the inclusion of different longitudinal population-based cohorts for this largest dichotomous genome-wide association study for anti–H pylori IgG titers to date.
IMPACT
A role for the TLR1/6/10 locus in host–H pylori interaction is likely, but alternative definitions for H pylori positivity may be required.
Funding
The Rotterdam Study I-II was supported by the Netherlands Organization of Scientific Research (NWO; 175.010.2005.011, 911–03-012), Research Institute for Diseases in the Elderly (RIDE; 014–93-015), Genomics Initiative/NWO (project no. 050–060-810), Erasmus Medical Center Rotterdam, Erasmus University Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), Ministry of Education, Culture, and Science and Ministry for Health, Welfare, and Sports, European Commission, and the Municipality of Rotterdam. GenerationR was supported by Erasmus Medical Center Rotterdam, Erasmus University Rotterdam, ZonMw (907.00303, 916.10159), NWO, and the Ministry for Health, Welfare and Sports. The Study of Health in Pomerania (SHIP) and SHIP-TREND were supported by Deutsche Krebshilfe/Dr Mildred-Scheel-Stiftung (109102), Deutsche Forschungsgemeinschaft (DFG GRK840-D2/E3/E4, MA 4115/1–2/3), Federal Ministry of Education and Research (BMBF GANI-MED 03IS2061A and BMBF 0314107, 01ZZ9603, 01ZZ0103, 01ZZ0403, 03ZIK012), the European Union (EU-FP-7-EPCTM and EU-FP7-REGPOT-2010–1), AstraZeneca (unrestricted grant), the Federal Ministry of Education and Research, Siemens Healthcare, the Federal State of Mecklenburg–West Pomerania, and the University of Greifswald. The Framingham Heart Study was supported by National Institutes of Health grants N01-HC-25195, HHSN268201500001I, and 75N92019D00031 (to Boston University) and the Division of Intramural Research, National Heart, Lung, and Blood Institute (NHLBI). The Multi-Ethnic Study of Atherosclerosis (MESA) and the MESA SHARe projects were supported by the NHLBI (75N92020D00001, HHSN268201500003I, N01-HC-95159, 75N92020D00005, N01-HC-95160, 75N92020D00002, N01-HC-95161, 75N92020D00003, N01-HC-95162, 75N92020D00006, N01-HC-95163, 75N92020D00004, N01-HC-95164, 75N92020D00007, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, and UL1-TR-001420. Funding for SHARe genotyping was provided by NHLBI grant N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, CA) and the Broad Institute of Harvard and MIT (Boston, MA) using the Affymetrix Genome-Wide Human SNP Array 6.0. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences grant UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The Epidemiological Investigations on Chances of Preventing Recognizing Early and Optimally Treating Chronic Diseases in an Elderly Population were supported by the State Ministry of Science, Research and Arts, Baden-Württemberg, Federal Ministry of Education and Research, and Federal Ministry of Family Affairs, Senior Citizens, Women and Youth. LATVIA was supported by the European Regional Development Fund (ERDF; 009/0220/1DP/1.1.1.2.0/09/APIA/VIAA/016), National Program for Research in Latvia, Ministry of Health (6–1396-2016), and Fundamental and Applied Research Projects Program in Latvia (project no. lzp-2018/1–0135).
Abbreviations used in this paper:
- CEUs
Utah residents from north and west Europe
- ELISA
enzyme-linked immunosorbent assay
- FCGR
Fc gamma receptor
- GWAS
genome-wide association study
- IL
interleukin
- LD
linkage disequilibrium
- PBMCs
peripheral blood mononuclear cells
- PMNs
polymorphonuclear cells
- PCR
polymerase chain reaction
- SNP
single-nucleotide polymorphism
- TLR
Toll-like receptor
- TNF
tumor necrosis factor
Footnotes
CRediT Authorship Contributions
Conceptualization: Linda Broer, Manon C.W. Spaander, Fabian Frost, Stefan Weiss, Georg Homuth, Henry Völzke, Markus M. Lerch, Ben Schöttker, Hermann Brenner, Daniel Levy, Shih-Jen Hwang, Alexis C. Wood, Stephen S. Rich, Jerome I. Rotter, Kent D. Taylor, Russell P. Tracy, Edmond K. Kabagambe, Marcis Leja, Janis Klovins, Raitis Peculis, Dace Rudzite, Liene Nikitina-Zake, Girts Skenders, Vita Rovite, André Uitterlinden, Ernst J. Kuipers, Maikel P. Peppelenbosch, and additional members of Rotterdam Study I-II, GenerationR, Study of Health in Pomerania, Framingham Heart Study, Multi-Ethnic Study of Atherosclerosis, Epidemiological Investigations on Chances of Preventing Recognizing Early and Optimally Treating Chronic Diseases in an Elderly Population, and LATVIA cohorts not directly involved in this manuscript.
Methodology: all authors.
Investigation: all authors.
Formal analysis of discovery: Linda Broer, Fabian Frost, Stefan Weiss, Georg Homuth, Henry Völzke, Markus M. Lerch, Daniel Levy, Shih-Jen Hwang, Alexis C. Wood, Stephen S. Rich, Jerome I. Rotter, Kent D. Taylor, Russell P. Tracy, and Edmond K. Kabagambe.
Formal analysis of replication: Yan Zhang, Hannah Stocker, Hermann Brenner, Marcis Leja, Janis Klovins, and Raitis Peculis.
Formal analysis of meta-analysis: Linda Broer.
Project administration: Suk Yee Lam and Gwenny M. Fuhler.
Resources: Fabian Frost, Stefan Weiss, Georg Homuth, Henry Völzke, Markus M. Lerch, Hermann Brenner, Daniel Levy, Shih-Jen Hwang, Alexis C. Wood, Stephen S. Rich, Jerome I. Rotter, Kent D. Taylor, Russell P. Tracy, Edmond K. Kabagambe, Marcis Leja, Janis Klovins, Dace Rudzite, Liene Nikitina-Zake, Girts Skenders, Vita Rovite, Ernst J. Kuipers, and Maikel P. Peppelenbosch.
Supervision: Manon C.W. Spaander, Fabian Frost, Stefan Weiss, Georg Homuth, Henry Völzke, Markus M. Lerch, Hermann Brenner, Daniel Levy, Shih-Jen Hwang, Alexis C. Wood, Stephen S. Rich, Jerome I. Rotter, Kent D. Taylor, Russell P. Tracy, Edmond K. Kabagambe, Marcis Leja, Janis Klovins, Gwenny M. Fuhler, Maikel P. Peppelenbosch, and André Uitterlinden.
Visualization: Suk Yee Lam, Michiel C. Mommersteeg, Bingting Yu, Linda Broer, and Gwenny M. Fuhler. Writing—original draft: Suk Yee Lam, Michiel C. Mommersteeg, and Gwenny M. Fuhler.
Writing—reviewing and editing: all authors.
Conflicts of interest
The authors declare no conflicts.
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2022.01.011.
References
- 1.Warren JR, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 1983;1:1273–1275. [PubMed] [Google Scholar]
- 2.Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1984;1:1311–1315. [DOI] [PubMed] [Google Scholar]
- 3.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Schistosomes, liver flukes and Helicobacter pylori. Lyon, 7–14 June 1994. IARC Monogr Eval Carcinog Risks Hum 1994;61:1–241. [PMC free article] [PubMed] [Google Scholar]
- 4.Kuipers EJ, Uyterlinde AM, Pena AS, et al. Long-term sequelae of Helicobacter pylori gastritis. Lancet 1995; 345:1525–1528. [DOI] [PubMed] [Google Scholar]
- 5.Helicobacter and Cancer Collaborative Group. Gastric cancer and Helicobacter pylori: a combined analysis of 12 case control studies nested within prospective cohorts. Gut 2001;49:347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Martel C, Georges D, Bray F, et al. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health 2020; 8:e180–190. [DOI] [PubMed] [Google Scholar]
- 7.Hooi JKY, Lai WY, Ng WK, et al. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology 2017;153:420–429. [DOI] [PubMed] [Google Scholar]
- 8.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process—First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res 1992;52:6735–6740. [PubMed] [Google Scholar]
- 9.Wong BC, Lam SK, Wong WM, et al. Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of China: a randomized controlled trial. Jama 2004; 291:187–194. [DOI] [PubMed] [Google Scholar]
- 10.Ford AC, Forman D, Hunt RH, et al. Helicobacter pylori eradication therapy to prevent gastric cancer in healthy asymptomatic infected individuals: systematic review and meta-analysis of randomised controlled trials. Bmj 2014;348:g3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee YC, Chiang TH, Chou CK, et al. Association between Helicobacter pylori eradication and gastric cancer incidence: a systematic review and meta-analysis. Gastroenterology 2016;150:1113–1124 e5. [DOI] [PubMed] [Google Scholar]
- 12.Leung WK, Wong IOL, Cheung KS, et al. Effects of Helicobacter pylori treatment on incidence of gastric cancer in older individuals. Gastroenterology 2018; 155:67–75. [DOI] [PubMed] [Google Scholar]
- 13.Savoldi A, Carrara E, Graham DY, et al. Prevalence of antibiotic resistance in Helicobacter pylori: a systematic review and meta-analysis in World Health Organization regions. Gastroenterology 2018;155:1372–1382.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuipers EJ, Pena AS, van Kamp G, et al. Seroconversion for Helicobacter pylori. Lancet 1993;342:328–331. [DOI] [PubMed] [Google Scholar]
- 15.Malaty HM, El-Kasabany A, Graham DY, et al. Age at acquisition of Helicobacter pylori infection: a follow-up study from infancy to adulthood. Lancet 2002; 359:931–935. [DOI] [PubMed] [Google Scholar]
- 16.Perez-Perez GI, Sack RB, Reid R, et al. Transient and persistent Helicobacter pylori colonization in Native American children. J Clin Microbiol 2003;41:2401–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowland M, Daly L, Vaughan M, et al. Age-specific incidence of Helicobacter pylori. Gastroenterology 2006; 130:65–72. [DOI] [PubMed] [Google Scholar]
- 18.Bardhan PK. Epidemiological features of Helicobacter pylori infection in developing countries. Clin Infect Dis 1997;25:973–978. [DOI] [PubMed] [Google Scholar]
- 19.Malaty HM, Engstrand L, Pedersen NL, et al. Helicobacter pylori infection: genetic and environmental influences. A study of twins. Ann Intern Med 1994; 120:982–986. [DOI] [PubMed] [Google Scholar]
- 20.Kuipers EJ. Review article: exploring the link between Helicobacter pylori and gastric cancer. Aliment Pharmacol Ther 1999;13(Suppl 1):3–11. [DOI] [PubMed] [Google Scholar]
- 21.Polk DB, Peek RM Jr. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer 2010;10:403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mommersteeg MC, Yu J, Peppelenbosch MP, et al. Genetic host factors in Helicobacter pylori–induced carcinogenesis: emerging new paradigms. Biochim Biophys Acta Rev Cancer 2018;1869:42–52. [DOI] [PubMed] [Google Scholar]
- 23.Tanikawa C, Urabe Y, Matsuo K, et al. A genome-wide association study identifies two susceptibility loci for duodenal ulcer in the Japanese population. Nat Genet 2012;44. 430–4, S1–2. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Gonzalez MA, Bujanda L, Quintero E, et al. Association of PSCA rs2294008 gene variants with poor prognosis and increased susceptibility to gastric cancer and decreased risk of duodenal ulcer disease. Int J Cancer 2015;137:1362–1373. [DOI] [PubMed] [Google Scholar]
- 25.Jiang J, Jia ZF, Kong F, et al. Association of polymorphism of PTPN 11 encoding SHP-2 with gastric atrophy but not gastric cancer in Helicobacter pylori seropositive Chinese population. BMC Gastroenterol 2012;12:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang FB, Li ZX, Wang YM, et al. Toll-like receptor 1 and 10 polymorphisms, Helicobacter pylori susceptibility and risk of gastric lesions in a high-risk Chinese population. Infect Genet Evol 2015;31:263–269. [DOI] [PubMed] [Google Scholar]
- 27.Rizzato C, Kato I, Plummer M, et al. Genetic variation in PSCA and risk of gastric advanced preneoplastic lesions and cancer in relation to Helicobacter pylori infection. PLoS One 2013;8:e73100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He CY, Sun LP, Xu Q, et al. PGC TagSNP and its interaction with H. pylori and relation with gene expression in susceptibility to gastric carcinogenesis. PLoS One 2014; 9:e115955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai M, Dai S, Chen W, et al. Environmental factors, seven GWAS-identified susceptibility loci, and risk of gastric cancer and its precursors in a Chinese population. Cancer Med 2017;6:708–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He BS, Sun HL, Xu T, et al. Association of genetic polymorphisms in the lncRNAs with gastric cancer risk in a Chinese population. J Cancer 2017;8:531–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sala N, Munoz X, Travier N, et al. Prostate stem-cell antigen gene is associated with diffuse and intestinal gastric cancer in Caucasians: results from the EPIC-EURGAST study. Int J Cancer 2012;130:2417–2427. [DOI] [PubMed] [Google Scholar]
- 32.Li M, Huang L, Qiu H, et al. Helicobacter pylori infection synergizes with three inflammation-related genetic variants in the GWASs to increase risk of gastric cancer in a Chinese population. PLoS One 2013;8:e74976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravishankar Ram M, Goh KL, Leow AH, et al. Polymorphisms at locus 4p14 of Toll-like receptors TLR-1 and TLR-10 confer susceptibility to gastric carcinoma in Helicobacter pylori infection. PLoS One 2015;10: e0141865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Study Group of Millennium Genome Project for Cancer, Sakamoto H, Yoshimura K, Saeki N, et al. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet 2008;40:730–740. [DOI] [PubMed] [Google Scholar]
- 35.Abnet CC, Freedman ND, Hu N, et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet 2010;42:764–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saeki N, Saito A, Choi IJ, et al. A functional single nucleotide polymorphism in mucin 1, at chromosome 1q22, determines susceptibility to diffuse-type gastric cancer. Gastroenterology 2011;140:892–902. [DOI] [PubMed] [Google Scholar]
- 37.Shi Y, Hu Z, Wu C, et al. A genome-wide association study identifies new susceptibility loci for noncardia gastric cancer at 3q13.31 and 5p13.1. Nat Genet 2011; 43:1215–1218. [DOI] [PubMed] [Google Scholar]
- 38.Companioni O, Bonet C, Munoz X, et al. Polymorphisms of Helicobacter pylori signaling pathway genes and gastric cancer risk in the European Prospective Investigation into Cancer—Eurgast cohort. Int J Cancer 2014; 134:92–101. [DOI] [PubMed] [Google Scholar]
- 39.Dargiene G, Streleckiene G, Skieceviciene J, et al. TLR1 and PRKAA1 gene polymorphisms in the development of atrophic gastritis and gastric cancer. J Gastrointestin Liver Dis 2018;27:363–369. [DOI] [PubMed] [Google Scholar]
- 40.Mayerle J, den Hoed CM, Schurmann C, et al. Identification of genetic loci associated with Helicobacter pylori serologic status. JAMA 2013;309:1912–1920. [DOI] [PubMed] [Google Scholar]
- 41.Sung H, Camargo MC, Yu K, et al. Association of 4p14 TLR locus with antibodies to Helicobacter pylori. Genes Immun 2015;16:567–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubicz R, Yolken R, Drigalenko E, et al. Genome-wide genetic investigation of serological measures of common infections. Eur J Hum Genet 2015;23:1544–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winkler TW, Day FR, Croteau-Chonka DC, et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc 2014;9:1192–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.den Hollander WJ, Holster IL, den Hoed CM, et al. Surveillance of premalignant gastric lesions: a multicentre prospective cohort study from low incidence regions. Gut 2019;68:585–593. [DOI] [PubMed] [Google Scholar]
- 46.Somasundaram R, Fernandes S, Deuring JJ, et al. Analysis of SHIP1 expression and activity in Crohn’s disease patients. PLoS One 2017;12:e0182308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng K, Wang X, Zhang S, et al. Discovery of small-molecule inhibitors of the TLR1/TLR2 complex. Angew Chem Int Ed Engl 2012;51:12246–12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lie M, van der Giessen J, Fuhler GM, et al. Low dose Naltrexone for induction of remission in inflammatory bowel disease patients. J Transl Med 2018;16:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson CM, Lyle EA, Omueti KO, et al. Cutting edge: a common polymorphism impairs cell surface trafficking and functional responses of TLR1 but protects against leprosy. J Immunol 2007;178:7520–7524. [DOI] [PubMed] [Google Scholar]
- 50.Hawn TR, Misch EA, Dunstan SJ, et al. A common human TLR1 polymorphism regulates the innate immune response to lipopeptides. Eur J Immunol 2007; 37:2280–2289. [DOI] [PubMed] [Google Scholar]
- 51.Wurfel MM, Gordon AC, Holden TD, et al. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. Am J Respir Crit Care Med 2008;178:710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uciechowski P, Imhoff H, Lange C, et al. Susceptibility to tuberculosis is associated with TLR1 polymorphisms resulting in a lack of TLR1 cell surface expression. J Leukoc Biol 2011;90:377–388. [DOI] [PubMed] [Google Scholar]
- 53.Leja M, Cine E, Rudzite D, et al. Prevalence of Helicobacter pylori infection and atrophic gastritis in Latvia. Eur J Gastroenterol Hepatol 2012;24:1410–1417. [DOI] [PubMed] [Google Scholar]
- 54.Feldman M, Cryer B, Lee E, et al. Role of seroconversion in confirming cure of Helicobacter pylori infection. Jama 1998;280:363–365. [DOI] [PubMed] [Google Scholar]
- 55.Correa P, Piazuelo MB. Evolutionary history of the Helicobacter pylori genome: implications for gastric carcinogenesis. Gut Liver 2012;6:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schumann RR, Tapping RI. Genomic variants of TLR1—it takes (TLR-)two to tango. Eur J Immunol 2007; 37:2059–2062. [DOI] [PubMed] [Google Scholar]
- 57.Mikacenic C, Schneider A, Radella F, et al. Cutting edge: genetic variation in TLR1 is associated with Pam3CSK4-induced effector T cell resistance to regulatory T cell suppression. J Immunol 2014;193:5786–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peek RM Jr, Fiske C, Wilson KT. Role of innate immunity in Helicobacter pylori–induced gastric malignancy. Physiol Rev 2010;90:831–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.