Abstract
Infected cell protein 0 (ICP0) of herpes simplex virus 1, a multifunctional ring finger protein, enhances the expression of genes introduced into cells by infection or transfection, interacts with numerous cellular and viral proteins, and is associated with the degradation of several cellular proteins. Sequences encoded by exon 2 of ICP0 (residues 20–241) bind the UbcH3 (cdc34) ubiquitin-conjugating enzyme, and its carboxy terminus expresses a ubiquitin ligase activity demonstrable by polyubiquitylation of cdc34 in vitro. We report that: (i) The physical interaction of cdc34 and ICP0 leads to its degradation. Thus, substitution of ICP0 aspartate 199 with alanine attenuates the degradation of cdc34 and its binding to the ICP0 ring finger domain. (ii) Substitution of residue 620 reported to abolish the interaction with a ubiquitin-specific protease has no effect on the function of ubiquitin ligase. (iii) ICP0 contains an additional distinct E3 ligase activity specific for the UbcH5a- and UbcH6 E2-conjugating enzymes mapping to the ring finger domain. This is, to our knowledge, the first identification of a viral protein with at least two physically separated E3 ligase activities with different E2 specificities. The results suggest that each activity may target different proteins.
Infected cell protein 0 (ICP0) of herpes simplex virus 1 (HSV-1) is essential for viral replication in cells infected at low multiplicity but is not essential in cells infected at high multiplicity (1–3). The 775-amino acid protein is translated from a spliced mRNA containing three exons encoding 19, 222, and 534 codons, respectively. The interaction of ICP0 with several diverse cellular proteins including the BMAL1 transactivator (4), the translation elongation factor 1δ (5), cyclin D3 (6), and the ubiquitin (Ub)-specific protease USP7 (7–9), suggests that its phenotype as a promiscuous transactivator reflects the sum of its multiple and diverse functions. Early in infection, ICP0 localizes with the promyelocytic leukemia protein and causes the disruption of ND10 structures (10–13). A zinc-binding RING finger (amino acids 106–149) characteristic of E3 Ub ligase enzymes has been identified in the domain encoded by exon 2 (14).
Analyses of ICP0 in the yeast two-hybrid system led to the discovery that ICP0 binds and stabilizes cyclin D3 (6). Mapping studies led to the identification of aspartate 199 as pivotal for both stabilization and binding of cyclin D3. Replacement of aspartate 199 with alanine (D199A) abolishes both binding and stabilization of cyclin D3 mediated by ICP0 and results in attenuated viral growth in quiescent human embryonic lung fibroblasts and reduced neuroinvasiveness (15, 16). These studies also revealed that ICP0 stabilizes cyclin D1 in a manner dependent on aspartate 199, even though it does not interact with it in vitro or in the yeast two-hybrid system (15, 16). Because HSV-1 replicates well in both dividing and stationary cells, these observations were pursued at several levels. The transcription of cyclin D3 or D1 is not induced after infection, but cyclin D3 was found to play a role in the translocation from the nucleus to the cytoplasm between 5 and 9 h after infection, and in independent experiments, substitution of aspartate 199 with alanine blocked the translocation of ICP0 (16). To account for the stabilization of cyclin D1, we examined the function of ICP0 as an E3 ligase in its interaction with cdc34, the E2 Ub-conjugating enzyme associated with proteasomal degradation of cyclin D (reviewed in ref. 14).
Several lines of evidence indicate that ICP0 functionally and physically interacts with the Ub–proteasomal degradation pathway. In addition to the interaction of USP7 with the carboxy terminus, ICP0 dynamically associates with the 26S proteasome in untreated cells but remains bound to proteasomes in cells treated with the proteasome inhibitor MG132 (17, 18). ICP0 similarly promotes the dynamic association of the E2 enzyme cdc34 with the proteasome in a manner dependent on aspartate 199 (17). Finally, a domain of ICP0 exon 3 (amino acids 543–768) functions as an E3 Ub ligase in conjunction with cdc34 in a substrate-independent in vitro ubiquitylation system, whereas the domain encoded by exon 2 (amino acids 20–241), which contains a RING finger motif, binds cdc34 (17). Thus, ICP0 differs from other RING finger E3 ligases in that the domain containing the RING finger binds E2, whereas the ligase activity maps to a different region of the protein. In vivo, ICP0 demonstrates attributes of an E3 ligase in that it mediates the proteasome-dependent degradation of multiple cellular proteins, including SUMOylated promyelocytic leukemia protein and other unidentified SUMOylated proteins (13), the catalytic subunit of DNA-dependent protein kinase (19, 20), centromeric proteins C and A (21, 22), and Sp100 (23, 24), and that foci of ICP0 observed in infected and transfected cells contain enhanced levels of conjugated Ub (25, 26).
Ub is recruited to ubiquitylation pathways (reviewed in ref. 14) via ATP-dependent activation by the E1 Ub-activating enzyme to form a thioester with a conserved active site cysteine. Activated Ub is then transesterified to a conserved cysteine of an E2 Ub-conjugating enzyme. E3 ligases represent the most diverse components of the system and bind both the E2 and the substrate while functioning to assemble Ub onto the substrate. The efficiency of Ub polymerization is increased by the E4 multi-Ub chain assembly protein (27).
The E2 family is diverse, and E2 enzymes promiscuously interact with E3 enzymes (reviewed in ref. 14). To determine whether ICP0 possessed E3 activity in conjunction with other E2 enzymes besides cdc34, we used an in vitro ubiquitylation system (17) containing recombinant Uba1 (E1), the indicated E2, ATP, an ATP regenerating system, and biotinylated Ub. The objective was to examine the ability of the ICP0 to promote substrate-independent Ub–protein ligation in the presence of UbcH6, UbcH5a, or UbcH7 E2 Ub-conjugating enzymes. We report that ICP0 encodes an additional distinct E3 ligase activity in residues 20–241 containing the RING finger. This site specifically functions in conjunction with a different set of E2 Ub-conjugating enzymes than the E3 activity mapping at the carboxyl-terminal domain of ICP0 (17). Additionally, ICP0 interacts with and promotes the degradation of the E2 enzyme cdc34 in a manner partially dependent on aspartate 199.
Materials and Methods
Cells and Viruses.
Telomerase-transformed human diploid foreskin fibroblasts (pHF) were obtained from Thomas Shenk (Princeton Univ., Princeton, NJ) and maintained as described elsewhere (28). HSV-1 strain F [HSV-1(F)] is the prototype HSV-1 strain used in this laboratory (29). The construction of the HSV-1 recombinant virus R7914 carrying the substitution of ICP0 aspartate 199 with alanine (D199A) was described elsewhere (15).
Plasmids and Glutathione S-Transferase (GST) Fusion Proteins.
pRB4994 containing ICP0 codons 20–241 inserted into pGEX 4T-1 in frame with GST and pRB4995 containing ICP0 codons 543–768 inserted into pGEX 4T-1 in frame with GST were described elsewhere (5). pRB5266 encodes ICP0 exon 2 (codons 20–241) containing the D199A substitution cloned in vector pGBT9 (15). The SphI/KpnI ICP0 fragment from pRB5266 was purified and ligated to a SphI-KpnI-digested pRB4994 plasmid. The resulting plasmid contained the D199A substitution in the ICP0 fragment encoded by pRB4994 and was designated pRB5840. pRB4988 was generated by amplifying the sequence encoding ICP0 amino acids 243–775 by PCR and cloning it into vector pGBT9 as previously described (5). Site-directed mutagenesis where complementary oligonucleotides containing the specific nucleotide substitution were annealed to pRB4988 DNA according to the manufacturer's instructions (Stratagene, catalog no. 200518) to yield the K620I substitution in the ICP0 fragment contained in pRB4988. The resulting plasmid was designated pRB5342. pRB3710 encodes a SacI-PstI fragment containing the entire ICP0 coding sequence in vector pUC18 (30). pRB5342 was digested with SalI and SnaBI. The resulting fragment was purified and ligated to a SalI-SnaB1-digested pRB3710 plasmid. The resulting plasmid, designated pRB5343, contained the K620I substitution in the full length ICP0 coding sequence. pRB5343 was digested with PpuMI and SalI, and the resulting fragment was purified and ligated to a PpuMI-SalI-digested pRB4995 plasmid. The resulting plasmid contained the K620I substitution in the ICP0 fragment encoded by pRB4995 and was designated pRB5841. pRB5840 and pRB5841 were sequenced at the University of Chicago Cancer Research Center DNA Sequencing Facility to confirm that the ICP0 fragments encoded by these plasmids contained the D199A and K620I mutations, respectively. GST fusion proteins encoded by pRB4994, pRB4995, pRB5840, and pRB5841 were designated GST-exon 2, GST-exon 3, GST-D199A, and GST-K620I, respectively. GST fusion proteins were expressed in Escherichia coli BL21 cells and purified as previously described (5).
Substrate-Independent in Vitro Ubiquitylation Reactions.
Substrate-independent in vitro ubiquitylation reactions (17) were performed in 30 μl of ubiquitylation buffer (50 mM Tris, pH 7.5/2.5 mM MgCl2/0.5 mM DTT) containing 40 ng of recombinant rabbit Ub-activating enzyme E1 (Calbiochem, catalog no. 662070), 2 μg of biotinylated Ub (Affiniti Research Products, Mamhead, Exeter, U.K., catalog no. UW8705), 0.2 mM ATP, along with an ATP regenerating system consisting of 1 mM creatine phosphate and 15 units of porcine heart creatine phosphokinase (Calbiochem, catalog no. 238395). Reaction mixtures also contained 40 ng of human recombinant Ub-conjugating enzymes [His6]-UbcH3 (Affiniti, catalog no. UW8730), [His6]-UbcH6 (Affiniti, catalog no. UW8710), GST-UbcH5a (Affiniti, catalog no. UW8670), or UbcH7 (Affiniti, catalog no. UW8455) in addition to 5 μg of purified GST, GST-Exon 2, GST-D199A, GST-Exon 3, or GST-K620I, as indicated. The mixtures were thoroughly mixed and allowed to react at 37°C for 90 min. The reaction was then stopped either by the addition of one part 4× disruption buffer (6.85% SDS/24% glycerol/3.3% β-mercaptoethanol/233 mM Tris, pH 6.8/0.008% bromophenol blue) to three parts reaction mixture or preparation for affinity capture, as described below.
Affinity capture was performed with glutathione-Sepharose 4B (Amersham Pharmacia) or talon resin (CLONTECH). For each sample, 15 μl from the in vitro ubiquitylation reactions described above was added to 485 μl of cold PBS on ice, mixed with 20 μl of resin and incubated overnight at 4°C. Sepharose pellets were collected by centrifugation and washed with PBS for 30 min three times at 4°C before addition of disruption buffer.
HSV-1 Infections.
Replicate cultures of pHF infected [10 plaque-forming units/cell], as described elsewhere (17), were harvested at the indicated time after infection, rinsed twice in PBS, pelleted by centrifugation, and solubilized at 4°C in PBS-A* (1% Nonidet P-40, 1% deoxycholate in PBS) in which complete, Mini, EDTA-free protease inhibitor mixture tablets (Roche Diagnostics) had been dissolved according to the manufacturer's instructions. The amount of total protein in the lysate was quantified using the Bio-Rad Protein Assay, according to the manufacturer's instructions.
Antibodies.
The antibodies used in these studies were as follows: The anti-GST antibody was made against the chimeric protein GST-ORF P (31) and used at a dilution of 1:2,000 for immunoblots. Mouse monoclonal antibody against ICP27 (32) was purchased from the Goodwin Cancer Research institute (Plantation, FL) and used at a dilution of 1:500. Rabbit polyclonal antibody against cdc34 (Neomarkers, Fremont, CA, catalog no. RB-043-P1) was used at a dilution of 1:500. Mouse monoclonal antibodies against cdc34 (catalog no. 610250), UbcH6 (catalog no. 611219), and UbcH7 (catalog no. 610853), all from BD Transduction Laboratories (San Diego, CA), were used at dilutions of 1:1,000, 1:1,000, and 1:250, respectively. Electrophoretic separations of total or precipitated proteins and immunoblotting of electrophoretically separated proteins were done as described elsewhere (17). Bands were scanned where indicated with a Canoscan FB1200S scanner (Canon USA, Lake Success, NY) and were quantified with imagequant software (Molecular Dynamics).
Results
ICP0 Promotes the Proteasome-Dependent Degradation of the E2 Enzyme cdc34.
In addition to being charged at cysteine, yeast cdc34 is ubiquitylated at multiple lysines in vitro and in vivo (33, 34). Earlier studies have shown that only polyubiquitylated forms of cdc34 are dynamically associated with proteasomes in wild-type virus-infected cells treated with the proteasome inhibitor MG132 (17). To test the hypothesis that the cdc34-dependent ICP0 E3 ligase activity promotes autoubiquitylation of cdc34 for proteasome-dependent degradation, we measured cdc34 protein levels in pHF that were mock-infected or infected with HSV-1(F) or the R7914 recombinant virus carrying the D199A substitution by reacting the electrophoretically separated proteins with monoclonal antibody directed against cdc34. Analyses of the results shown in Fig. 1A indicate that cdc34 was degraded more rapidly in cells infected with HSV-1(F) that in R7914 (D199A) mutant infected cells. Cells treated for 1 h with MG132 5 h after infection with wild-type virus accumulated 4-fold more cdc34 than untreated infected cells. In contrast, treated D199A infected cells accumulated 1.8-fold more cdc34 than untreated infected cells, whereas treated mock-infected cells accumulated only 1.2-fold more cdc34 than untreated mock-infected cells. Because the viral early protein infected cell protein 27 (ICP27) accumulated in comparable amounts in HSV-1(F)- and in R7914-infected cells (Fig. 1B), the observed differences in cdc34 accumulation cannot be attributed to exposure of cells to different amounts of HSV-1 virus. Furthermore, these results show that HSV-1 infection does not generally promote proteasome-dependent protein degradation, as MG132 has no effect on ICP27 levels (Fig. 1B, lanes 3 and 4 and lanes 5 and 6), indicating it is not degraded by the proteasome. These experiments were repeated with polyclonal rabbit anti-cdc34 with the same results (data not shown). We conclude from these experiments that cdc34 is targeted for destruction by the Ub–proteasomal pathway and that the process depends in part on the D199 locus in ICP0.
Figure 1.
ICP0 mediates proteasome-dependent degradation of the E2 cdc34 in HSV-1-infected cells. (A) pHF infected with 10 plaque-forming units of HSV-1(F) (lanes 3 and 4) or R7914 (lanes 5 and 6) per cell were mock-treated or treated with MG132 for 1 h before harvest at 5 h after infection. Cells were solubilized, and lysates containing 100 μg of total protein were subjected to electrophoresis in denaturing polyacrylamide gels and reacted with a mouse monoclonal antibody against cdc34. (B) pHF were infected with HSV-1 and treated with MG132 as in A. The harvested cells were processed as above but probed with anti-ICP27 mouse monoclonal antibody. The bands were scanned and quantified as described in Materials and Methods.
The D199A Substitution Attenuates but Does Not Ablate the Ability of the Polypeptide Encoded by ICP0 Exon 2 to Interact with cdc34.
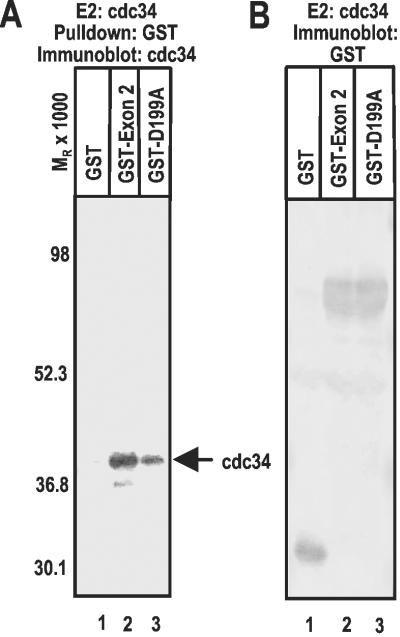
Because the D199A substitution blocks the ICP0-dependent dynamic association of cdc34 with proteasomes in HSV-1-infected cells and cdc34 interacts with ICP0 exon 2 encoded amino acid sequence in vitro (17), we used a GST pull-down experiment to examine the effect of the D199A substitution on the interaction between ICP0 and cdc34 in the in vitro system (Fig. 2A). The results (Fig. 2A) were as follows: cdc34 was not pulled down by GST (Fig. 2A, lane 1). Elsewhere, we have shown that cdc34 is not pulled down by sequences encoded by ICP0 exon 3 (17). Sequences encoding ICP0 exon 2 carrying the D199A substitution (GST-D199A) pulled down ≈30% of the amount of cdc34 pulled down by sequences encoded by HSV-1(F) ICP0 exon 2. Chimeric proteins GST-exon 2 and GST-D199A included in the reactions were comparable in amount and were of predicted molecular weight (Fig. 2B); therefore, the differences in the affinity of GST-exon 2 and GST-D199A for cdc34 could not be attributed to differences in the amounts of chimeric proteins in the reaction mixtures. We conclude from this experiment that aspartate 199 plays a role in the interaction of ICP0 with cdc34, and that this interaction plays a role in the Ub–proteasome pathway-dependent degradation of cdc34.
Figure 2.
Characterization of the interaction between cdc34 and sequences encoded by ICP0 exon 2. In vitro ubiquitylation reactions were performed as previously described (17). (A) GST (lane 1) or the indicated GST fusion protein (lanes 2 and 3) was pulled down with glutathione Sepharose beads from reactions containing cdc34, electrophoretically separated in a denaturing polyacrylamide gel, and reacted with anti cdc34 rabbit polyclonal antibody. (B) In vitro ubiquitylation reactions were performed by using cdc34 as in A. The proteins separated as above were reacted with anti-GST antibody. The bands were scanned and quantified as described in Materials and Methods.
ICP0 Encodes a Second Discrete E3 Ligase Function in Exon 2.
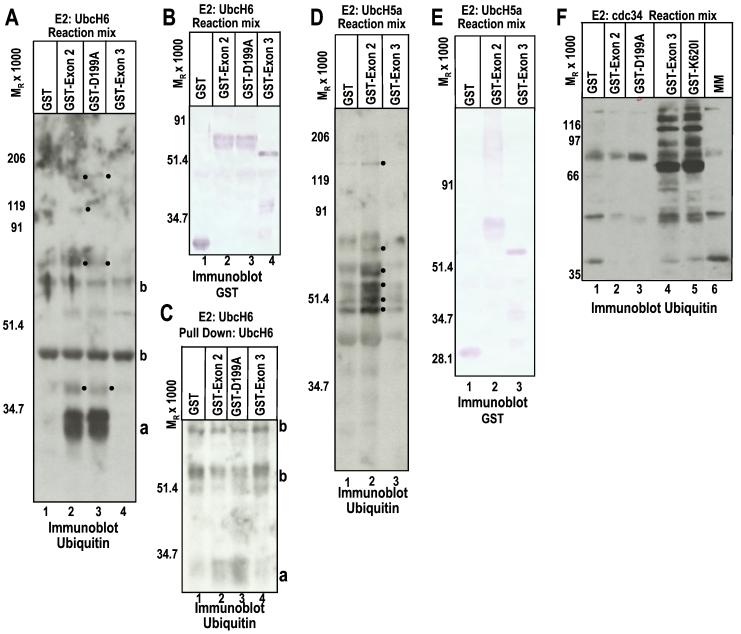
The objective of this series of experiments was to determine whether ICP0 functionally interacts with other E2 Ub-conjugating enzymes. Substrate-independent in vitro ubiquitylation reactions were carried out as described in Materials and Methods according to the design reported elsewhere (17). In such reactions, the ability of a protein to promote Ub–protein ligation is indicative of E3 activity (14, 17). Thus, addition of an E3 enzyme to the reaction would be expected to cause the accumulation of increased levels of Ub–protein conjugates, additional Ub–protein conjugates, and/or higher molecular weight Ub–protein conjugates in the reaction mixture, as compared with reactions where an E3 enzyme was not added. It is important to note that a basal level of Ub–protein ligation mediated by the E2 occurs in vitro in the absence of an E3. The results (Fig. 3) were as follows: Streptavidin–horseradish peroxidase (HRP) reacted with multiple protein conjugates with biotinylated Ub from electrophoretically separated mixtures containing UbcH6 and either GST-exon 2 of wild-type or D199A-substituted ICP0 (Fig. 3A, lane 2 or 3), which were not present in lanes containing GST or GST-exon 3 (Fig. 3A, lane 1 or 4), suggesting that ICP0 exon 2 promotes Ub–protein ligation mediated by UbcH6. Fig. 3B shows that the mixtures contain comparable amounts of GST-exon 2, GST-D199A, GST, and GST-exon 3 proteins. Finally, Fig. 3C shows that Streptavidin–HRP reacted with prominent bands containing ubiquitylated His6-UbcH6 pulled down from reactions containing GST-exon 2 and GST-D199A with talon resin beads (bands designated a with Mr 30,000; Fig. 3C, lanes 2 and 3). However, this ubiquitylated form of His6-UbcH6 was not present to a significant extent in reactions containing GST or GST-exon 3 (Fig. 3C, lanes 1 and 4). The ubiquitylated UbcH6 bands seen in Fig. 3C (a bands) correspond to the prominent bands seen in lanes 2 and 3 in Fig. 3A. The molecular weights of these bands (Mr 30,000) are consistent with that of monoubiquitylated His6-UbcH6. E3 activity has often been shown to promote autoubiquitylation of the E2 enzyme in substrate-independent in vitro ubiquitylation reactions (14, 17, 35).
Figure 3.
ICP0 sequences encoded by exon 2 express an E3 ligase function. (A) In vitro ubiquitylation reactions were performed as previously described (17). The mixtures containing GST or the indicated GST fusion protein, E1, the E2 UbcH6, biotinylated Ub, ATP, and an ATP regenerating system were allowed to react for 30 min, then were subjected to electrophoresis in denaturing polyacrylamide gels and reacted with streptavidin HRP to detect biotinylated Ub. The dots to the right of the bands indicate polyubiquitylated species, which are specifically present or augmented in reactions containing GST-exon 2, either wild-type or carrying the D199A substitution. a, monoubiquitylated UbcH6; b, proteins present in reactions or ubiquitylated species formed independently of E3 activity, which reacted with streptavidin–HRP. (B) The same as A, except that the electrophoretically proteins were reacted with anti-GST antibody. The data serve as a loading control. (C) [His6]-UbcH6 was pulled down with talon resin from reaction mixtures containing UbcH6, GST, or the indicated GST fusion protein to determine whether the E2 was being ubiquitylated in the reactions (17), separated as above, and probed with streptavidin–HRP conjugate. a, monoubiquitylated UbcH6. Note the absence of polyubiquitylated UbcH6. (D) In vitro ubation reactions were done as in A but with E2 UbcH5a. The reaction mixture was probed for ubiquitylated species with streptavidin–HRP conjugate. The dots to the right of the bands indicate polyubiquitylated species, which are present or augmented in reactions containing GST-exon 2. (E) The electrophoretically separated reaction mix was reacted with anti-GST antibody as a loading control. (F) The experiment was set up as above but with the E2 cdc34. The objective was to test whether a mutation reported to abolish interaction with USP7 (36) affects the polyubiquitylation of cdc34 by ICP0 sequences encoded by exon 3. The reaction mixtures were separated on denaturing polyacrylamide gels and reacted with streptavidin–HRP conjugate to detect Ub. Note the presence of polyubiquitylated species in lanes 4 and 5.
Essentially similar results were obtained with mixtures containing UbcH5a. Again, specific ubiquitylated bands were present in mixtures containing GST-exon 2 but not GST alone or GST-exon 3, and Ub–protein conjugates accumulated to higher levels in reactions containing GST-exon 2 than in those where GST, GST-exon 3, or no GST fusion protein was added (Fig. 3D and data not shown). Fig. 3E shows that the mixtures contained GST, GST-exon 2, and GST-exon 3 in comparable amounts.
Because the RING finger of many E3 ligases binds the E2, and E3 enzymes are expected to colocalize E2 enzymes with their substrates, we tested whether UbcH6 interacted with GST-exon 2 or GST-exon 3. In contrast to results obtained with cdc34 (ref. 17 and Fig. 2A), GST-exon 2 or GST-exon 3 failed to pull down UbcH6 (data not shown). Inasmuch as ICP0-exon 2 promotes Ub–protein ligation, it must interact with UbcH6 transiently or with low affinity.
We conclude from these studies the following: (i) ICP0 exon 2 but not ICP0 exon 3 has E3 Ub ligase activity in conjunction with the E2 enzymes UbcH6 and UbcH5a, and this E3 function promotes the autoubiquitylation of the UbcH6 E2 Ub-conjugating enzyme in vitro. This is in contrast to the results obtained with UbcH3 (cdc34), which acts in conjunction with an E3 function encoded by ICP0 exon 3 (Fig. 3F and ref. 17). (ii) The D199A substitution in ICP0 exon 2 has no apparent effect on ICP0 exon 2 E3 activity (Fig. 3A), suggesting that the residues encoded by exon 2 and associated with the interactions of ICP0 with UbcH6 and cdc34 are different.
ICP0 Exon 3 E3 Ub Ligase Activity Is Independent of the Ability to Bind USP7.
The USP7-binding site (amino acids 594–646) (36) is contained in the domain of exon 3 that has E3 activity with cdc34. The question arose, therefore, whether ICP0 exon 3 E3 activity depended on its ability to bind USP7. Because it has been reported that the K620I substitution in exon 3 abrogates the interaction of ICP0 with USP7 (36), we compared mutant and wild-type GST-exon 3 for its ability to act as the E3 ligase for cdc34 in the in vitro ubiquitylation system described above. As shown in Fig. 3F, the K620I substitution had no effect on the ability of GST-exon 3 to promote Ub–protein ligation, whereas, as expected, neither GST-exon 2 wild-type nor GST-exon 2 D199A promoted Ub polymerization in reactions containing the E2 cdc34. These results indicate that the E3 activity residing in exon 3 was independent of its ability to bind USP7 and reaffirmed the conclusion that ICP0 contains at least two nonoverlapping E3 domains.
Functional Interactions Between ICP0 E3 Ligase Domains and E2 Enzymes Are Specific.
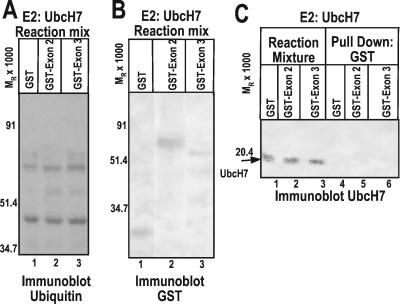
The results of experiments designed to test the interaction of UbcH7 E2 Ub-conjugating enzyme with ICP0 are shown in Fig. 4. In these experiments, neither GST-exon 2 nor GST-exon 3 promoted Ub–protein ligation over a basal level (Fig. 4A), and no interaction between UbcH7 and any of the chimeric proteins was detected (Fig. 4C). GST and GST fusion proteins were added to the reaction mixtures in comparable amounts (Fig. 4B). Thus, neither of the ICP0 E3 domains functioned in conjunction with UbcH7. These results demonstrated the specificity of the functional interaction between the E2 enzymes and ICP0.
Figure 4.
ICP0 does not functionally or physically interact with the E2 enzyme UbcH7. (A) In vitro ubiquitylation reactions containing E2 UbcH7 were performed as in Fig. 3, then separated by SDS/PAGE and probed with streptavidin–HRP conjugate. (B) In vitro ubiquitylation reactions were performed and electrophoretically separated as in A and reacted with antibody directed against GST. (C) GST or GST fusion protein was pulled down with glutathione Sepharose beads from reaction mixtures containing UbcH7, separated as above, and probed with anti-UbcH7 mouse monoclonal antibody (lanes 4–6). Total reaction mixtures were also electrophoretically separated and reacted with mouse monoclonal antibody directed against UbcH7 (lanes 1–3).
Discussion
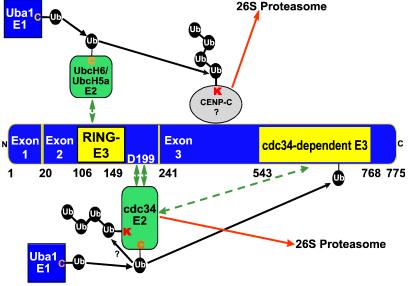
We present the unprecedented finding that ICP0 encodes two discrete E3 Ub ligase functions in different portions of the protein. Each of these sites specifically acts in conjunction with a particular E2 or subset of E2 enzymes. Although other unimolecular RING finger E3 ligases such as c-Cbl (reviewed in ref. 14) act in conjunction with multiple E2 enzymes in in vitro reactions, the E3 activity remains localized to a specific domain of the protein. This novel arrangement raises the possibility that ICP0 promotes the degradation of proteins whose normal turnover is mediated by different classes of E3 ligases. Thus, we propose a model where the cdc34-dependent E3 function in ICP0 exon 3 promotes the autoubiquitylation of cdc34, whereas the UbcH6-dependent E3 in exon 2 would promote polyubiquitylation of other proteins degraded in an ICP0-dependent manner (Fig. 5). The model illustrated in Fig. 5 takes cognizance of the observation that although ubiquitylated ICP0 has not been detected in lysates of infected cells, recent findings indicate that the GST-exon 3 polypeptide is ubiquitylated in the presence of ATP and cdc34 (R.H. and B.R., unpublished results). These results suggest the possibility that ubiquitin is transferred to a lysine residue in cdc34 from exon 3 via a mechanism analogous to that of HECT E3 ligases.
Figure 5.
Model for multiple specific E3 Ub ligase functions encoded by ICP0. The E3 Ub ligase function encoded by ICP0 exon 2 promotes Ub–protein ligation in conjunction with the E2 enzymes UbcH6 and UbcH5a. As ICP0 exon 2 contains the RING finger domain (amino acids 106–149), it is likely that this domain encodes a classical RING finger unimolecular E3 ligase (14, 20, 21). The accumulation of conjugated Ub and the degradation of cellular proteins by ICP0 require the RING finger (13, 20–22, 24, 25), suggesting it is required for the E3 function involved in these processes. UbcH6 is presumed to transiently or weakly interact with the RING finger domain or other sequences within exon 2. ICP0 exon 3 encodes an unconventional RING finger-independent E3 ligase that functions in conjunction with the E2 cdc34. cdc34 interacts with ICP0 exon 2, and aspartate 199 is required for optimal interaction. The model proposes that the interaction tethers cdc34 to exon 3, which promotes the autoubiquitylation and degradation of cdc34. It is also possible that the cdc34-dependent E3 activity in exon 3 may act on additional substrates, although the degradation of cdc34 mediated by ICP0 would render such activity inefficient. This may provide the reason the virus evolved two discrete E3 activities in ICP0 to ensure the efficient degradation of cellular proteins. cdc34 is presumed to transiently or weakly interact with ICP0 exon 3. Transient or weak interactions are indicated by a double arrow. Strong interactions are indicated by two double green arrows. Numbers indicate the residue position in ICP0.
The UbcH6-dependent E3 ligase activity maps to exon 2, which contains the ICP0 RING finger. As the RING finger domain is required for E3 activity in all known RING finger E3 ligases hitherto described with the exception of the ICP0 exon 3 E3 (14, 17, 35), it is likely that the RING finger domain mediates the UbcH6- and UbcH5a-dependent E3 activities in ICP0 exon 2. The cdc34-dependent E3 activity in ICP0 exon 3 is unconventional among RING finger E3 ligases, because catalytic activity occurs in vitro independent of the RING finger domain in exon 2 (14, 17). In addition to RING finger E3 ligases, the other major catalytic class of E3 enzymes contains a HECT domain in the carboxy terminus, which covalently binds Ub and does not contain a RING finger. Available data, however, appear to exclude the possibility that the ICP0 exon 3 E3 may be a HECT E3. Sequence comparisons show no significant similarity between ICP0 exon 3 and the prototypic HECT domain in the carboxy terminus of E6-AP (37), suggesting the E3 encoded by the carboxy terminus of ICP0 is not part of the HECT class of E3 ligases but rather operates by a novel catalytic mechanism. In addition, sequence comparisons also show no significant similarity between ICP0 exon 3 and the recently described U box family of E3 ligases (38). Elsewhere, we hypothesized that the interaction of cdc34 with ICP0 exon 2 would increase the efficiency of cdc34 autoubiquitylation promoted by the E3 catalytic activity in exon 3 by tethering cdc34 to exon 3 (17). As RING fingers are required for some E3s to bind E2s (14), the RING finger may play a role in this interaction. The observation that the D199A substitution impairs this interaction is concordant with the less pronounced reduction of cdc34 in cells infected with the R7914 mutant carrying this substitution and suggests that, in vivo, cdc34 must be firmly tethered to the ring finger domain to enable the polyubiquitylation of the protein by the exon 3 ligase mapping near the carboxy terminus of ICP0.
Our results may provide a solution to the puzzle of the mechanism by which ICP0 mediates the stabilization of cyclin D1 without binding it. Cdc34 is the major E2 that interacts with the Skp1-cdc53-F-box (SCF) E3 complex (14). SCF complexes containing the F-box protein Skp2 promote the ubiquitylation and degradation of cyclin D1 (39, 40). Thus defects in the Ub-mediated proteolysis of cyclin D1 by proteins such as Skp2 lead to the stabilization of cyclin D1 in several cancer cell lines (40, 41). Therefore, aspartate 199-dependent stabilization of cyclin D1 mediated by ICP0 (16) may result from the degradation of cdc34. Significantly, inactivation of cdc34 would not lead to a total defect in the degradation of SCF substrates. SCF complexes containing UbcH5b and UbcH5c have been described and shown to mediate the degradation of IκBα, which is also a substrate for SCF complexes containing cdc34 (42). In addition, the SCF substrate E2F1 can be ubiquitylated by other E3 ligases that do not act in conjunction with cdc34 (43). It is also possible that the ICP0 exon 2 E3 ligase activity could compensate for SCF for some substrates. Thus, only substrates specifically targeted for degradation by SCF and cdc34 would be expected to be stabilized by ICP0. In addition, cyclin D3 has been shown to interact with the SCF component Skp2 and is stabilized in some cancer cell lines (41). Thus, it is unclear whether ICP0 aspartate 199-dependent stabilization of cyclin D3 is mediated by interaction of ICP0 with cyclin D3 or degradation of cdc34 effected by ICP0 or a combination of these two functions of ICP0.
ICP0 is among the first gene products made during HSV-1 infection. These early products regulate viral replication and the environment of the infected cell to ensure orderly expression of viral genes and evasion of cellular responses to infection. To efficiently attain these objectives, these proteins are multifunctional (1). The dual E3 ligase functions of ICP0 are representative of this strategy.
Acknowledgments
R.H. is a Howard Hughes Medical Institute Predoctoral Fellow. These studies were aided by grants from the National Cancer Institute (CA78766, CA71933, CA83939, CA87661, and CA88860).
Abbreviations
- ICP0
infected cell protein 0
- HSV-1
herpes simplex virus 1
- Ub
ubiquitin
- pHF
telomerase-transformed human diploid foreskin fibroblasts
- GST
glutathione S-transferase
- HRP
horseradish peroxidase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Roizman B, Knipe D M. In: Fields Virology. 4th Ed. Knipe D M, Howley P, Griffin D E, Lamb R A, Martin M A, Roizman B, Straus S E, editors. New York: Lippincott–Williams & Wilkins; 2001. pp. 2399–2459. [Google Scholar]
- 2.Sacks W R, Schaffer P A. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stow N D, Stow E C. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 4.Kawaguchi Y, Tanaka M, Yokoymama A, Matsuda G, Kato K, Kagawa H, Hirai K, Roizman B. Proc Natl Acad Sci USA. 2001;98:1877–1882. doi: 10.1073/pnas.041592598. . (First Published February 6, 2001; 10.1073/pnas.041592598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawaguchi Y, Bruni R, Roizman B. J Virol. 1997;71:1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawaguchi Y, Van Sant C, Roizman B. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everett R D, Meredith M R, Orr A, Cross A, Kathoria M, Parkinson J. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meredith M R, Orr A, Everett R D. Virology. 1994;200:457–469. doi: 10.1006/viro.1994.1209. [DOI] [PubMed] [Google Scholar]
- 9.Meredith M R, Orr A, Elliott M, Everett R D. Virology. 1995;209:174–187. doi: 10.1006/viro.1995.1241. [DOI] [PubMed] [Google Scholar]
- 10.Maul G G, Guldner H H, Spivack J G. J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 11.Everett R D, Maul G G. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maul G G, Everett R D. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 13.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson P K, Eldridge A G, Freed E, Furstenthal L, Hsu J Y, Kaiser B K, Reimann J D R. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- 15.Van Sant C, Kawaguchi Y, Roizman B. Proc Natl Acad Sci USA. 1999;96:8184–8189. doi: 10.1073/pnas.96.14.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Sant C, Lopez P, Advani S J, Roizman B. J Virol. 2001;75:1888–1898. doi: 10.1128/JVI.75.4.1888-1898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Sant C, Hagglund R, Lopez P, Roizman B. Proc Natl Acad Sci USA. 2001;98:8815–8820. doi: 10.1073/pnas.161283098. . (First Published July 10, 2001; 10.1073/pnas.161283098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez P, Van Sant C, Roizman B. J Virol. 2001;75:3832–3840. doi: 10.1128/JVI.75.8.3832-3840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lees-Miller S P, Long M C, Kilvert M A, Lam V, Rice S A, Spencer C A. J Virol. 1996;70:7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkinson J, Lees-Miller S P, Everett R D. J Virol. 1999;73:650–657. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everett R D, Earnshaw W C, Findlay J, Lomonte P. EMBO J. 1999;18:1526–1538. doi: 10.1093/emboj/18.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lomonte P, Sullivan K F, Everett R D. J Biol Chem. 2001;276:5829–5835. doi: 10.1074/jbc.M008547200. [DOI] [PubMed] [Google Scholar]
- 23.Chelbi-Alix K M, de The H. Oncogene. 1999;18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 24.Parkinson J, Everett R D. J Virol. 2000;74:10006–10017. doi: 10.1128/jvi.74.21.10006-10017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Everett R D. J Virol. 2000;74:9994–10005. doi: 10.1128/jvi.74.21.9994-10005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parkinson J, Everett R D. J Virol. 2001;75:5357–5362. doi: 10.1128/JVI.75.11.5357-5362.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koegl M, Hoppe T, Schlenker S, Ulrich H D, Mayer T U, Jentsch S. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 28.Bresnahan W A, Hultman G E, Shenk T. J Virol. 2000;74:10816–10818. doi: 10.1128/jvi.74.22.10816-10818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ejercito P M, Kieff E D, Roizman B. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 30.McKnight J L C, Kristie T M, Roizman B. Proc Natl Acad Sci USA. 1987;84:7061–7065. doi: 10.1073/pnas.84.20.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagunoff M, Randall G, Roizman B. J Virol. 1996;70:1810–1817. doi: 10.1128/jvi.70.3.1810-1817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ackermann M, Braun D K, Pereira L, Roizman B. J Virol. 1984;52:108–118. doi: 10.1128/jvi.52.1.108-118.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerjee A, Gregori L, Xu Y, Chau V. J Biol Chem. 1993;268:5668–5675. [PubMed] [Google Scholar]
- 34.Goebl M G, Goetsch L, Byers B. Mol Cell Biol. 1994;14:3022–3029. doi: 10.1128/mcb.14.5.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joaziero C A P, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y-C. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 36.Everett R D, Meredith M, Orr A. J Virol. 1999;73:417–426. doi: 10.1128/jvi.73.1.417-426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huibregtse J M, Scheffner M, Beaudenon S, Howley P M. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama K-I. J Biol Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 39.Zhong-Kang Y, Gervais J L M, Zhang H. Proc Natl Acad Sci USA. 1998;95:11324–11329. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ganiatsas S, Dow R, Thompson A, Schulman B, Germain D. Oncogene. 2001;20:3641–3650. doi: 10.1038/sj.onc.1204501. [DOI] [PubMed] [Google Scholar]
- 41.Russell A, Hendley J, Germain D. Oncogene. 1999;18:6454–6459. doi: 10.1038/sj.onc.1203030. [DOI] [PubMed] [Google Scholar]
- 42.Gonen H, Bercovich B, Orian A, Carrano A, Takizawa C, Yamanaka K, Pagano M, Iwai K, Ciechanover A. J Biol Chem. 1999;274:14823–14830. doi: 10.1074/jbc.274.21.14823. [DOI] [PubMed] [Google Scholar]
- 43.Ohta T, Xiong Y. Cancer Res. 2001;61:1347–1353. [PubMed] [Google Scholar]