Abstract
Abietadiene synthase catalyzes the committed step in resin acid biosynthesis, forming a mixture of abietadiene double-bond isomers by two sequential, mechanistically distinct cyclizations at separate active sites. The first reaction, protonation-initiated cyclization, converts the universal diterpene precursor geranylgeranyl diphosphate to the stable bicyclic intermediate copalyl diphosphate. In the second, magnesium ion-dependent reaction, diphosphate ester ionization-initiated cyclization generates the tricyclic perhydrophenanthrene-type backbone and is coupled, by intramolecular proton transfer within a transient pimarenyl intermediate, to a 1,2-methyl migration that generates the C13 isopropyl group characteristic of the abietane structure. Alternative deprotonations of the terminal abietenyl carbocation provide a mixture of abietadiene, levopimaradiene, and neoabietadiene, and this product profile varies as a function of pH. Mutational analysis of amino acids at the active site of a modeled structure has identified residues critical for catalysis, as well as several that play roles in specifying product formation, apparently by ligation of a magnesium ion cofactor. These results strongly suggest that choice between alternatives for deprotonation of the abietenyl intermediate depends more on the positioning effects of the carbocation–diphosphate anion reaction partners than on the pKa of multiple participating bases. In one extreme case, mutant N765A is unable to mediate the intramolecular proton transfer and aborts the reaction, without catalyzing 1,2-methyl migration, to produce only sandaracopimaradiene, thereby providing supporting evidence for the corresponding stereochemistry of the cryptic pimarenyl intermediate of the reaction pathway.
Keywords: geranylgeranyl diphosphate‖copalyl diphosphate‖abietane diterpenes‖ resin acids‖diterpene synthase
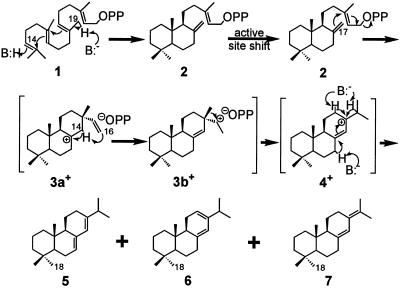
A primary response of conifers to physical wounding is secretion of oleoresin (pitch), a mixture of roughly equal amounts of monoterpene olefins (turpentine) and diterpene resin acids (rosin) (1). Evaporation of the volatile turpentine carrier results in solidification of the resin acids to form a physical barrier that seals the wound (2). The oleoresin of grand fir (Abies grandis) contains resin acids derived largely from the abietane family of diterpene olefins (Fig. 1), which undergo oxidation of the C18-methyl group to the corresponding carboxylic acids (3, 4). Abietadiene synthase (AS) of grand fir performs the committed step of resin acid biosynthesis by catalyzing the cyclization and rearrangement of the universal diterpene precursor (E,E,E)-geranylgeranyl diphosphate (GGPP; 1) to a mixture of abietadiene double-bond isomers (Fig. 1; ref. 5). AS is bifunctional in catalyzing two sequential, mechanistically different cyclizations at separate active sites occurring in structurally distinct domains (6). Protonation across the terminal 14–15 double bond of GGPP, followed by bicyclization and deprotonation, produces the stable intermediate (+)-copalyl diphosphate (CPP; 2), in a reaction similar to that catalyzed by (−)-copalyl diphosphate synthase (kaurene synthase A) of gibberellin biosynthesis (7, 8). This reaction occurs in an N-terminal active site with an acid/base catalytic mechanism that may serve as a general model for such protonation-initiated cyclizations (9). CPP diffuses from the N-terminal domain to a C-terminal active site (6) where, in a reaction similar to that mediated by kaurene synthase B (7, 8), AS ionizes the diphosphate ester (CPP) to promote cyclization to the tricyclic perhydrophenanthrene backbone. However, this cyclization by AS is further coupled to a 1,2-methyl migration, by means of intramolecular proton transfer within a pimarenyl intermediate (10), to generate the C13 isopropyl group characteristic of the abietane skeleton. Finally, deprotonation of the resulting abietenyl carbocation at one of three alternative positions (C7, C12, or C15) leads to the three principal olefin products abieta-7(8),13(14)-diene (abietadiene, 5), abieta-8(14),12(13)-diene (levopimaradiene, 6), and abieta-8(14)-13(15)-diene (neoabietadiene, 7), respectively (5).
Figure 1.
Proposed reaction mechanism for abietadiene synthase. GGPP (1) is bound at the N-terminal active site and protonated at C14 to initiate A/B-ring closure followed by deprotonation at C19 (9) to yield (+)-CPP (2). (+)-CPP then diffuses (6) to the C-terminal active site where ionization of the diphosphate ester initiates anti-SN′ cyclization (11) to the C8-sandaracopimerenyl cation (3a+) which undergoes intramolecular proton transfer from C14 to C16 (9) to afford the C15-sandaracopimarenyl carbocation (3b+) (11). 1,2-Methyl migration in 3b+ generates the abietenyl carbocation (4+), from which deprotonation at the indicated positions yields the observed abietadiene products (5-7) (5). In the N765A mutant, intramolecular proton transfer does not occur, and carbocation 3a+ is deprotonated at C14 to yield sandaracopimaradiene.
The stereochemistry at C13 and double-bond placement in the pimarenyl intermediate of the reaction catalyzed by AS are eliminated on methyl migration; however, studies on the stereospecific cyclization of 8α-hydroxy-17-nor-CPP to 17-nor manoyl oxide (11) and on the relative potency of 15-aza-15,16-dihydropimarene inhibitors (M. M. Ravn, R.J.P., R. M. Coates, and R.B.C., unpublished work) indicate a 13β-methyl configuration and 8,14-double-bond placement corresponding to sandaracopimaradiene. Additional experiments revealed that conversion of CPP to abietadiene occurs by initial anti-SN′ cyclization to form a sandaracopimar-15-en-8-yl carbocation (Fig. 1, 3a+), followed by stereospecific intramolecular transfer of the pro-E hydrogen at C14 (10) to the si-face of the vinyl group and syn-related methyl migration (11). AS mediates an energetically unfavorable conversion of a tertiary to a secondary carbocation (Fig. 1, 3a+ → 3b+) (11), with the latter stabilized by ionic interaction with the paired diphosphate anion (M. M. Ravn, R.J.P., R. M. Coates, and R.B.C., unpublished work).
Figure 3.
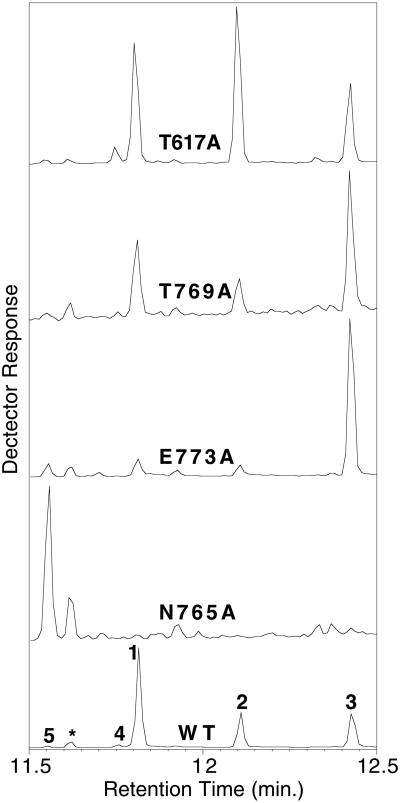
GC-MS analysis (total ion chromatograms) of the products of wild-type rAS and those of T617A, T769A, E773A, and N765A. The indicated products are levopimaradiene (1), abietadiene (2), neoabietadiene (3), palustradiene (4), sandaracopimaradiene (5), and geranyllinalool (*) arising from substrate solvolysis during the extended incubations required for analysis of severely impaired mutants. WT, wild type
In an initial attempt to define structure–function relationships in the complex cyclization cascade catalyzed by AS, the enzyme was modeled against the crystal structure of the sequiterpene cyclase 5-epi-aristolochene synthase (5-EAS) from tobacco (12). The similar fold (13) and location of the DDXXD motif (14, 15) involved in the binding and ionization of the prenyl diphosphate substrate of other terpenoid synthases (16) and mechanistically related prenyltransferases (17) allowed placement of the putative second active site, for the ionization-dependent step, at a central cavity in the C-terminal helical barrel domain. Mutational analysis of the amino acids comprising this presumptive active site in AS has suggested specific roles for several of these residues. Mutational analysis of the N-terminal active site involved in the first step of the reaction cycle (protonation-initiated cyclization of GGPP to CPP) has been described (9).
Materials and Methods
Materials and General Procedures.
Liquid scintillation counting and product analysis by GC-MS were performed as described (4, 18). The preparations of (E,E,E)-[1-3H]geranylgeranyl diphosphate (120 Ci/mol; 1 Ci = 37 GBq; ref. 4) and of (+)-[1-3H]CPP (120 Ci/mol) (5) have also been described elsewhere. A modeled structure for AS was generated by using tobacco 5-EAS, the only plant terpene cyclase structure currently available (12), as the scaffold, with energy minimization performed by using GROMOS96 (19).
Enzyme Assays.
Kinetic assays with freshly prepared enzyme were performed as described (5), except that reaction times and rAS concentrations were scaled to permit measurement of mutant enzymatic activity. With severely impaired mutant enzymes, the reported kinetic constants are accurate to within ±50% on the basis of replicated assays. To examine product distribution, 50-μl reactions were performed with 0.1–0.5 μM rAS and 50 μM GGPP in 0.3-ml conical vials, and the reaction products were extracted with pentane. These reactions were run for several days to obtain sufficient product with kinetically impaired mutants and at kinetically compromised pH values. Product analysis was performed by GC-MS to permit unambiguous product identification, rather than by GC with flame ionization detection as used in earlier studies (5). Because quantification depends on the type of detection (electron impact ionization versus flame ionization), reference assays with wild-type rAS were included in all experiments. To determine the effects of pH on product distribution, 50 mM 1,3-bis[Tris(hydroxymethyl)methylamino]propane was substituted for the original Hepes buffer to increase the effective buffering range. To determine the requirement for a divalent metal ion cofactor, 2.5 mM EDTA was substituted for MgCl2 and MnCl2 in the normal assay buffer, and kinetic analysis was performed with CPP as substrate in the presence of EDTA.
The influence of mutations on the second, diphosphate ester ionization-dependent reaction was determined by using CPP as substrate. Because CPP freely diffuses between the first (N-terminal) and second (C-terminal) active sites, a mutant (D404A) specifically deficient in the protonation-initiated cyclization of GGPP to CPP can complement mutants defective in the ionization-dependent reaction (CPP to abietadienes), as described (6). Therefore, a coupled assay was used to examine the effects of the mutants described here on the first, protonation-initiated cyclization step. In brief, a 3 nM concentration of D404A was combined with a 3 nM concentration of a particular mutant and assayed with 5 μM GGPP as substrate; the resulting activity was compared with that obtained with a 3 nM concentration of the wild-type enzyme in a parallel control assay.
Mutant Construction and Expression.
Point mutants were constructed from the wild-type rAS gene by using an overlapping fragment PCR strategy as described (6), in which complementary mutagenic primers were used to produce two overlapping fragments containing the introduced mutation, which were then recombined to produce the full-length mutated gene. Mutant and wild-type rAS were expressed in Escherichia coli BLR (Stratagene) at 15°C in 1 L NZY cultures, and were purified as described (5). The concentration of purified rAS was determined by absorbance at 280 nm with the calculated extinction coefficient (138,350 M−1⋅cm−1).
Results
Factors Affecting Abietadiene Synthase Product Distribution.
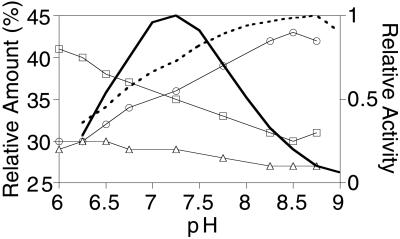
Previous analyses, at the optimum pH of 7.2 for GGPP as substrate (4), indicated that AS produces nearly equal amounts of abietadiene, levopimaradiene, and neoabietadiene as the result of deprotonation of the terminal abietenyl carbocation at C7, C12, or C15, respectively (Fig 1; ref. 5). Analysis of the influence of pH in 1,3-bis[Tris(hydoxymethyl)methylamino]propane buffer on product distribution from GGPP revealed a subtle but reproducible alteration, the most notable feature of which was a decrease in the proportion of abietadiene and a corresponding increase in levopimaradiene with increasing pH (Fig. 2). These changes in product outcome were not caused by variation in ionic strength, because increasing salt concentration (0–0.7 M NaCl) did not significantly alter product distribution, suggesting that the observed influence of pH is a direct effect of changes in the protonation state of active-site residues. However, subtle alteration in product distribution was also observed with different buffers, as indicated by the difference in product distribution at pH 7.2 between 1,3-bis[Tris(hydoxymethyl)methylamino]propane and Hepes (cf. Figs. 2 and 3).
Figure 2.
Change in relative proportion of the major products of abietadiene synthase (abietadiene, □; levopimaradiene, ○; neoabietadiene, ▵) as a function of pH. Also shown (thick lines) is the change in relative activity of AS with GGPP (solid line) or CPP (dotted line) as substrate as a function of pH.
Mutational Analysis of Putative Active-Site Residues.
A structure for AS was generated by sequence alignment and modeling (19) against that of 5-EAS from tobacco, a sequiterpene cyclase that was crystallized with various substrate analogs to assist in assigning possible catalytic roles for several active-site residues (12). The location of these substrate analogs, as well as the presence of the highly conserved DDXXD motif involved in diphosphate binding and ionization (16), in the central cavity of the C-terminal helical barrel domain clearly identified this location as the active site for the diphosphate ester ionization-dependent cyclization reaction mediated by 5-EAS. The DDXXD motif has been shown previously to be involved in the diphosphate ionization-dependent step of the AS reaction cycle (6), and its similar location in the central cavity of the C-terminal helical barrel domain of the modeled structure demonstrated that this central cavity is the location of the active site for this second reaction catalyzed by AS. Many of the residues proposed to have catalytic roles in 5-EAS are conserved in AS and in other plant terpene cyclases (14), and these residues, along with other charged and polar residues that line the putative C-terminal active site of AS, were mutated, in general to alanine, to evaluate their potential role(s) in the ionization-dependent cyclization and rearrangement of CPP to abietadienes.
Kinetic analysis of these C-terminal domain mutations, with either GGPP or CPP as substrate, revealed highly selective influence on the second (diphosphate ionization-dependent) reaction step. The initial cyclization of GGPP to CPP (protonation-dependent) conducted in the N-terminal domain was not affected by these mutations, all of which exhibited rates equivalent to the wild-type enzyme for this first reaction step. Furthermore, effects on the ionization-dependent cyclization and rearrangement reaction were exerted largely on catalysis, and not substrate binding. Thus, mutagenic changes in KM were less than 6-fold, whereas changes in kcat exhibited decreases of up to five orders of magnitude (Table 1).
Table 1.
Kinetic data for mutations in the diphosphate ester ionization-dependent active site of abietadiene synthase
| Variant | KM (μM) | kcat (s−1) |
|---|---|---|
| WT + Mg2+ | 0.4 ± 0.2 | 2.2 ± 0.3 |
| WT − Mg2+ | 10 | 8 × 10−6 |
| R584A | 1 | 6 × 10−4 |
| R586A | 2 | 1 × 10−2 |
| E589A | 1 | 0.1 |
| T617A | 0.5 | 0.4 |
| D621A | 0.7 | 8 × 10−6 |
| D625A | 2 | 3 × 10−3 |
| E699A | 0.7 | 4 × 10−2 |
| R762A | 0.7 | 1 × 10−5 |
| N765A | 0.5 | 9 × 10−5 |
| D766A | 0.4 | 2 × 10−3 |
| T769A | 2 | 3 × 10−4 |
| E773A | 0.3 | 5 × 10−4 |
| E778A | 0.3 | 0.2 |
| S721A | 3 | 4 × 10−2 |
| Y841F | 0.8 | 8 × 10−2 |
| D845A | 2 | 1 × 10−4 |
| T848A | 0.8 | 4 × 10−2 |
The influence of site-directed mutations on product distribution was determined by comparison with the distribution of wild-type enzyme at the pH 7.2 optimum for overall activity (from GGPP). Four mutant enzymes (of the 17 evaluated) exhibited changes in product distribution (Fig. 3), whereas most mutant enzymes yielded product distributions similar to that of the wild-type enzyme. Thus, T617A produced relatively more abietadiene at the expense of levopimaradiene, whereas T679A produced a higher proportion of neoabietadiene at the expense of both levopimaradiene and abietadiene. Mutant E773A provided a significant increase in the relative amount of neoabietadiene and of sandaracopimaradiene. N765A was completely unable to mediate the methyl migration necessary for formation of the abietane skeleton, and this mutant produced only sandaracopimaradiene. T679A, E773A, and N765A were all also severely compromised in catalysis.
Discussion
Many terpene cyclases produce multiple products; however, few investigations have been made into the determinants of such catalytic promiscuity (16). With AS, changes in reaction pH, but not salt concentration, influence product distribution, indicating that changes in the protonation state of active-site residues can alter product distribution. Such effects could arise from positioning differences of the terminal abietenyl carbocation intermediate with respect to the base(s) responsible for deprotonation, or reflect the presence of multiple participating bases with different pKa values. The observation that product distribution also varies as a function of buffer composition at the same pH indicates that changes in product distribution are not caused solely by multiple bases with different pKa values, and suggests that positioning effects on the abietenyl carbocation are more important. This suggestion is consistent with that of Starks et al. (12), who, in noting that residues lining the active site of 5-EAS are conserved in vetispiradiene synthase, suggested that the difference in product outcome between these two cyclases must arise from secondary structural effects caused by distal residues. However, to determine how product distribution is controlled in AS it was necessary to obtain structural information regarding the second, ionization-dependent active site.
To gain insight to the architecture of this second active site, a model of AS was constructed (19) based on the structure of 5-EAS, the only available structure for a plant terpene cyclase (12). On the basis of the relative positions of various substrate analogs crystallized with 5-EAS, specific catalytic roles have been suggested for active-site residues in this sequiterpene cyclase (12). Many of these residues and their relative locations are conserved in AS and other plant terpene synthases (14), thereby implying conserved function (16). On the basis of the 5-EAS precedent (12), the first and last aspartates (D621 and D625) in the canonical DDXXD motif of this second active site of AS chelate a magnesium ion (Mga2+) along with the assistance of the highly conserved E699. A second magnesium ion (Mgb2+) is bound by N765 (almost always an aspartate in other synthases), a nonconserved polar residue (in this case, T769), and the highly conserved E773. On substrate binding, a third magnesium ion (Mgc2+) binds to the diphosphate moiety and is further chelated by D621 (which then ligates both Mga2+ and Mgc2+). R586 then assists in organizing the enzyme to exclude water from the active site. This residue is part of a highly conserved RXR motif in which the absolutely conserved first arginine, R584, is thought to play a role in binding and stabilizing the released diphosphate anion, along with the assistance of R762, another absolutely conserved arginine. In the 5-EAS–substrate complex, however, only R584 seems to interact directly with the diphosphate; R762 is hydrogen-bonded to the highly conserved D766. A tyrosine residue of 5-EAS acted as an acid in reprotonating the germacrene A intermediate to permit formation of the final aristolochene product (20). The equivalent Y841F mutant was constructed to test the involvement of this corresponding residue in the similar proton transfer (Fig. 1, 3a+ → 3b+) mediated by AS. Conserved aspartate D845 was also mutated; the equivalent residue in 5-EAS is five rather than four residues upstream of the aforementioned tyrosine, and was originally suggested to act as a base in deprotonating the carbocation that forms the germacrene A intermediate of the 5-EAS reaction cycle (12, 20). Alanine substitutions were made for all the residues indicated above, and for several other polar and/or charged residues that line the active site of the modeled AS structure.
Kinetic analysis of these mutants generally supports the functioning of the targeted residues at the second, diphosphate ester ionization-dependent active site, because many of these mutations profoundly and specifically influence the conversion of CPP to abietadienes, with no detectable effect on the CPP synthase activity of the N-terminal (first) active site of the bifunctional AS. The expression levels for all these soluble mutant proteins were equivalent to that of the wild-type enzyme, suggesting that the mutations had no gross detrimental effects on overall folding or stability. Furthermore, the effects of these mutations on the second (ionization-dependent) reaction step are exerted largely on catalysis and not substrate binding, confirming that the C-terminal domain active site is able to tightly bind the substrate and is therefore also functionally folded.
Mutagenesis of five residues selected on the basis of their presence in the modeled active site (E589, T617, E778, S721, and T848) had relatively small effects on kcat (5–50 fold decrease); however, substitution of alanine for E589 and S721 had significant influence on KM, suggesting a role in substrate binding. T617 corresponds to an alanine found in the closely related levopimaradiene synthase from Ginko biloba (21) (of the 17 residues examined here, only T617 and T848 differ between these two diterpene cyclases). T617A did exhibit a subtle effect on product distribution; however, rather than increasing the proportion of levopimaradiene, this mutation increased the relative proportion of abietadiene formed at the expense of levopimaradiene (Fig. 3). The ability of T617A to alter product distribution supports the earlier suggestion that it is the dynamics and/or binding of the terminal abietenyl carbocation that most influences the choice between alternative sites for deprotonation.
As found in previous investigations with other terpene synthases and related prenyltransferases, mutation of the first aspartate of the highly conserved DDXXD motif has a much greater effect than substitution for the last aspartate (16, 17). In fact, D621 is essential for catalysis (Table 1); alanine substitution reduces kcat to the level observed for wild-type enzyme in the absence of the required divalent metal ion, likely reflecting its role in ligating both Mga2+ and Mgc2+. In contrast, alanine substitution for D625 and E699, which are involved in ligating Mga2+ only, have relatively smaller effects (i.e., reduction in kcat by 2 to 3 orders of magnitude).
Perhaps the most surprising kinetic observation was the absolute requirement for R762; R762A is essentially inactive, exhibiting kcat = 10−5 s−1 (Table 1). This effect was significantly greater than that of R584A (kcat = 6 × 10−4 s−1), a residue postulated to act with R762 in binding and stabilizing the departing diphosphate anion. Clearly, R762 plays a much more active role in the reaction, perhaps because of its interaction with D766, which is itself important for catalysis (kcat ≈ 10−3 s−1 for D766A); no role in catalysis has yet been suggested for the aspartate residue corresponding to D766. Substitution of the second arginine in the RXR motif (R586) had relatively minor influence, and, perhaps more surprisingly, mutation of this residue did not lead to the abortive formation of diterpenol products, as might have been expected from its postulated role in organizing a “lid” over the active site to exclude water.
The three residues thought to be involved in ligating Mgb2+ (N765, T769, and E773) all seem important for catalysis, because alanine substitution for each leads to an approximately four-orders-of-magnitude decrease in turnover (Table 1). This observation suggests that Mgb2+, although it only interacts with the substrate diphosphate through an intermediary water in 5-EAS, is important in AS and may be of greater catalytic significance than the other magnesium ions. Furthermore, mutations at these three positions all affect product distribution (Fig. 3). Although T769A exhibits only a slight increase in the proportion of neoabietadiene, this effect is dramatically more pronounced with E773A, which also produces more sandaracopimaradiene. N765A exhibits an extreme phenotype in producing only sandaracopimaradiene, and this mutant thus seems incapable of mediating the intramolecular proton transfer required for the 1,2-methyl migration step that creates the abietane skeleton (10). The selective production of sandaracopimaradiene by N765A supports previous indirect evidence for a sandaracopimarenyl intermediate in the cyclization and rearrangement reaction (Fig. 1; refs. 10 and 11; M. M. Ravn, R.J.P., R. M. Coates, and R.B.C., unpublished work). In contrast, mutant Y841F [corresponding to Y520 of 5-EAS involved in a crucial reprotonation reaction (20) which is similar to that mediated by AS] had very little effect on catalysis, and no influence on product distribution. D845 (the corresponding residue in 5-EAS has been postulated to act as a base) is clearly catalytically important (kcat = 10−4 s−1 for D845A) but does not influence product distribution. These results indicate that, although D845 is catalytically important, AS does not use Y841 or D845 in acid/base catalysis.
To generate the abietane skeleton with its characteristic C13 isopropyl group, AS mediates an intramolecular proton transfer to promote 1,2-methyl migration within the sandaracopimarenyl intermediate (10). This reaction sequence (Fig. 1) entails an energetically unfavorable transition from a tertiary to a secondary carbocation (11) in which the latter species is stabilized in part by ionic interaction with the paired diphosphate anion (M. M. Ravn, R.J.P., R. M. Coates, and R.B.C., unpublished work). Therefore, it is suggested that Mgb2+ may be involved in binding and positioning the diphosphate anion. Thus, elimination of Mgb2+ ligands would lead to mispositioning of the diphosphate and decreased stabilization of the secondary sandaracopimarenyl carbocation to promote formation of sandaracopimardiene. The changes in product distribution observed with T679A and E773A might also result from misalignments of the paired diphosphate anion, which favors formation of neoabietadiene.
In conclusion, we have presented a detailed mutational analysis of the AS active site responsible for the cyclization and rearrangement of CPP to abietadienes. Based on kinetic analysis of these mutants, it is suggested that D621, the first aspartate in the canonical DDXXD motif, and R762, a completely conserved arginine, play particularly critical roles in catalysis. D621 may be involved in ligating two of the required Mg2+ ions, which could account for this critical influence in catalysis. R762 may be involved in interactions with D766, another catalytically important residue, and in stabilizing and positioning the diphosphate leaving group. Although reaction pH affects the product distribution of AS, the similar influence of buffer composition at fixed pH and the ability of the T617A mutation to change product distribution suggest that choice between alternatives for deprotonation of the abietenyl carbocation depends largely on positioning effects rather than on the pKa of multiple participating bases. All three residues presumed to chelate Mgb2+ have some effect on product distribution. In particular, N765A produces only sandaracopimaradiene, thereby providing strong evidence for this stereochemistry of the pimarenyl intermediate of the reaction pathway. Finally, the results presented suggest a role for Mgb2+ in binding and positioning the paired diphosphate anion to stabilize the secondary carbocation arising from intramolecular proton transfer, and in promoting secondary effects on deprotonation alternatives in the final abietenyl carbocation.
Acknowledgments
We thank Dr. E. Davis for assistance in modeling AS and for productive discussions. This work was supported by National Institutes of Health Grant GM31354 (to R.B.C.) and by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research (to R.J.P.).
Abbreviations
- AS
abietadiene synthase
- rAS
the recombinant “pseudomature” AS
- CPP
copalyl diphosphate
- GGPP
(E,E,E)-geranylgeranyl diphosphate
- 5-EAS
5-epi-aristolochene synthase
References
- 1.Phillips M A, Croteau R B. Trends Plant Sci. 1999;4:184–190. doi: 10.1016/s1360-1385(99)01401-6. [DOI] [PubMed] [Google Scholar]
- 2.Johnson M, Croteau R. Am Chem Soc Symp Ser. 1987;209:76–92. [Google Scholar]
- 3.Funk C, Croteau R. Arch Biochem Biophys. 1994;308:258–266. doi: 10.1006/abbi.1994.1036. [DOI] [PubMed] [Google Scholar]
- 4.LaFever R E, Stofer Vogel B, Croteau R. Arch Biochem Biophys. 1994;131:139–149. doi: 10.1006/abbi.1994.1370. [DOI] [PubMed] [Google Scholar]
- 5.Peters R J, Flory J E, Jetter R, Ravn M M, Lee H-J, Coates R M, Croteau R B. Biochemistry. 2000;39:15592–15602. doi: 10.1021/bi001997l. [DOI] [PubMed] [Google Scholar]
- 6.Peters R J, Ravn M M, Coates R M, Croteau R B. J Am Chem Soc. 2001;123:8974–8978. doi: 10.1021/ja010670k. [DOI] [PubMed] [Google Scholar]
- 7.Saito T, Abe H, Yamane H, Sakurai A, Murofushi N, Takio K, Takahashi N, Kamiya Y. Plant Physiol. 1995;109:1239–1245. doi: 10.1104/pp.109.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan J D, West C A. Plant Physiol. 1981;68:1128–1134. doi: 10.1104/pp.68.5.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters, R. J. & Croteau, R. B. (2002) Biochemistry, in press. [DOI] [PubMed]
- 10.Ravn M M, Coates R M, Jetter R, Croteau R. J Chem Soc Chem Commun. 1998;1998:21–22. [Google Scholar]
- 11.Ravn M M, Coates R M, Flory J, Peters R J, Croteau R. Org Lett. 2000;2:573–576. doi: 10.1021/ol991230p. [DOI] [PubMed] [Google Scholar]
- 12.Starks C M, Back K, Chappell J, Noel J P. Science. 1997;277:1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 13.Lesburg C A, Zhai G, Cane D E, Christianson D W. Science. 1997;277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- 14.Bohlmann J, Meyer-Gauen G, Croteau R. Proc Natl Acad Sci USA. 1998;95:4126–4133. doi: 10.1073/pnas.95.8.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trapp S C, Croteau R B. Genetics. 2001;158:811–832. doi: 10.1093/genetics/158.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis E M, Croteau R. Top Curr Chem. 2000;209:53–95. [Google Scholar]
- 17.Koyama T, Ogura K. In: Isoprenoids Including Carotenoids and Steroids. Cane D E, editor. Vol. 2. Oxford: Elsevier Science; 1999. pp. 69–96. [Google Scholar]
- 18.Stofer Vogel B, Wildung M R, Vogel G, Croteau R. J Biol Chem. 1996;271:23262–23268. doi: 10.1074/jbc.271.38.23262. [DOI] [PubMed] [Google Scholar]
- 19.Peitsch M C, Guex N. Biochem Soc Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- 20.Rising K A, Starks C M, Noel J P, Chappell J. J Am Chem Soc. 2000;122:1861–1866. [Google Scholar]
- 21.Schepmann H G, Pang J, Matsuda S P. Arch Biochem Biophys. 2001;392:263–269. doi: 10.1006/abbi.2001.2438. [DOI] [PubMed] [Google Scholar]