Abstract
We discovered a thermostable enzyme from the archaeon Pyrococcus furiosus (Pfu), which increases yields of PCR product amplified with Pfu DNA polymerase. A high molecular mass (>250 kDa) complex with PCR-enhancing activity was purified from Pfu extracts. The complex is a multimer of two discrete proteins, P45 and P50, with significant similarity to bacterial dCTP deaminase/dUTPase and DNA flavoprotein, respectively. When tested in PCR, only recombinant P45 exhibited enhancing activity. P45 was shown to function as a dUTPase, converting dUTP to dUMP and inorganic pyrophosphate. Pfu dUTPase improves the yield of products amplified with Pfu DNA polymerase by preventing dUTP incorporation and subsequent inhibition of the polymerase by dU-containing DNA. dUTP was found to accumulate during PCR through dCTP deamination and to limit the efficiency of PCRs carried out with archaeal DNA polymerases. In the absence of dUTP inhibition, the combination of cloned Pfu DNA polymerase and Pfu dUTPase (PfuTurbo DNA polymerase) can amplify longer targets in higher yield than Taq DNA polymerase. In vivo, archaeal dUTPases may play an essential role in preventing dUTP incorporation and inhibition of DNA synthesis by family B DNA polymerases.
The use of high-fidelity DNA polymerases in the polymerase chain reaction (PCR) is important for minimizing amplification errors in products that will be cloned, sequenced, and expressed. Several archaeal DNA polymerases with 3′- to 5′-exonuclease-dependent proofreading activity have been commercialized for high-fidelity PCR amplification. Of the enzymes characterized to date, Pyrococcus furiosus (Pfu) DNA polymerase exhibits the highest fidelity, with an average error rate estimated to be 2- to 60-fold lower than other proofreading enzymes and 6- to 100-fold lower than Taq DNA polymerase (1–4), which lacks proofreading activity.
Although inherent high fidelity and other unique biochemical properties of Pfu DNA polymerase have been exploited in numerous molecular biology procedures, including blunt-end cloning (5), amplification of GC-rich targets (6), mutation detection (7), ligation-independent cloning (8), site-directed mutagenesis (9), genotyping (10), and trinucleotide repeat analyses (11), it has been reported that proofreading DNA polymerases tend to perform less reliably than Taq DNA polymerase (4, 12). PCRs conducted with Taq are typically more efficient and require less optimization than reactions carried out with proofreading DNA polymerases. Lower product yields with Pfu DNA polymerase have been attributed to the enzyme's relatively slow polymerization rate (550 vs. 2,800 nucleotides per min for Taq) and interference from its associated 3′- to 5′-exonuclease activity (13).
Improvements in the performance of Pfu DNA polymerase have been achieved by increasing PCR extension times (2 min/kb of target; Stratagene Pfu DNA polymerase manual), incorporating additives such as DMSO (6, 14), and mixing Pfu with Taq DNA polymerase (4). Although DNA polymerase mixtures synthesize higher yields of product and allow amplification of longer templates than is possible with single-enzyme formulations (4), long-range blends exhibit lower fidelity than Pfu DNA polymerase because of the relatively high proportion of Taq used (1).
In addition to Taq, there are other proteins that might be expected to enhance the performance of Pfu DNA polymerase. For example, polymerase accessory proteins have been shown to dramatically increase the processivity of replicative DNA polymerases from eukaryotes, Escherichia coli, and bacteriophages (15–17). The discovery of thermostable polymerase accessory factors has been reported in thermophilic bacteria (18) and archaea (19), but with the exception of single-stranded DNA binding protein (20, 21), the potential effect of such proteins on PCR amplification is presently unknown. In addition, proteins that remove inhibitors would also be expected to serve as PCR enhancers. For example, pyrophosphatase facilitates DNA polymerase-catalyzed reactions by eliminating inorganic pyrophosphate (PPi) generated during incorporation of nucleoside triphosphates (22).
In an effort to further improve the performance of Pfu DNA polymerase, we screened Pfu extracts for factors or proteins with PCR-enhancing activity. We report our success in identifying and cloning a dUTPase that increases yields of product amplified with Pfu DNA polymerase. As we will show, Pfu dUTPase enhances PCRs by preventing incorporation of dUTP, which arises from dCTP deamination during temperature cycling.
Materials and Methods
Reagents.
All molecular biology reagents were from Stratagene, unless otherwise noted. VentR and Deep VentR DNA polymerases were purchased from New England Biolabs. Individual dNTPs were obtained from Amersham Pharmacia, whereas dNDPs and dNMPs were from Sigma.
PCR Assays.
Pfu extracts or purified proteins were added to amplifications of 5- to 8-kb genomic targets conducted with cloned Pfu DNA polymerase, using enzyme concentrations (≤2.5 units/100 μl) and/or extension times (≤1 min/kb of target), which were insufficient for significant product synthesis. Polymerase-enhancing activity was identified by a visible increase in product yield.
Purification/Analysis of Native PCR-Enhancing Activity.
Polymerase-enhancing activity was isolated from Pfu extracts by sequential purification over Q, SP, and heparin Sepharose columns, followed by a Sephacryl S-200 gel filtration column. Fractions were analyzed by SDS/PAGE, and 45 kDa and 50 kDa bands were subject to N-terminal and internal sequence analysis.
Cloning and Preparation of Recombinant Proteins.
To isolate DNA sequences encoding Pfu P50 and P45, a Pfu genomic library was constructed in the Lambda ZAP II vector and screened by hybridization. Probes were prepared by Vectorette technology (P45; Genosys Biotechnologies) or by PCR using degenerate primers designed from chemically determined N-terminal and internal sequences (P50). Positives were excised and inserts were analyzed by DNA sequencing. Coding regions were subcloned into the pCAL-n-EK vector (Affinity Protein Expression and Purification System), which contains an upstream, in-frame calmodulin-binding peptide (CBP) tag for purifying fusion proteins with calmodulin-agarose.
A fragment encoding human dUTPase (GenBank accession no. U62891) was amplified from reverse-transcribed placental RNA (cDNA), purified, and cloned into the pCAL-n-EK vector. Positives were analyzed by DNA sequencing. Human dUTPase (CBP-fusion protein) was purified and assayed for dUTPase activity (see below).
HPLC Analysis of P45 Reaction Products.
P45 samples were incubated with 10 mM dNTP in 1× cloned Pfu buffer (as defined by Stratagene). The reaction products were chromatographed (TriLink BioTechnologies, San Diego) on a Waters Delta-pak C-18 column (3.9 × 250 cm; 300 Å/15 μm), equilibrated with 50 mM triethylammonium acetate, pH 7.0 (A). Products were eluted (2 ml/min) with acetonitrile (B) using the gradient 0% B for 5 min, 0–10% B for 20 min. Absorbance at 260 nm was monitored with a photodiode array detector, and peak areas were integrated. Nucleotide standards were prepared and chromatographed in a similar fashion.
dUTPase Assay.
PPi production was quantified by using the enzymatic determination kit from Sigma.
Results
PCR-Enhancing Activity.
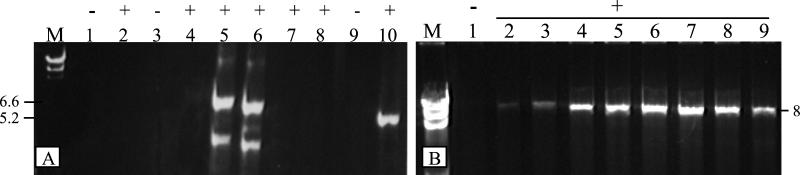
We tested Pfu fractions for the presence of PCR-enhancing activity. Samples were added to amplifications of relatively long (5–8 kb) targets, carried out with cloned Pfu DNA polymerase under limiting conditions, such that product synthesis was barely detectable. The addition of active fractions brought about significant increases in PCR product yield, irrespective of the primers and template used (Fig. 1). Increased product synthesis was not due to adding contaminating DNA template or native Pfu DNA polymerase, as PCR-enhancing fractions failed to generate product when added to reactions lacking exogenous DNA template or cloned Pfu DNA polymerase (Fig. 1A, lanes 1–4). Titration experiments with Sephacryl S-200-purified protein indicated that optimal enhancement was achieved by using a fairly broad (≈200-fold) range of protein amounts (Fig. 1B).
Figure 1.
PCR-enhancing activity from Pfu. Fractions eluted from heparin-Sepharose (A) or Sephacryl S-200 (B) columns were added to PCRs using Pfu DNA polymerase. (A) A 6.6-kb (lanes 1–8) or 5.2-kb (lanes 9–10) genomic target was amplified in the presence (+) of the following amount of eluate: lanes 1, 3, and 9, none (−); lanes 2, 4, 5, and 10, 1 μl of undiluted; lane 6, 1 μl of diluted 10−1; lane 7, 1 μl of diluted 10−3; and lane 8, 1 μl of diluted 10−5. As negative controls, either Pfu DNA polymerase (A, lanes 1 and 2) or genomic DNA (A, lanes 3 and 4) was omitted. The 6.6-kb PCR system produces a second nonspecific band (lower band in lanes 5 and 6) irrespective of Pfu P45 addition. (B) The following amount (ng) of S-200-purified protein was added (+) to amplifications of an 8-kb genomic target: lane 1, none (−); lane 2, 0.1; lane 3, 0.2; lane 4, 0.5; lane 5, 1; lane 6, 5; lane 7, 25; lane 8, 100; or lane 9, 200.
Composition of Purified PCR-Enhancing Activity.
Peak PCR-enhancing activity eluted from a Sephacryl S-200 column within the void volume (>250-kDa cut-off; data not shown). SDS/PAGE analysis revealed that the active pool consisted of two discrete proteins with apparent molecular masses of 50 kDa and 45 kDa (data not shown), which will be referred to here as P50 and P45. Further analysis showed that P45 migrates with an apparent molecular mass of 18 kDa when the complex is denatured with heat and trichloroacetic acid, suggesting that P45 is a dimer or trimer of 18-kDa subunits or simply migrates anomalously when incompletely denatured. The molar ratio of P50 to P45 in the native complex could not be estimated from SDS/PAGE gels because of differential staining with various protein stains (data not shown).
PCR-Enhancing Activity of Recombinant Proteins.
As a first step toward elucidating the mechanism of action of the Pfu PCR-enhancing complex, we cloned the genes encoding P50 and P45 (GenBank accession nos. AY066005 and AY066006). Recombinant proteins were expressed and purified as N-terminal CBP fusions. When tested in PCR, only recombinant P45 exhibited polymerase-enhancing activity (data not shown). In contrast, recombinant P50 had no effect on PCR product yields when added alone or used in conjunction with various amounts of P45. We concluded that P45 was the active component of the native PCR-enhancing complex.
Identification of Proteins Related to P45.
The calculated molecular mass (18.6 kDa) of the translated amino acid sequence of the p45 clone agrees with its apparent molecular mass (18 kDa) under acidic denaturation conditions. A blastx search conducted in multiple databases revealed that the P45 sequence exhibited the greatest similarity to bacterial dCTP deaminase (dcd gene) and to dUTPases (dut genes) (data not shown).
The biochemical properties of dCTP deaminase have been extensively studied in bacteria, where dCTP deaminase is a homotetramer that catalyzes the conversion of dCTP to dUTP and NH3 (23). DNA sequences exhibiting similarity to the E. coli dcd gene have also been identified in archaea, but dCTP deaminase is not thought to be present in eukaryotes (24, 25). dUTPase, on the other hand, is a ubiquitous enzyme, found in bacteria, eukaryotes, eukaryotic viruses, and archaea, that converts dUTP to dUMP and PPi (25–28). The initial description of an archaeal dUTPase was an enzyme cloned from Sulfolobus islandicus virus (SIRV) (25). Like Pfu P45, SIRV dUTPase exhibited striking sequence similarity to bacterial dCTP deaminase and to dUTPases. However, enzyme activity studies showed that recombinant SIRV dUTPase hydrolyzes dUTP to dUMP and lacks detectable dCTP deaminase activity (25).
Identification of Proteins Related to P50.
A blastx search revealed that the P50 sequence exhibited very strong homology to the DNA flavoprotein (Dfp), implicated in DNA synthesis and pantothenate metabolism in E. coli (29, 30). In vitro, E. coli Dfp promotes decarboxylation of the terminal cysteine moiety of R-4′-phospho-N-pantothenoylcysteine to R-4′-phosphopantetheine in CoA biosynthesis (31). Sequences related to E. coli Dfp have been identified previously in archaea (24), but the properties of archaeal homologs have not been studied.
HPLC Analysis of P45 Substrates.
Sequence similarity of P45 to dCTP deaminase and dUTPase prompted us to test whether dCTP and dUTP were substrates of P45. The native PCR-enhancing complex was incubated with various nucleotides at 72°C, and reaction products were analyzed by reverse-phase HPLC. Under these conditions, native P45 used both dCTP and dUTP as substrates (data not shown). Based on retention times relative to standards and λmax (272 nm), the dCTP reaction product was identified as dCMP, rather than dUTP as expected for a dCTP deaminase. In addition, the P45 + dUTP reaction product was identified as dUMP, based on retention time, λmax (263 nm), and mass spectroscopy (data not shown). Additional studies showed that UTP, UMP, dNMPs, dNDPs, dATP, TTP, and dGTP do not serve as substrates for P45 (data not shown).
Recombinant P45 was found to use the same substrates as native P45, converting dUTP to dUMP and dCTP to dCMP (data not shown). dUTP was the preferred substrate for both native and recombinant P45. Under the same conditions that result in 100% conversion of dUTP to dUMP, native P45 converted only 16.6% of dCTP to dCMP. Moreover, when dCTP and dUTP were incubated together with native P45, dUMP was the only product detected (data not shown).
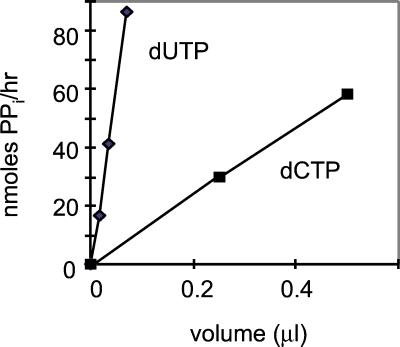
P45 Produces PPi.
We confirmed that PPi is generated as a byproduct of dUTP to dUMP and dCTP to dCMP conversion. PPi formation was detected in reactions of native (Fig. 2) and recombinant (data not shown) P45 with dUTP and dCTP. At 85°C, the amount of PPi produced from dUTP was 10- to 20-fold higher than the amount of PPi generated from dCTP for both native and recombinant P45. No PPi was detected in control reactions lacking enzyme or containing dATP instead of dUTP.
Figure 2.
PPi formation by native P45. The indicated amounts of native P45 (700 ng/μl) were incubated with 10 mM dUTP or dCTP for 1 h at 85°C. PPi production was quantified as described in Material and Methods.
Inhibition of PCR with dUTP.
Inherent dUTPase activity suggested that Pfu P45 enhances PCR amplifications by eliminating dUTP (through conversion to dUMP) and preventing incorporation of dU into PCR products. Lasken et al. (32) have shown that archaeal DNA polymerases are inhibited by dU-containing DNA. Although dUTP is not intentionally added to PCR amplification reactions, it is possible that low levels of dUTP are present in commercial nucleotide preparations or are generated by dCTP deamination during PCR.
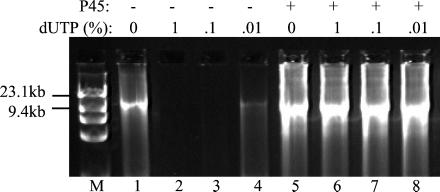
To address the possibility that P45 enhances PCR by eliminating dUTP, we measured the effects of dUTP on PCRs using Pfu DNA polymerase and P45. In Fig. 3, amplification of a 10-kb target was almost completely inhibited by as little as 0.02 μM dUTP (0.01% TTP + dUTP pool). dUTP inhibition also has been observed when amplifying other primer-template systems with Pfu DNA polymerase (ref. 33 and data not shown). The dUTP concentration that completely inhibited PCR varied with each amplification system, such that those using long or low-copy-number targets appeared to be the most sensitive to the lowest amounts (0.02–0.2 μM) of dUTP tested (data not shown). Unlike dUTP, the dUMP byproduct had no effect on PCR product yields (data not shown).
Figure 3.
P45 prevents inhibition of PCR by dUTP. A 10-kb target was amplified from λ phage DNA by using 2.5 units of Pfu DNA polymerase in the absence (−, lanes 1–4) or presence (+, lanes 5–8) of native P45 (1 ng). dUTP was present in the amounts indicated (percentage TTP + dUTP pool) in reactions run in lanes 2–4 and 6–8. M, HindIII-digested λ phage DNA and HaeIII-digested φX174 phage DNA markers.
No inhibition was observed in PCR mixtures containing P45. In the example shown, 1 ng of native P45 was sufficient to prevent inhibition caused by 2 μM dUTP (1% TTP pool) (Fig. 3, lane 6). In other PCR systems, similar amounts of native and recombinant P45 overcame the inhibitory effects of up to 20 μM dUTP (data not shown). Additional experiments showed that P45 was ineffective in preventing the inhibition of PCRs by dU-oligonucleotides (conducted as in ref. 32; data not shown). Thus, P45 can significantly increase PCR product yields in cases in which inhibitory concentrations of dUTP, but not dU-DNA (incorporated dUTP), are present.
dUTP Accumulation During PCR.
The potent inhibitory effects of dUTP, coupled with the discovery that dUTPase exhibits PCR-enhancing activity, imply that dUTP is present during PCR cycling. We looked for evidence that sufficient dUTP is generated as a by-product of dCTP deamination (34) during temperature cycling to inhibit Pfu DNA polymerase. dCTP solutions were heated at 95°C (PCR denaturation temperature) or subject to PCR cycling in the presence of cloned Pfu buffer. Heat-generated products were analyzed by HPLC and identified on the basis of retention time and λmax relative to standards.
Heating 10 mM dCTP solutions (1 h, 95°C) produced 25% dCDP, 3.5% dCMP, and 0.1% dUTP (Table 1). Cycling dCTP with or without Pfu DNA polymerase produced 0.064% dUTP under the same PCR conditions used in Fig. 3 (30 cycles, 10.5-min extension time). Assuming similar deamination rates for 10 mM and 0.2 mM dCTP solutions (Table 1 legend), the percentage of dCTP converted to dUTP during temperature cycling (0.06% accumulated after 30 cycles; Table 1) is within the range of dUTP concentrations found to inhibit amplification of long targets (≥0.01–0.1%; Fig. 3). When dCTP is cycled in the presence of native P45, dUTP accumulation is not detected (Table 1). This result is consistent with the proposal that P45 eliminates (by conversion to dUMP) the small amount of dUTP that arises during thermal cycling.
Table 1.
Products generated by heating/cycling dCTP
| Reaction conditions | Product yield, %
|
|||
|---|---|---|---|---|
| dCTP (8.1–8.7 min; 272 nm) | dCDP (5.1–5.3 min; 272 nm) | dCMP (3.3 min; 272 nm) | dUTP (11.9–12.2 min; 263 nm) | |
| dCTP (+ Pfu) | ||||
| None | 99.0 | 1.0 | 0 | 0 |
| Cycled* | 79.0 (79.0) | 19.1 (19.2) | 1.8 (1.7) | 0.064 (0.065) |
| 95°C/1 h | 71.6 (71.4) | 24.6 (24.8) | 3.5 (3.6) | 0.104 (0.081) |
| 95°C/2.5 h | 48.5 (47.6) | 37.7 (38.3) | 13.5 (13.9) | 0.115 (0.133) |
| dCTP + P45 | ||||
| Cycled* | 73.7 | 18.7 | 7.9 | 0 |
| 95°C/1 h | 70.0 | 24.6 | 5.3 | 0 |
| 95°C/2.5 h | 45.7 | 37.3 | 16.9 | 0 |
The retention times and λmax are given in parentheses for each nucleotide. The 50× PCR conditions are as follows: 10 mM dCTP heated/cycled alone or with 0.125 unit/μl Pfu DNA polymerase (dCTP + Pfu DNA polymerase product yields in parentheses) or 0.5 ng/μl native P45.
Thirty cycles: 95°C for 1 min, 60°C for 1 min, 72°C for 10.5 min.
Cytosine nucleotide levels are also affected by cycling dCTP in the presence of P45. In Table 1, dCTP and dCMP concentrations were 5% lower and 6% higher, respectively, in cycled dCTP + P45 samples as compared with dCTP samples. These data indicate that the dCTPase activity associated with P45 can further reduce dCTP levels during PCR cycling, albeit slightly. The small drop in dCTP levels (5%) is not expected to reduce product yield or polymerase fidelity when relatively low P45 concentrations are used. In fact, we found no difference in fidelity when the error rate of Pfu DNA polymerase was measured (1) in the presence of P45, at the same [P45]/[nucleotide] ratios used in Table 1 (data not shown).
In addition to dUTP arising from dCTP deamination, dUTP also may be present in PCRs as a contaminant of commercial nucleotide preparations. The nucleotide mixes used here were produced from dCTP lots lacking detectable dUTP (<0.01% dUTP by HPLC).
Human dUTPase Has PCR-Enhancing Activity.
We ruled out the possibility that factors other than inherent dUTPase activity were responsible for, or at least contributed to, the PCR-enhancing activity of P45. For example, P45 may interact directly with Pfu DNA polymerase and simply enhance polymerase stability during temperature cycling. Alternatively, archaeal dCTP deaminase/dUTPase homologs may possess additional enzymatic activities that have not yet been identified.
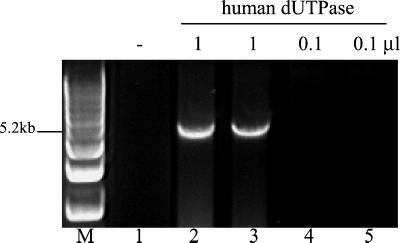
To eliminate these possibilities, we tested whether a distantly related dUTPase could also enhance PCRs carried out with Pfu DNA polymerase. For this study, we chose human dUTPase (34% amino acid identity to P45) because, unlike Pfu P45, it is relatively thermolabile and more closely related to dUTPases than to dCTP deaminase. Aliquots of CBP-tagged human dUTPase were added manually at each annealing step (58°C) of a 30-cycle PCR. Enzyme assays showed that human dUTPase retained detectable activity at 58°C for at least 2 min (data not shown). In the presence of human dUTPase, Pfu DNA polymerase successfully amplified a 5.2-kb genomic target, which was too difficult for the enzyme to amplify alone (Fig. 4).
Figure 4.
Human dUTPase exhibits PCR-enhancing activity. A 5.2-kb target was amplified with 5 units of Pfu DNA polymerase. At each annealing step (58°C), 0.5 μl of the following was added: buffer (lane 1) or human dUTPase preparation, undiluted (lanes 2 and 3), or diluted 10−1 (lanes 4 and 5).
P45 Enhances Other Archaeal DNA Polymerases.
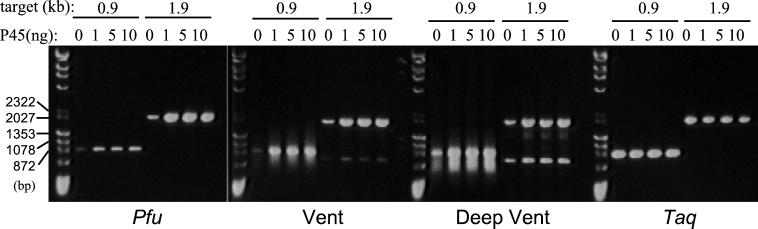
Because dU-DNA inhibition appears to be a property of all archaeal family B DNA polymerases (32), we tested whether Pfu P45 could improve PCRs by using enzymes other than Pfu DNA polymerase. As shown in Fig. 5, PCR product yields were improved by adding P45 to reactions conducted with VentR and Deep VentR (from the archaea Thermococcus litoralis and Pyrococcus sp. GB-D, respectively). Additional studies showed that P45 also increases the yield of products amplified with Pwo (identical to Pfu DNA polymerase; ref. 35) and KOD DNA polymerases (data not shown). In contrast, P45 had no effect on PCRs performed with Taq (from the eubacterium Thermus aquaticus) (Fig. 5), consistent with the relative insensitivity of eubacterial DNA polymerases to dU-DNA (32).
Figure 5.
Pfu P45 enhances PCRs using archaeal DNA polymerases. Native P45 (0, 1, 5, or 10 ng) was added to amplifications of 0.9-kb and 1.9-kb genomic targets. PCRs were carried out with 2.5 units of Pfu DNA polymerase or 1 unit of VentR, Deep VentR, or Taq DNA polymerase in each enzyme's optimal PCR buffer.
P45 Improves Long PCR.
The amount of dUTP produced by dCTP deamination during PCR should increase with cycle number and extension time (typically 1 min/kb of target). Thus, dUTP poisoning is expected to significantly limit Pfu DNA polymerase reactions that use extended cycling times (e.g., PCRs of long targets) or reach exponential phase in later cycles when dUTP levels are highest (e.g., PCRs of low-copy-number targets). Although dU-DNA sensitivity is likely to impair amplification of long or complex targets when Pfu DNA polymerase is used, insensitivity to dU-DNA may contribute to the reliability of Taq (4, 12).
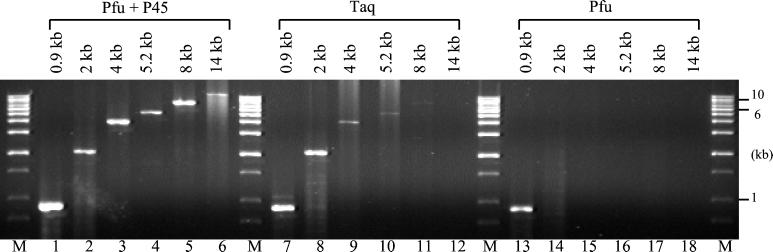
To assess the contribution of dUTP poisoning to PCR performance, we amplified a series of long targets by using Taq or Pfu DNA polymerase, with and without P45 (Fig. 6). The mixture of Pfu DNA polymerase and P45 used in these studies has a composition similar to PfuTurbo DNA polymerase (Stratagene). As expected, P45 dramatically improved amplification of long targets when Pfu DNA polymerase was used. In the absence of P45, Pfu DNA polymerase failed to amplify targets >1 kb in length when using 1 min/kb extension times (2 min/kb recommended by Stratagene). In the presence of P45, Pfu DNA polymerase successfully amplified 0.9- to 8-kb genomic targets and a 14-kb λ DNA target (Fig. 6). In comparison, Taq exhibited difficulty amplifying genomic targets >2 kb in length (Fig. 6). Therefore, in the absence of dUTP poisoning, Pfu DNA polymerase exhibits greater target-length capability compared with Taq DNA polymerase.
Figure 6.
Pfu P45 improves amplification of long targets by Pfu DNA polymerase. PCRs were performed with Taq or Pfu DNA polymerase, in the absence or presence of P45, in each enzyme's recommended buffer. The following PCR products were amplified: lanes 1–4, 7–10, and 13–16, 0.9- to 5.2-kb fragments of the human α-1-antitrypsin gene; lanes 5, 11, and 17, 8-kb fragment from mouse genomic DNA; and lanes 6, 12, and 18, 14-kb fragment from λ phage DNA. PCR amplifications were conducted by using 2.5 units (lanes 4, 10, and 16) or 5 units of each enzyme and an extension time of 1 min/kb of target.
Discussion
We have demonstrated that dUTPase enhances the performance of Pfu DNA polymerase in PCR. During the course of these experiments, we identified dUTP as a potent proinhibitor (precursor to dU-DNA) of PCRs carried out with Pfu DNA polymerase (Fig. 3) and related archaeal DNA polymerases (Fig. 5). dUTP was shown previously to inhibit PCRs using Pfu and Vent DNA polymerases under conditions in which TTP was completely replaced by dUTP (33). We found that when amplifying relatively long targets, product yields can be significantly reduced by as little as 0.02 μM dUTP. Pfu P45 was shown to prevent the inhibitory effects of added dUTP (Fig. 3) by converting dUTP to dUMP and PPi.
The discovery that dUTPases exhibit PCR-enhancing activity, and that dUTP is a potent inhibitor of PCRs using archaeal DNA polymerases, implies that dUTP is present during PCR amplification. We identified hydrolytic deamination of dCTP as a source of dUTP in PCRs. In a mock PCR, we found that the amount of dUTP produced from dCTP (≈0.06% dUTP, 30 cycles) was within the range that inhibited amplification of a 10-kb target (≥0.01% dUTP) when added before cycling.
The deleterious effect of dUTP is presumably related to the strong inhibition of archaeal DNA polymerases by single- and double-stranded dU-containing DNA. Unlike eubacterial, eukaryotic, and bacteriophage DNA polymerases, archaeal family B DNA polymerases (Pfu, Vent, Deep Vent) exhibit the unique property of tightly binding dU-containing oligonucleotides (32). Greagg et al. (36) have shown that archaeal DNA polymerases recognize dU residues in the template strand and stall DNA synthesis. This read-ahead function may represent the first step in a pathway to repair premutagenic lesions caused by cytosine deamination in archaeal DNA (36). During PCR, cytosine deamination could lead to dU-DNA formation/inhibition through a combination of two mechanisms: (i) dCTP deamination to dUTP, followed by incorporation (identified in this study), and (ii) dC deamination in DNA. Deamination rates, measured under PCR-like conditions (95°C, pH 7.4), indicate that cytosine residues in single-stranded DNA (e.g., primers, denatured template, or PCR products) are deaminated only 15% slower than dCMP (2.6 × 10−7 s−1; dCTP deamination not measured; ref. 34). Further studies are needed to address the potential contribution of dC-DNA deamination (dU-DNA formation) to the efficiency of PCRs carried out with archaeal DNA polymerases.
Relatively little is known about dU-DNA repair in archaea. In bacteria and eukaryotes, uracil is recognized and excised from DNA through a base excision repair pathway initiated by uracil DNA glycosylase or dU/T⋅dG-mismatch-specific DNA glycosylases (37–39). Similar processes for dU-DNA repair are likely to be used in archaea based on the recent discoveries of a uracil DNA glycosylase homolog in Archaeoglobus fulgidus (40) and dU/T⋅dG-mismatch-specific DNA glycosylases in Methanobacterium and Pyrobaculum species (41, 42).
In addition to dU-DNA repair enzymes, dUTPases play an essential role in preventing dUTP incorporation by maintaining low dUTP levels (26, 27). In bacteria and yeast lacking dUTPase, uracil DNA glycosylase and other components of the cellular excision repair process act upon dU-DNA, leading to DNA fragmentation and cell death. The discovery of dUTPases in Pfu and SIRV (25) suggests that archaea also rely on dUTPases to minimize dUTP incorporation and prevent initiating dU-DNA repair processes.
In bacteria, dUTP is generated by dCTP deaminase in the biosynthetic pathway for thymidine nucleotides (23). In contrast, eukaryotes appear to lack dCTP deaminase, and instead, produce dUMP (thymidylate synthetase substrate) primarily through deamination of dCMP by dCMP deaminase (43). dCTP deaminase/dUTPase homologs have been identified in the genomes of several archaea. Interestingly, two homologs were identified in the whole genome sequences of Methanococcus jannaschii (24) and Methanobacterium thermoautotrophicum (44), whereas only one homolog has been identified in blast searches of SIRV, Desulfurolobus ambivalens, Pfu, and Pyrococcus horikoshii genomes (ref. 25 and data not shown). In cases in which there are two homologs, it is not yet known whether one functions as a dUTPase and the other as a dCTP deaminase. Lack of an apparent dCTP deaminase in Pfu indicates that dUTP arises from other sources, such as hydrolytic deamination of dCTP at high temperatures or reduction of UTP by ribonucleotide reductase (45).
E. coli dUTPase exhibits a trimeric structure (50). The subunit composition of Pfu dUTPase is unknown, although both native and recombinant P45 migrate as apparent aggregates in nondenaturing SDS/PAGE (data not shown). In this study, native Pfu dUTPase was isolated in association with Dfp, migrating as an apparent complex in SDS/PAGE and gel filtration. Curiously, the genes encoding dUTPase and Dfp are located next to each other in the E. coli genome, although there has been no evidence of linked gene expression or biochemical association in bacteria (29, 30). The dfp and dut genes do not appear to be located near each other in the genomes of Pfu and other archaea (data not shown). The discovery of Dfp in archaea, and its possible biochemical association with dUTPase, has not further elucidated the physiological role of Dfp, which was shown previously to play an essential but unknown role in DNA replication and pantothenate metabolism in E. coli (29, 30).
The PCR-enhancing activity of Pfu dUTPase has obvious commercial applications. Stratagene has made available a more robust version of Pfu DNA polymerase containing Pfu dUTPase (PfuTurbo DNA polymerase), which allows high-fidelity amplification of a much broader range of target lengths and complexities. Moreover, dUTP poisoning has consequences for long-range PCR performed with Taq blends containing an archaeal DNA polymerase (e.g., Pfu) as the minor proofreading component (4). dUTP poisoning is expected to limit the contribution of archaeal proofreading enzymes to the fidelity and performance of blends, especially in later cycles when dUTP concentrations are highest. dU-DNA inhibition may help explain the reduced fidelity (3- to 6-fold) exhibited by Taq-based blends compared to Pfu DNA polymerase (1) and the observed restrictions on the percentage of Pfu DNA polymerase in blends (4).
Acknowledgments
We thank Frances Bai for technical assistance.
Abbreviations
- Pfu
Pyrococcus furiosus
- P45
45-kDa protein with PCR-enhancing/dUTPase activities
- P50
50-kDa protein in native PCR-enhancing complex
- CBP
calmodulin-binding peptide
- dU
deoxyuridine
- dC
deoxycytidine
- PPi
inorganic pyrophosphate
- SIRV
Sulfolobus islandicus virus
- Dfp
DNA flavoprotein
Footnotes
References
- 1.Cline J, Braman J C, Hogrefe H H. Nucleic Acids Res. 1996;24:3546–3551. doi: 10.1093/nar/24.18.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flaman J-M, Frebourg T, Moreau V, Charbonnier F, Martin C, Ishioka C, Friend S H, Iggo R. Nucleic Acids Res. 1994;22:3259–3260. doi: 10.1093/nar/22.15.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cha R S, Thilly W G. In: PCR Primer. Dieffenbach C W, Dveksler G S, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. [Google Scholar]
- 4.Barnes W M. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa G L, Weiner M P. Nucleic Acids Res. 1994;22:2423. doi: 10.1093/nar/22.12.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong S S, Eichler E E, Nelson D L, Hughes M R. Am J Med Genet. 1994;51:522–526. doi: 10.1002/ajmg.1320510447. [DOI] [PubMed] [Google Scholar]
- 7.Andre P, Kim A, Khrapko K, Thilly W G. Genome Res. 1997;7:843–852. doi: 10.1101/gr.7.8.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aslanidis C, de Jong P J. Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W, Malcolm B A. BioTechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- 10.Hu Y W, Balaskas E, Kessler G, Issid C, Scully L J, Murphy D G, Rinfret A, Giulivi A, Scalia V, Gill P. Nucleic Acids Res. 1998;26:5013–5015. doi: 10.1093/nar/26.21.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe M, Abe K, Akoi M, Kameya T, Itoyama Y, Shoji M, Ikeda M, Iizuka T, Hirai S. Neurol Res. 1996;18:16–18. doi: 10.1080/01616412.1996.11740370. [DOI] [PubMed] [Google Scholar]
- 12.Foord O S, Rose E A. PCR Methods Appl. 1994;3:S1490–S161. doi: 10.1101/gr.3.6.s149. [DOI] [PubMed] [Google Scholar]
- 13.Skerra A. Nucleic Acids Res. 1992;20:3551–3554. doi: 10.1093/nar/20.14.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith K T, Long C M, Bowman B, Manos M M. Amplifications. 1990;5:16–17. [Google Scholar]
- 15.Stillman B. Annu Rev Cell Biol. 1989;5:197–245. doi: 10.1146/annurev.cb.05.110189.001213. [DOI] [PubMed] [Google Scholar]
- 16.Kuriyan J, O'Donnell M. J Mol Biol. 1993;234:915–925. doi: 10.1006/jmbi.1993.1644. [DOI] [PubMed] [Google Scholar]
- 17.Modrich P, Richardson C C. J Biol Chem. 1975;250:5508–5514. [PubMed] [Google Scholar]
- 18.McHenry C S, Seville M, Cull M G. J Mol Biol. 1997;272:178–189. doi: 10.1006/jmbi.1997.1238. [DOI] [PubMed] [Google Scholar]
- 19.Cann I K O, Ishino S, Hayashi I, Komori K, Toh H, Morikawa K, Ishino Y. J Bacteriol. 1999;181:6591–6599. doi: 10.1128/jb.181.21.6591-6599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz K, Hansen-Hagge T, Bartram C. Nucleic Acids Res. 1990;18:1079. doi: 10.1093/nar/18.4.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang H, Keohavong P. DNA Cell Biol. 1996;15:589–594. doi: 10.1089/dna.1996.15.589. [DOI] [PubMed] [Google Scholar]
- 22.Kornberg A. In: Horizons in Biochemistry. Kasha H, Pullman P, editors. New York: Academic; 1962. pp. 251–264. [Google Scholar]
- 23.Wang L, Weiss B. J Bacteriol. 1992;174:5647–5653. doi: 10.1128/jb.174.17.5647-5653.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 25.Prangishvili D, Klenk H P, Jakobs G, Schiechen A, Hanselmann C, Holz I, Zillig W. J Biol Chem. 1998;273:6024–6029. doi: 10.1074/jbc.273.11.6024. [DOI] [PubMed] [Google Scholar]
- 26.El-Hajj H H, Zhang H, Weiss B J. J Bacteriol. 1988;170:1069–1075. doi: 10.1128/jb.170.3.1069-1075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gadsden M H, McIntosh E M, Game J C, Wilson P J, Haynes R H. EMBO J. 1993;12:4425–4431. doi: 10.1002/j.1460-2075.1993.tb06127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elder J H, Lerner D L, Hasselkus-Light C S, Fontenot D J, Hunter E, Luciw P A, Montelaro R C, Phillips T R. J Virol. 1992;66:1791–1794. doi: 10.1128/jvi.66.3.1791-1794.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spitzer E D, Weiss B. J Bacteriol. 1985;164:994–1003. doi: 10.1128/jb.164.3.994-1003.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spitzer E D, Jiminez-Billini H E, Weiss B. J Bacteriol. 1988;170:872–876. doi: 10.1128/jb.170.2.872-876.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kupke T, Uebele M, Schmid D, Jung G, Blaesse M, Steinbacher S. J Biol Chem. 2000;275:31838–31846. doi: 10.1074/jbc.M004273200. [DOI] [PubMed] [Google Scholar]
- 32.Lasken R S, Schuster D M, Rashtchian A. J Biol Chem. 1996;271:17692–17696. doi: 10.1074/jbc.271.30.17692. [DOI] [PubMed] [Google Scholar]
- 33.Slupphaug G, Alseth I, Eftedal I, Volden G, Krokan H E. Anal Biochem. 1993;211:164–169. doi: 10.1006/abio.1993.1248. [DOI] [PubMed] [Google Scholar]
- 34.Lindahl T, Nyberg B. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- 35.Dabrowski S, Kur J. Protein Expression Purif. 1998;14:131–138. doi: 10.1006/prep.1998.0945. [DOI] [PubMed] [Google Scholar]
- 36.Greagg M A, Fogg M J, Panayotou G, Evans S J, Connolly B A, Pearl L H. Proc Natl Acad Sci USA. 1999;96:9045–9050. doi: 10.1073/pnas.96.16.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindahl T. Proc Natl Acad Sci USA. 1974;71:3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neddermann P, Jiricny J. J Biol Chem. 1993;268:21218–21224. [PubMed] [Google Scholar]
- 39.Gallinari P, Jiricny J. Nature (London) 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]
- 40.Sandigursky M, Franklin W A. J Biol Chem. 2000;275:19146–19149. doi: 10.1074/jbc.M001995200. [DOI] [PubMed] [Google Scholar]
- 41.Horst J-P, Fritz H-J. EMBO J. 1996;15:5459–5469. [PMC free article] [PubMed] [Google Scholar]
- 42.Yang H, Fitz-Gibbon S, Marcotte E M, Tai J H, Hyman E C, Miller J H. J Bacteriol. 2000;182:1272–1279. doi: 10.1128/jb.182.5.1272-1279.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiner K X, Weiner R S, Maley F, Maley G F. J Biol Chem. 1993;268:12983–12989. [PubMed] [Google Scholar]
- 44.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riera J, Robb F T, Weiss R, Fontecave M. Proc Natl Acad Sci USA. 1997;94:475–478. doi: 10.1073/pnas.94.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larsson G, Svensson L A, Nyman P O. Nat Struct Biol. 1996;3:532–538. doi: 10.1038/nsb0696-532. [DOI] [PubMed] [Google Scholar]