Abstract
Background
The metabolism of stearoyl-GPE plays a key role in the liver metastasis of gastric cancer. This investigation delves into the mechanisms underlying the intricate tumor microenvironment (TME) heterogeneity triggered by stearoyl metabolism in gastric cancer with liver metastasis (LMGC), offering novel perspectives for LMGC.
Objective
Utilizing Mendelian randomization, we determined that stearoyl metabolism significantly contributes to the progression of gastric cancer (GC). Following this, bulk transcriptome analyses and single-cell multiomics techniques to investigate the roles of stearoyl-GPE metabolism-related genes, particularly NCOA4, in regulating LMGC TME.
Results
Our analysis highlights the crucial role of stearoyl metabolism in modulating the complex microenvironment of LMGC, particularly impacting monocyte cells. Through single-cell sequencing and spatial transcriptomics, we have identified key metabolic genes specific to stearoyl metabolism within the monocyte cell population, including NCOA4. Regarding the relationship between ferroptosis, stearoyl metabolism, and LMGC findings, it is plausible that stearoyl metabolism and LMGC pathways intersect with mechanisms involved in ferroptosis. Ferroptosis, characterized by iron-dependent lipid peroxidation, represents a regulated form of cell death. The activity of Stearoyl-CoA desaturase (SCD), a critical enzyme in stearoyl metabolism, has been associated with the modulation of lipid composition and susceptibility to ferroptosis. Furthermore, the LMGC is integral to cellular processes related to oxidative stress and lipid metabolism, both of which are significant factors in the context of ferroptosis.
Conclusion
This study enhances the understanding of the relationship between stearoyl metabolism and ferroptosis in promoting liver metastasis of gastric cancer and its role in the regulation of tumor heterogeneity. In addition, this study contributes to a deeper understanding of the dynamics of gastric cancer tumor microenvironment (TME) and provides a basis for the development of better interventions to combat cancer metastasis.
Keywords: LMGC, Ferroptosis, Stearoyl metabolism, NCOA4, Spatial transcriptomics, Single-cell RNA sequencing, Multi-omics data, Tumor heterogeneity, Tumor microenvironment (TME)
Introduction
Liver metastasis from gastric cancer is an indication of a further progressive condition of the disease that becomes common each year [1–4]. This negative trend is especially concerning for cases of locally metastatic gastric cancer (LMGC) due to its significant impact on mortality rates. Although surgical resection is the only potentially curative option, its effectiveness is strongly dependent on early tumor detection. Unfortunately, in LMGC, approximately 80% of diagnoses are made at an advanced metastatic stage, resulting in a grim 5-year survival rate of less than 5% [5, 6]. The TME exhibits significant metabolic and immune heterogeneity, which facilitates tumor evasion and resistance to standard treatments. In response to these challenges, immunotherapy has gained attention as an innovative cancer treatment approach. It aims to boost the body’s natural anti-tumor immune responses while minimizing the side effects commonly associated with chemotherapy and radiotherapy [7–9].
Single-cell multi-omics technologies surpass traditional methodologies in sensitivity and resolution, emerging as pivotal tools in the elucidation of the cancer TME, as well as in pharmacology, molecular diagnostics, and prognosis [10–12]. In recent decades, the rapid evolution of omics technologies has profoundly enriched our understanding of cellular composition and the intricate functions of genes, proteins, and metabolites [13–15]. While thousands of gene sequences have been sequenced to date, earlier research often concentrated on the aggregate functions of cell populations, thereby neglecting the critical insights provided by cellular subpopulations. The inherent heterogeneity within cells and genetic profiles was challenging to discern with conventional techniques [16–18]. However, single-cell multi-omics technologies have successfully bridged this gap [19, 20]. By employing these sophisticated techniques, we have unveiled the complex and nuanced immune microenvironment of LMGC.
Moreover, contemporary research from the American Cancer Center has unveiled a significant escalation in lipid biosynthesis, accumulation, and metabolic activity across a diverse array of cancer types. This intricate reprogramming of lipid metabolism now stands recognized as a critical contributor to the dysregulation of the TME [21]. The metabolic shift notably includes variations in the metabolism of stearoyl. Recent studies have firmly established a profound linkage between lipid metabolism dynamics and the aggressive phenotypes of cancer cells, encompassing their rapid proliferation, survival, migration, invasion, and metastatic capabilities. Comprehensive analyses reveal that patients with gastric cancer exhibit markedly elevated lipid levels in both plasma and tissue samples compared to healthy controls. Furthermore, genetic mutations or aberrant expression of lipid metabolism-related genes have been conclusively associated with the pathogenesis of LMGC [22].
NcoA4 (Nuclear receptor coactivator 4), a pivotal protein of ferroptosis, operates as a transporter protein. Originally identified within the RetFused Gene in a subset of papillary thyroid carcinomas [23], NcoA4 has emerged as a focal point in recent oncological research. Our investigations have elucidated that the downstream target genes of Stearoyl metabolites exhibit a profound association with ferroptosis. This underscores the significance of ferroptosis as a crucial mechanism for impeding liver metastasis in gastric cancer. Recent studies indicate that the absence of NCOA4 disrupts cellular ferroptosis pathways by depleting intracellular free iron, glutathione, and reactive oxygen species (ROS). This perturbation is intricately linked to the initiation and progression of various malignancies, including prostate, ovarian, and breast cancers. Moreover, the enhancement of ferroptosis has been demonstrated to amplify the anti-tumor efficacy of immunotherapy, revealing a synergistic interplay between ferroptosis and immunotherapeutic strategies in the eradication of cancer cells [24]. NCOA4’s involvement in tumor proliferation and metastasis positions it as a critical element in the oncogenic paradigm, underscoring its importance in the mechanisms driving cancer progression and development [25–27]. Despite the heightened focus on ferroptosis research, the role of stearoyl metabolism in the context of gastric cancer (GC) remains inadequately explored, warranting further in-depth investigation.
Recent advances in technology have significantly propelled our ability to explore stearoyl metabolism in LMGC, such as multi-omics single-cell sequencing [28–30]. However, their effects on the complex mechanisms of tumorigenesis are not clear. In our investigation, we aim to elucidate genomic patterns associated with genes linked to monocyte cells and stearoyl metabolism within LMGC. Despite the capabilities of advanced tools like single-cell sequencing, achieving a spatially integrated understanding of this disease has been challenging. Our research, therefore, integrates single-cell and spatial transcriptomics to gain an in-depth insight into the interactions of monocyte cells within the TME of LMGC [31–33]. Moreover, we also integrate BULK transcriptome technology to construct a prognostic model, which provides a solid foundation for LMGC treatment.
Methods and materials
Analysis of GEO and TCGA database progression
In our study, we leveraged data from the GC TCGA-STAD and TCGA-LIHC datasets, complemented by clinical data sourced from the TCGA databases accessible at https://portal.gdc.cancer.gov [34]. The TCGA-STAD and TCGA-LIHC datasets were meticulously analyzed to elucidate the expression patterns of key genes. In addition our research further encompassed a cohort of patients diagnosed with gastric cancer (GC) alongside healthy control subjects from the GEO databases (www.ncbi.nlm.nih.gov/geo). Specifically, the datasets included GSE79973, GSE62254, GSE54129, GSE34942, and GSE5118986 for GC, and GSE76427, GSE45267, GSE148851, and GSE112790 for hepatocellular carcinoma (HCC). This comprehensive approach enabled a robust comparative analysis and enhanced our understanding of the gene expression landscapes in both cancerous and healthy tissues.
Standardization of GEO data
(1) Data Acquisition: Obtain the required gene expression data or other bioinformatics data from the GEO database. (2) Data Quality Control: Assess the quality of the raw data, including checking for missing values, outliers, and data distributions. (3) Data Preprocessing: Preprocess the raw data, including background correction, normalization, and transformation, to ensure comparability across different samples. (4) Data Normalization: Normalize the data using appropriate normalization methods (such as Z-score normalization, quantile normalization, etc.) to remove technical variations and ensure comparability of data across different samples. (5) Data Integration: Integrate different datasets into a unified data matrix for further analysis. (6)) Statistical Analysis: Perform bioinformatics analyses such as differential expression analysis, clustering analysis, survival analysis, etc., using the standardized data to reveal biological insights within the data.
scRNA-Seq data processing
We have procured the GSE163558 dataset from the GEO database (https://www.ncbi.nlm.nih.gov/), which includes samples from three primary cases of gastric cancer (GSM5004180, GSM5004181, GSM5004182), one adjacent non-tumoral sample (GSM5004183), and two gastric cancer liver metastasis samples (GSM5004188, GSM5004189). For single-cell RNA sequencing (scRNA-seq) analysis, we employed six samples following the 10 × Genomics scRNA-seq protocol for subsequent analysis.
Utilizing the “Seurat 4.0” R package, we constructed a Seurat object to integrate all sample data. Each sample underwent rigorous quality control, applying screening conditions of a minimum of 200 genes, a maximum of 4000 genes, and a mitochondrial gene percentage (pctMT) below 10%. Post-quality control, normalization was performed using the “LogNormalize” method. Principal Component Analysis (PCA) was conducted on the top 1000 genes to reduce dimensionality, followed by Uniform Manifold Approximation and Projection (UMAP) to visualize the spatial arrangement of cells. This approach allowed for describing various intricacies of the cellular organization of gastric cancer and its metastasis at length.
Spatial transcriptomic data processing
We retrieved the gastric cancer (GC) spatial transcriptomic data (GSM7990475) from the GEO database (https://www.ncbi.nlm.nih.gov/). For the spatial transcriptomic sequencing (stRNA-seq) analysis, the GSM7990475 dataset utilized the 10X Genomics Spatial Transcriptomics platform. To maintain the accuracy of data collected, genes expressed in ten or fewer spots were eliminated from the study.
After this preprocessing step, stringency and accuracy was enhanced through the application of the dimensionality reduction techniques and clustering algorithms in order to determine the most dominant cell types contained in the GC spatial transcriptomic data. This approach made it easier to get a better grasp of the tumor microenvironment (TME) and the various cell types that are involved, which helped improve the knowledge of the spatial heterogeneity of gastric cancer.
GWAS data sources
The summary level across immune phenotypes of genomic variants from the genome-wide association studies (GWAS) were easily retrievable from the GWAS Catalog using accession numbers beginning with GCST90002017. Consequently, the analysis of metabolite traits was done with the data from the same Catalog with the accession codes starting with GCST90199772. These datasets provide important information about genes and linkages to immune features and metabolites for further research to better understand the parts that such traits and profiles play within numerous biological functions and pathologies. Further, upon the use of the data available in the GWAS Catalog (accession numbers starting from GCST90274820), we proceeded to analyze the aforementioned inflammatory traits.
Finally, the analyzed study comprised of 731 immunophenotypes, 1400 metabolite traits and 91 inflammation traits. More specifically, immune traits are described as the medium fluorescence intensity of monocytes, the inflammatory traits are described LIFR, while the metabolite traits are stearoyl metabolism. Consequently, high-density arrays were used to type around 22 million SNPs; this analysis employed a reference database developed using a population of Sardinian origin. The listing of subsequent evaluations of associations incorporate consideration of relevant covariates to minimize effects of confounding that improves the credibility of the results. This sort of approach helped in increasing the reliability and robustness of the genetic associations that were established by the study.
Two-sample MR
The MR approach was used to determine the effect estimate of exposure by analyzing the instrumental variable (IV) relationships among cancer-immune genes IV, immune genes-metabolites, and metabolites-inflammatory factors. Consequently, we deployed two-sample MR to examine the causal circulating immune-metabolites-inflammation-disease relationship that underlies the involvement of the exposure in driving certain outcomes.
Statistical analysis
Data processing and analysis of the data was done using R software version 3. 6. 1. and GraphPad Prism. For comparison between two groups, the student’s t-test was used while comparison of more than two groups was determined by one-way ANOVA. A p-value below 0.05 was used used to test for significance differences between various groups being compared out. These statistical procedures and tests are widely applied in biomedical research in order to evaluate the relevance of tested effects and compare measurable values.
Result
Exploration of the causal effect of CCR2 positive monocyte in GC
In an effort to shed more light on the relationship between immune factors and gastric cancer (GC), our aim was to determine the effects of immune cells in this specific type of cancer. Therefore, utilizing the FDR approach to adjust multiple test corrections, it was concluded that none of the immune traits reached the significance level of 0.05. This suggests that, indeed, none of the immune characteristics assessed could be seen to have been statically related to their respective counterparts at the predefined alpha level of significance while accounting for multiple comparisons in the study as a way of controlling for inflated statistical significance. This stringent approach reduces the likelihood of false discoveries in such large-scale genetic association investigations.
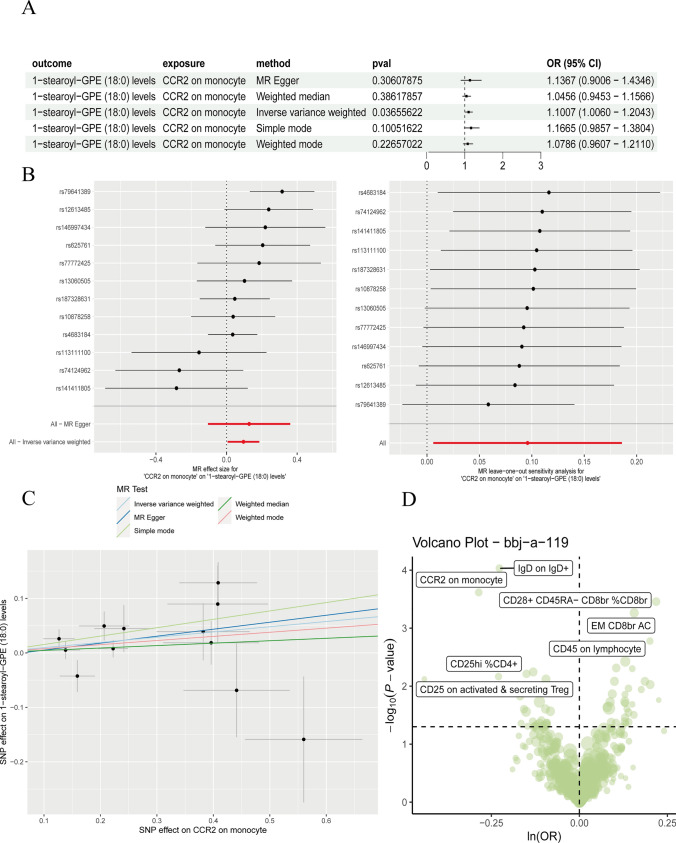
However, in the study conducted at same level of significance (p < 0.05), CCR2 positive monocytes were found in GC. These CCR2-positive monocytes exhibited differential expression within the immune cells associated with GC (as referenced in bbj-a-119, Fig. 1d). Using the inverse-variance weighted (IVW) method, two-sample Mendelian Randomization (MR) analysis revealed that CCR2-positive monocytes exhibit specific expression patterns during the development of GC (β = − 0.285895, P = 0.0002419). Furthermore, at a p-value threshold of 0.05, we identified 70 metabolites, one of which plays a role in regulating lipid metabolism, specifically stearoyl. Additionally, it was found that CCR2-positive monocytes could inhibit the levels of stearoyl (β = − 0.087576, 95% CI = 1.0060 ~ 1.2043, P = 0.03655622, Fig. 1a). To validate the robustness of these causal associations, additional methods and sensitivity analyses, such as MR-Egger and the MR global test, were employed (Fig. 1b, c). These comprehensive analyses provided deeper insights into the interplay between immune factors, metabolite regulation, and the pathogenesis of gastric cancer.
Fig. 1.
CCR2 positive monocyte were found to restrain stearoyl metabolism accelerated progression of GC. a Forest plot display then shows key immune cell with the risk of GC; b The validity and robustness of Mendelian Randomization (MR) are assessed through c the analysis results of MR Heterogeneity Test, which evaluate the coherence of genetic associations across various single nucleotide polymorphisms (SNPs). d The Volcano plot depicts the correlation between crucial immune cell factors and the susceptibility to gastric cancer (GC)
Exploration of the causal effect of stearoyl metabolism level in GC
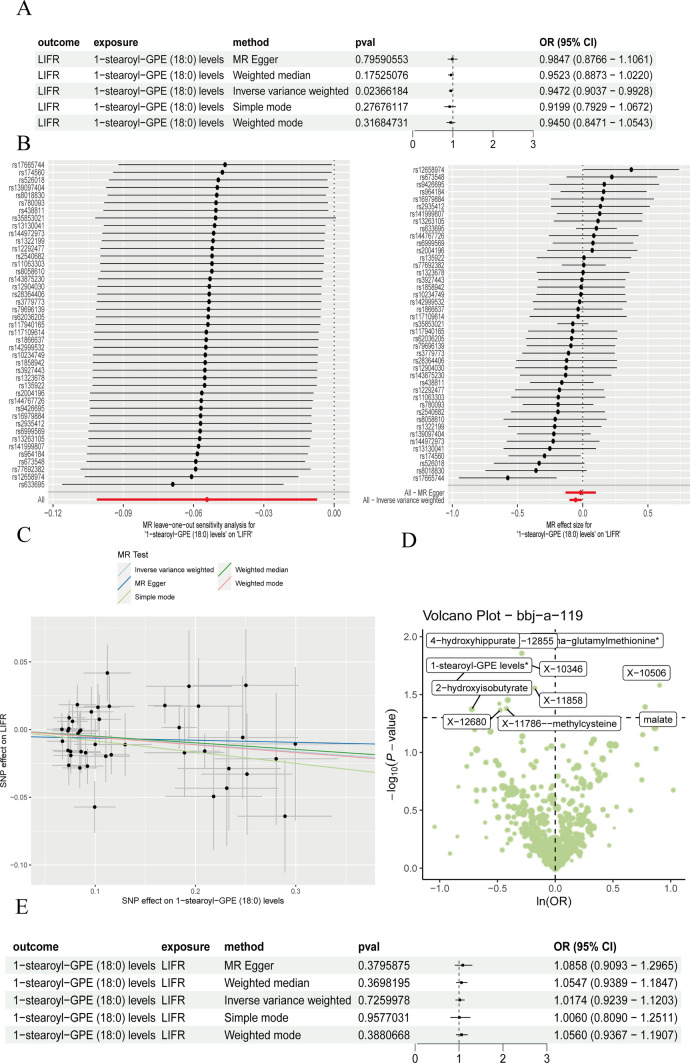
Our preliminary findings underscored the pivotal role of CCR2-positive monocytes in suppressing stearoyl metabolism levels. Subsequent research revealed that the interplay between stearoyl metabolism levels and inflammatory factors significantly influences the progression of GC. To delve deeper into the significance of stearoyl metabolism in GC, Mendelian Randomization (MR) analysis elucidated that stearoyl metabolism levels are associated with the inhibition of LIFR expression, as demonstrated via the inverse-variance weighted (IVW) method (β = − 0.087576, 95% CI = 0.9037 ~ 0.9928, P = 0.02366184, Fig. 2a). The robustness of these causal relationships was further corroborated through additional validation techniques (Fig. 2b, c). We found that GPE metabolic levels exhibited differential expression in GC (Fig. 2d). When inflammatory cytokines were introduced, the reverse MR method showed no alteration in metabolic levels (as depicted in Fig. 2e). This indicates that the influence of metabolism on inflammation is unidirectional within the context of GC.
Fig. 2.
Stearoyl metabolism level restrained inflammatory factor LIFR accelerated progression of GC. a Forest plot display shows key stearoyl metabolism with the risk of GC; b The effectiveness and strength of Mendelian Randomization (MR) are examined through c the analysis outcomes of the MR Heterogeneity Test, which assess the uniformity of genetic connections across various SNPs. d The Volcano plot visually represents the relationship between critical stearoyl metabolism and the susceptibility to gastric cancer (GC); e Forest plot display shows inflammatory factor LIFR with the risk of GC
Identifying the cell types within the GC and LMGC TME
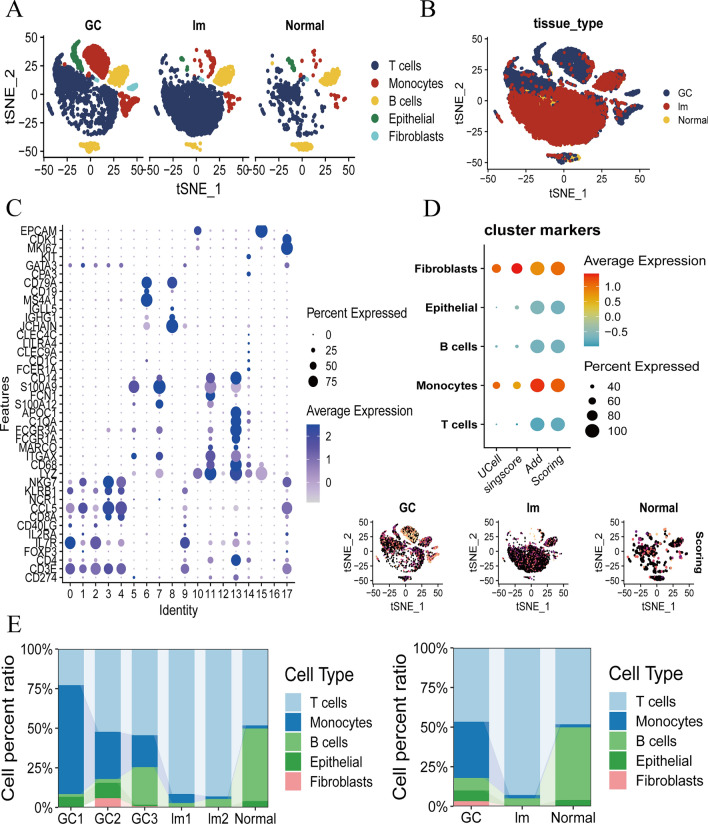
Given the high mortality rate associated with gastric cancer (GC) liver metastasis and the unclear mechanisms underlying this process, our study aimed to elucidate the fundamental processes and functions governing liver metastasis. Utilizing single-cell RNA sequencing (scRNA-seq) data from the GEO database (GSE163558), we identified distinct cell types present in both primary GC tumors and their liver metastases, which contribute to a supportive tumor microenvironment that facilitates metastasis.
Our study revealed five predominant cell types in GC, characterized by high expression markers identified through the Marky annotation method. These cell types include T cells, monocytes, B cells, epithelial cells, and fibroblasts. Notably, monocytes were significantly abundant in primary GC sites but exhibited a marked decrease in abundance within gastric cancer liver metastases (Fig. 3a, c). Additionally, we observed that all cell types in GC liver metastasis samples were highly abundant across all specimens, among them, fibroblasts and monocyte cells were more significantly enriched (Fig. 3b, d). Furthermore, while monocytes constituted a considerable proportion of cells in primary GC, their percentage decreased significantly in liver metastases, highlighting their critical role in the metastatic process (Fig. 3e). These findings provide valuable insights into the cellular dynamics and mechanisms that facilitate GC liver metastasis, potentially informing future therapeutic strategies.
Fig. 3.
ScRNA-seq analysis of normal sample, GC sample and GC liver metastasis sample. a, b. Identification of TME cells type expression in all sample, including T cells, Monocytes cells, B cells, Epithelial cells, and Fibroblasts cells in GSE163558. c The expression of key annotation marker in all sample. d The TME cells type scoring. e Differential ratios of TME cells type. On the left were the ratios for each sample, on the right were the ratios for the Normal, GC and GC liver metastasis
Stearoyl metabolism aggravated the progression of cancer metastasis by NF-KB signaling pathways
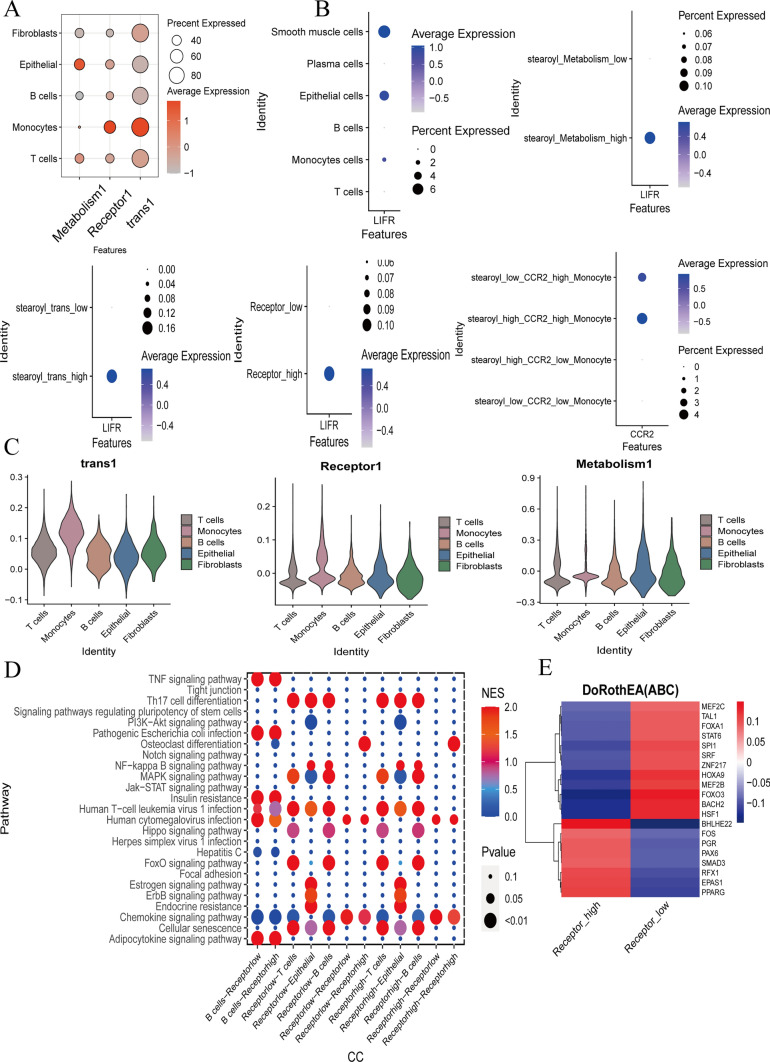
To elucidate the regulatory role of stearoyl metabolism in monocytes within the contexts of GC and GC liver metastasis, we isolated monocytes from both primary GC tumors and liver metastases. Our investigation revealed that the NF-κB signaling pathway plays a pivotal transcriptional role in regulating stearoyl metabolism levels. Furthermore, lipid metabolism emerged as a key factor in the progression of GC to liver metastases. Our findings indicate that stearoyl metabolism facilitates the transformation of GC to liver metastasis in monocytes (Fig. 4a).
Fig. 4.
Stearyl metabolism participated in the regulation of Lipid metabolism by NF-KB signaling pathways. a, c. The different expressions of receptor, transcription, and metabolism in TME cell type. b. Relationship between the Inflammatory factor LIFR, Stearyl metabolism level and Lipid metabolism receptor. d. KEGG pathways enrichment show that key pathways were found in Lipid metabolism receptor. Positive regulation is represented by the hue of crimson, whereas negative regulation is depicted by the hue of azure. e The expression of transcription factors varies in lipid metabolism receptor regulation
Additionally, our research identified the presence of the LIFR inflammatory factor in monocyte cells. One of the critical findings was that increased stearoyl transcription, metabolism, and receptor activity were significantly correlated with elevated expression of the LIFR inflammatory factor. We further discovered that variations in stearoyl levels distinctly influenced the phenotype of CCR2-positive monocytes. Specifically, higher levels of stearoyl metabolism were linked to an increase in CCR2-positive immune cells, whereas lower levels corresponded to a decrease in their presence. This highlights the role of stearoyl metabolism in modulating immune cell phenotypes and inflammatory responses during the progression of gastric cancer (Fig. 4b).
To further investigate the impact of stearoyl metabolism on cellular functions, we examined its transcription, metabolism, and receptor activity across different cell types. Our data reveal significant interactions between cells, notably the interactions between recipient cells and B cells. Moreover, we found that the NF-κB signaling pathway is highly enriched in receptor-high B cells (Fig. 4c, d) regulated by the transcription factor FOS (Fig. 4e). In conclusion, elevated stearoyl metabolism is integral to cellular functions, closely associated with the regulation of the NF-κB signaling pathway, stearoyl transcription levels, and high lipid metabolism receptor levels. These findings underscore the importance of stearoyl metabolism in the regulatory mechanisms underlying gastric cancer progression and metastasis.
Identification of monocyte stearoyl receptor key genes in algorithm model
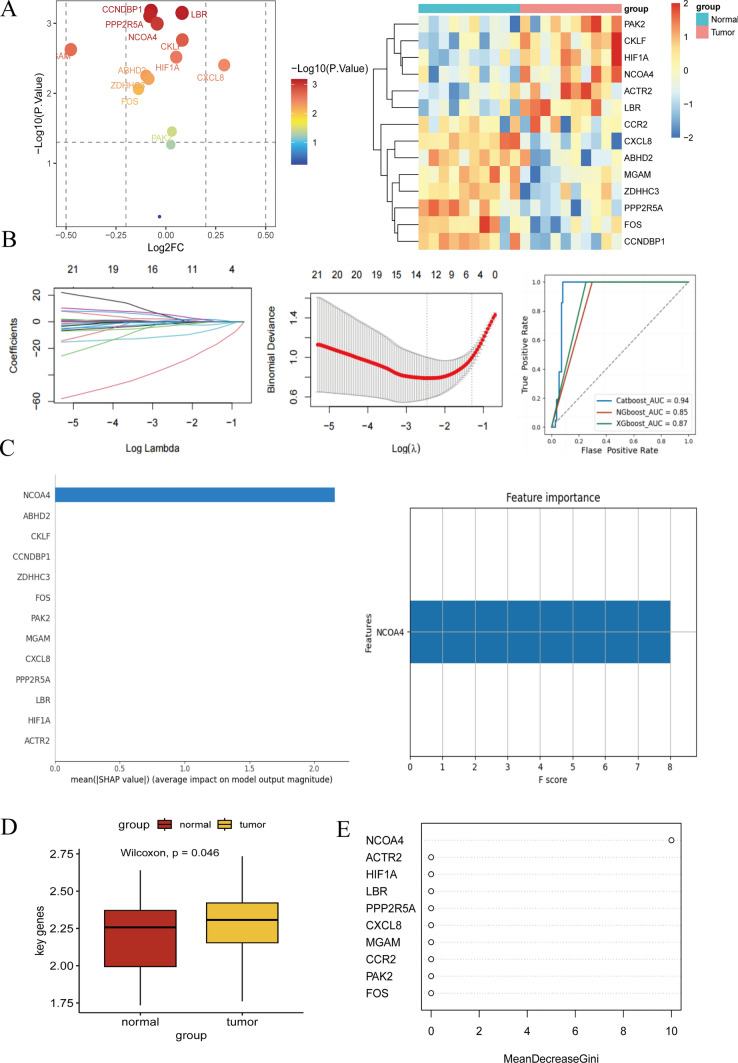
Initially, we extracted monocyte cells from tumor samples, including those from primary gastric cancer (GC) and liver metastases. Following this procedure, we identified differentially expressed genes (P < 0.05, log2FC > 1) in these cells. To ensure the reliability of our findings, we conducted a validation set analysis using GEO datasets GSE79973 and GSE54129. Within these datasets, we constructed algorithmic models and observed that three models—XGBoost (AUC 0.87), NGBoost (AUC 0.85), and CatBoost (AUC 0.94)—exhibited robust performance (Fig. 5b).
Fig. 5.
Bulk transcriptomic analysis of monocyte stearoyl receptor key genes expressed in tumor tissue. a The differential expression of monocyte stearoyl receptor genes in the GC transcriptome were analyzed. In the heatmap and volcano plot, we found that the key genes were found, Intense expression is depicted by the crimson hue, while subdued expression is denoted by the azure shade. b The machine algorithm model was constructed in GSE79973 and GSE54129. AUC > 0.8 indicates high sensitivity and specificity. c, e The expression of NCOA4 in the differential genes. d key genes entirety assessment of the situation
Subsequently, our analysis identified NCOA4 as a significant protein regulating the progression of both GC and its liver metastases, as highlighted by SHAP values, which reflect the average effect on model output magnitude (Fig. 5c). Additionally, we noted differential expression of all key genes within the GSE79973 and GSE54129 datasets (Fig. 5a). Our integrated assessment of all proteins revealed that these key genes facilitated the transformation of GC to liver metastasis (Fig. 5d). Moreover, the results from the MeanDecreaseGini method further corroborated our previous conclusions (Fig. 5e). In summary, our comprehensive analysis underscores the critical regulatory role of NCOA4 and other key genes in the progression of gastric cancer and its liver metastasis, validated through robust algorithmic models and differential gene expression assessments.
Expression prospect of NCOA4, an intermediate hub gene for stearoyl metabolism and ferroptosis
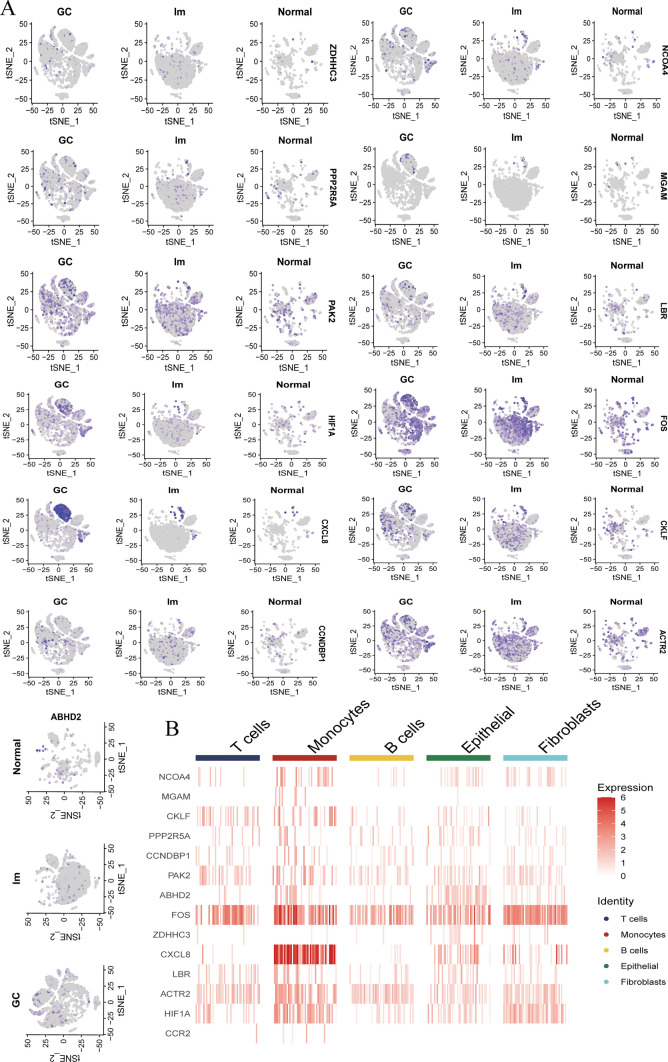
To further substantiate the reliability of our findings, we employed single-cell RNA sequencing (scRNA-seq) data from the GEO dataset GSE163558 to investigate the differential expression of key genes in both GC and GC liver metastasis samples. Our analysis revealed that all identified differential genes were expressed across all samples. Notably, NCOA4 proteins exhibited high enrichment in GC monocyte cells but demonstrated lower enrichment in monocyte cells from liver metastases (Fig. 6a). Moreover, the temporal expression patterns indicated that the migration of NCOA4 expression was a critical factor driving the liver metastasis of GC, particularly evident during the transition from early to late stages of the disease. Additionally, we observed significant variations in the expression of pivotal genes across different cell types within the TME (Fig. 6b). In conclusion, our comprehensive analysis underscores the crucial role of NCOA4 as an intermediate hub gene for stearoyl metabolism and ferroptosis in the progression of gastric cancer to liver metastasis, validated through rigorous scRNA-seq data examination and temporal expression pattern assessments.
Fig. 6.
ScRNA-seq verifies monocyte stearoyl receptor key genes expressed in tumor. a, b. The expression of stearoyl receptor key genes expressed in GC, liver metastasis and normal tissue
Identification of NCOA4 + mono cell types expressed in GC and HCC spatial transcriptomics
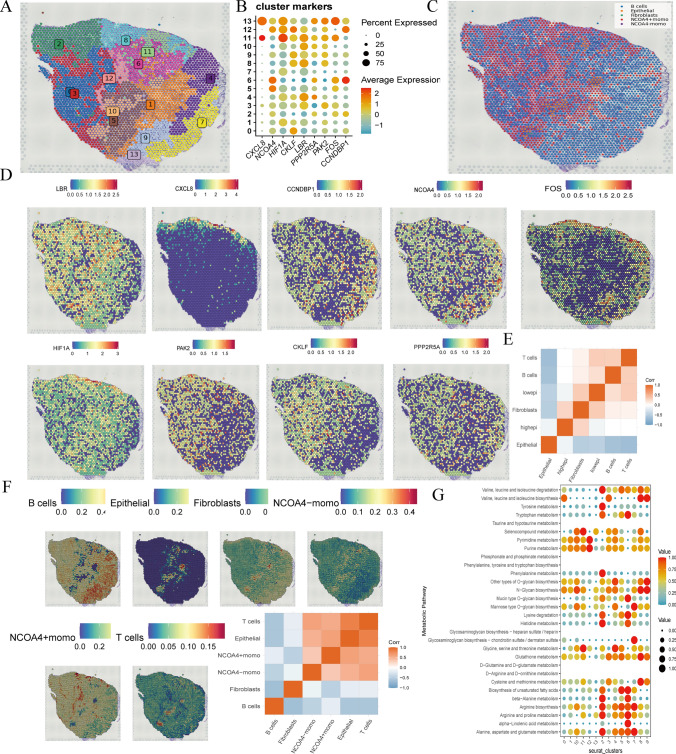
To further validate the expression of key genes, we employed spatial transcriptomics (10X Genomics) to investigate gene expression within GC samples. Due to the lack of available GC liver metastasis samples, we utilized spatial transcriptomics data from GC (GSM7990475) and hepatocellular carcinoma (HCC) (GSM6177612) for comprehensive analysis. This approach led to the identification of 13 distinct cell clusters within the GC samples (Fig. 7a). Among these clusters, key genes were highly expressed in clusters 5 and 6 (Fig. 7b).
Fig. 7.
GC spatial transcriptomics verify monocyte stearoyl receptor key genes expressed. a. Identification of different cell clusters in GC spatial transcriptomics. b. The monocyte stearoyl receptor key genes were expressed in cell cluster. Red signifies high expression, while blue indicates low expression. c, f. The major cell types were identification, including B cells, NCOA4 + mono and NCOA4-mono, Epithelial, Fibroblasts and so on. d, e. The monocyte stearoyl receptor key genes expressed in GC spatial transcriptomics. g. Spatial transcriptomics metabolic shown involved in GC cell metabolic state in all cluster. Elevated expression is represented by the color red, while subdued expression is depicted by the color blue
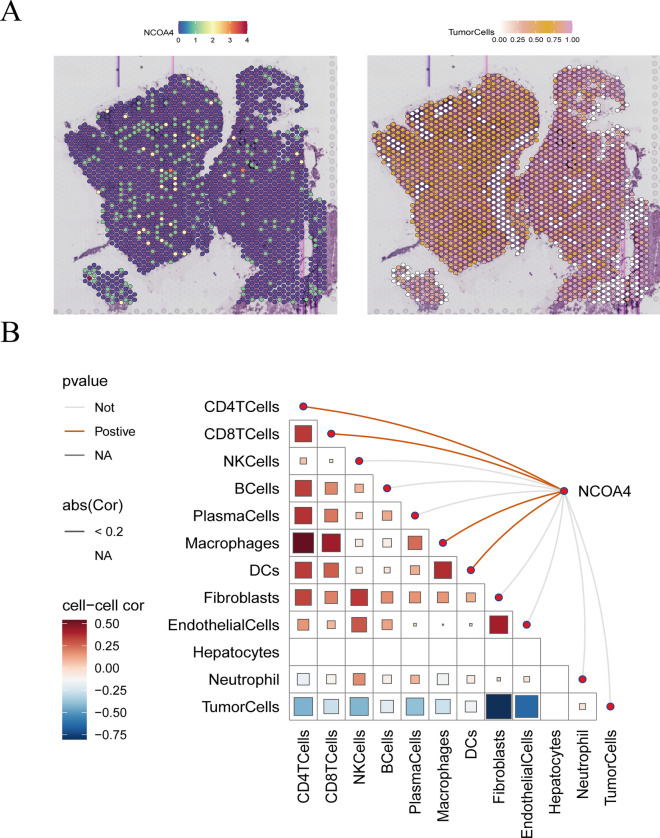
Building upon the outcomes of our previous validations, we focused on the NCOA4 gene. Our analysis revealed the presence of B cells, NCOA4-positive monocytes (NCOA4 + mono), NCOA4-negative monocytes (NCOA4- mono), epithelial cells, and fibroblasts within the GC spatial transcriptomics data (Fig. 7c, f). Notably, high key gene expression levels were closely associated with fibroblast types (Fig. 7e). Further investigation showed that NCOA4-positive monocytes (NCOA4 + mono) exhibited elevated NCOA4 expression levels in GC samples. Additionally, NCOA4 expression was also observed in HCC regions (Fig. 8a). These NCOA4 + monocytes demonstrated significant associations with T cells and epithelial cell types within the spatial transcriptomics data from both GC and HCC tissues (Figs. 7d and 8b).
Fig. 8.
HCC spatial transcriptomics verify NCOA4 expressed (a). Spatial transcriptomics approach to identify NCOA4 expressed in HCC cancer regions (b). The major cell types were identification
Finally, metabolic analysis revealed a close relationship between alpha-linolenic acid metabolism and GC (Fig. 7g). These results suggest a strong interplay between NCOA4 + monocytes, epithelial cells, and B cells. The increased expression of NCOA4 in these cells may mediate inflammatory damage to epithelial and B cells through alpha-linolenic acid metabolism pathways. In summary, our findings underscore the critical role of NCOA4 and its expression in various cell types within the tumor microenvironment. These insights highlight potential mechanisms of inflammatory damage mediated by alpha-linolenic acid metabolism in the progression of gastric cancer and hepatocellular carcinoma.
NCOA4 immunotherapy and prognosis by bulk transcriptome analysis in GC
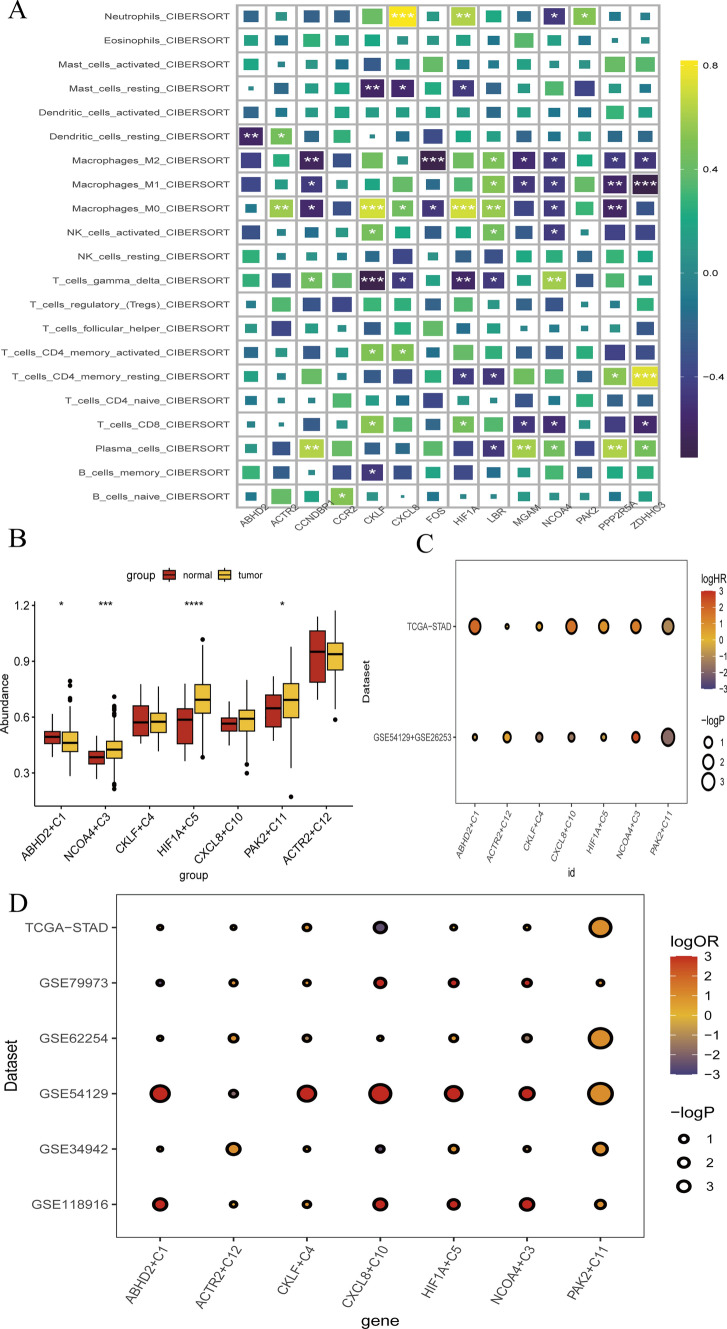
We investigated the association between NCOA4 and various immune cell populations in gastric carcinoma. Our findings indicated that NCOA4 expression was correlated with altered immune cell levels, showing a positive association with several immune cells, including plasma cells, regulatory T cells, CD8 + T cells, macrophages-M0, macrophages-M1, and macrophages-M2 (Fig. 9a). Furthermore, we observed that elevated NCOA4 expression was linked to poorer prognosis (Fig. 9b). Subsequent analysis of bulk transcriptome data revealed that NCOA4 plays a crucial role in promoting gastric cancer (GC) progression, acting as a significant driver in the disease’s advancement (Fig. 9c). Notably, these results were consistent with our previous observations. Additionally, we discovered that NCOA4 proteins were associated with improved prognosis for GC based on immune checkpoint blockade (ICB) therapy outcomes (Fig. 9d). In summary, our comprehensive analysis highlights the pivotal role of NCOA4 in modulating immune cell populations and its significant impact on the prognosis and progression of gastric cancer. These insights underscore the potential of NCOA4 as a critical biomarker and therapeutic target in GC, particularly in the context of ICB therapy.
Fig. 9.
ICB immunotherapy restrained the progress of GC a Immune infiltration of monocyte stearoyl receptor key genes in GC. b The expression of monocyte stearoyl receptor key genes in GC. c. bulk transcriptome analysis of the expression of monocyte stearoyl receptor key genes and prognosis by TCGA-STAD, GSE54129 and GSE26253 in GC. Red represents the injure factor, blue represents the protection factor. d. Evaluation of the prognosis of ICB immunotherapy by TCGA-STAD, GSE79973, GSE62254, GSE54129, GSE34942 and GSE5118986 in GC. Red indicates a better ICB response, blue indicates poor response
Immunotherapy and prognosis in liver metastasis and primary liver cancer
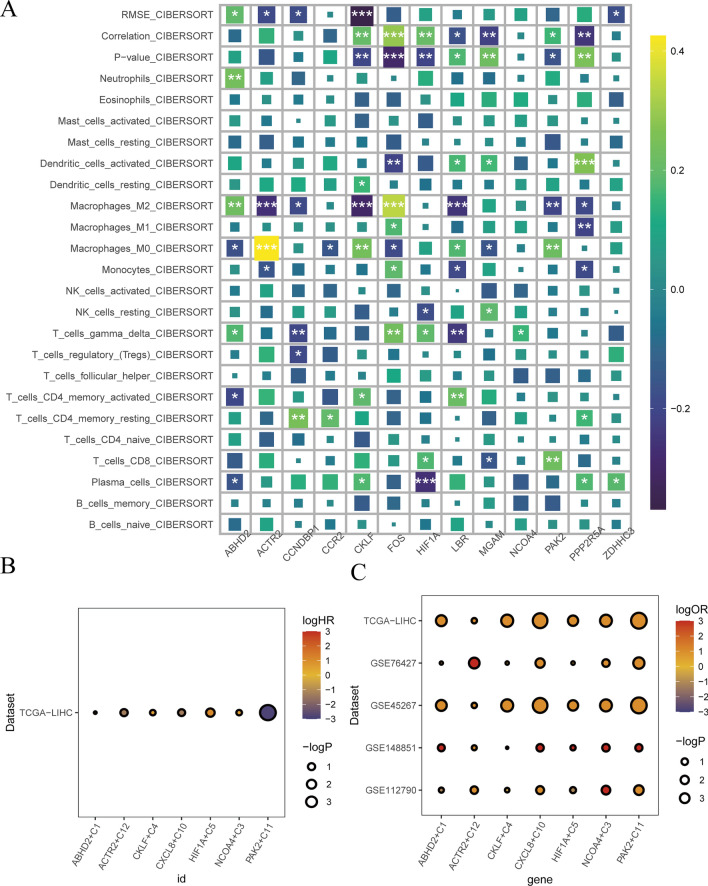
In order to delve deeper into the correlation between NCOA4 proteins and different immune cells, additional studies should focus on liver metastasis from primary sites, such as GC, as well as primary liver cancer. Our data revealed that the expression of NCOA4 was leading to T cells function disorder (Fig. 10a). This is consistent with the results previously described. The results showed that NCOA4 as an key factor promoted progression of liver cancer (Fig. 10b). Meanwhile, we used primary liver cancer data (GSE76427, GSE45267, GSE112790 and TCGA-LIHC) and liver metastasis data (GSE148851) analysis, this results also revealed that the NCOA4 proteins presents a better prognosis for GC based on ICB (Fig. 10c).
Fig. 10.
ICB immunotherapy restrain the progress of liver metastasis and primary liver cancer. a Immune infiltration of monocyte stearoyl receptor key genes in liver cancer. b bulk transcriptome analysis the expression of monocyte stearoyl receptor key genes and prognosis by TCGA-LIHC in liver cancer. Red represents the injure factor, blue represents the protection factor. c. Evaluation of the prognosis of ICB immunotherapy by TCGA-LIHC, GSE76427, GSE45267, GSE148851 and GSE112790 in liver metastasis and primary liver cancer. Red indicates a better ICB response, blue indicates poor response. Of these, GSE148851 was liver metastases sample, the TCGA-LIHC, GSE76427, GSE45267 and GSE112790 were primary liver cancer samples
Discussion
Previous studies have identified cancer immune escape as a significant risk factor contributing to the high incidence of cancer [35, 36]. Tumor-infiltrating immune cells, such as T cells [37], B cells [38, 39], and monocytes [40, 41], can compromise the body’s natural defense and surveillance mechanisms. Emerging research has illuminated that monocytes, previously considered a uniform cell group, actually encompass a heterogeneous array of cells with diverse responses to various stimuli. These cells contribute to anti-tumor immunity through multiple cellular mechanisms, including the release of tumoricidal substances, regulation of B and T cell activities, recruitment of lymphocytes, and differentiation into tumor-associated macrophages and dendritic cells [42]. In our research focusing on cancer immune escape, we discovered that the dysregulation of CCR2-positive monocyte surveillance significantly promotes cancer progression, particularly in late-stage liver metastasis. Furthermore, we observed a notable relationship between CCR2-positive monocytes and metabolic processes, including stearoyl metabolism. It emerges from the study that CCR2-positive monocytes suppress stearoyl metabolism and exacerbate cancer development. These results emphasize the possible applicability of immunotherapy as a new and effective treatment strategy to use the body’s immune response to combat tumor cells. As immunotherapy tends to focus on particular subsets of immune cells and the metabolic circuits in this regard, it potentially defines new approaches to combating malignant growth.
Recent studies have indicated that stearoyl is important for modulating lipid metabolism, affecting cancer advancement [43]. Imbalance of lipid metabolism synthesis has been identified as a potential oncogenic factor for different types of cancer, such as lung cancer [44], liver cancer [45], and prostate cancer [46]. Previous works have exposed the knowledge that lipid metabolism is a fundamental part of T cell signaling and inflammation, activity [47, 48]. We have identified stearoyl as the key molecule that controls inflammation process, indicating that stearoyl promotes the expression of inflammatory factor LIFR, hence, slowing down the gastric cancer (GC) and its liver metastasis process. On the other hand, decreased stearoyl expression leads to the downregulation of LIFR levels, thus, enhancing cell cancer growth to progression. Furthermore, Mendelian randomization (MR) could support that stearoyl metabolism has a negative association with inflammatory factors, which is consistent with our conclusions. What this implies is that low stearoyl metabolism impairs energy and nutrient uptake which sets the stage for the growth and spread of the cancers. Concisely, our study emphasizes the exceptional role of stearoyl in lipid profiles and inflammation and their collective functions in cancer development, pointing towards the possibility of stearoyl as a promising anti-cancer drug target.
In a study finding of this research, we have established that lipid metabolism especially involving the stearoyl receptor can significantly contribute to cell regulatory mechanisms. In our present work, we examined NCOA4, which mainly relates to the stearoyl receptor; we confirmed this gene as a high-risk factor for GC and LM and its effect on physiological and pathological changes. It is worth mentioning that the intertumor heterogeneity of NCOA4 appears to be especially manifested with the progression of advanced GC and liver metastasis. Stearoyl stands out as an influential factor affecting inflammation, to which our research primarily attributes. Namely, the rise in stearoyl levels leads to the activation of LIFR, the anti-inflammatory factor, which slows down the development of GC and its liver metastasis. Serving as the link of GC and liver metastasis, NCOA4, a nuclear receptor coactivator, promotes the process of GC liver metastasis through lipid metabolism related pathway. Nonetheless, owing to the variety of regulatory genes and the complexity of liver metastasis, the exact function of NCOA4 gene on the regulation is still elucidated. Thus, our research indicates that NCOA4 affects the T-cell activity through the NF-κB signaling pathways leading to exacerbation of the disease. The results offer a new avenue for understanding the development and formation of liver metastasis in GC, and identify NCOA4 as a potential therapeutic target for intervention.
Immune checkpoint blockade (ICB) immunotherapy has demonstrated significant success in enhancing the immune response across various cancers [49, 50], including liver cancer [51, 52], lung cancer [53], and colorectal cancer [54, 55]. It is crucial to emphasize that immune evasion, often resulting from T cell depletion and dysfunction, plays a pivotal role in cancer progression [56, 57]. Our recent observations further underscore the importance of T cell dysfunction as a connecting factor between gastric cancer (GC) and GC liver metastasis, suggesting a strong association between T cell depletion, dysfunction, and the metastatic process.
In our analysis of NCOA4 in the context of ICB, we stratified GC from GC liver metastasis based on survival rates. The results indicated a favorable prognosis, suggesting the therapeutic potential of NCOA4 in this setting. These findings open new possibilities for the treatment of liver metastasis in gastric cancer and present opportunities to enhance the efficacy of ICB immunotherapy in combating this malignancy.
Expanding the discussion on NCOA4 in relation to ICB to explore potential combination therapies and resistance mechanisms could offer valuable insights. A detailed examination of how NCOA4 and stearoyl metabolism influence immune cell exhaustion and response to ICB could enrich the narrative on immunotherapy. Understanding the specific pathways through which NCOA4 and stearoyl metabolism impact immune cell function, such as T cell exhaustion and regulatory T cell activity, is essential for comprehensively grasping their roles in modulating the immune response in the tumor microenvironment.
Furthermore, exploring potential combination therapies targeting NCOA4 or stearoyl metabolism alongside ICB may reveal novel strategies to overcome resistance and enhance treatment effectiveness. By integrating these aspects into our discussion, we can delve deeper into the intricate interactions among stearoyl metabolism, NCOA4, and immune cell exhaustion within the context of ICB, offering valuable insights for advancing cancer immunotherapy research.
Our study provides significant insights into the transition from GC to its liver metastasis, with a focus on the critical role of monocyte immune dysfunction in regulating stearoyl metabolite levels. Notably, the involvement of lipid metabolism through the stearoyl metabolite receptor indicates a contribution to disturbances within the tumor microenvironment and inflammation via the NF-κB signaling pathway. Additionally, our findings highlight the upregulation of the key protein NCOA4, associated with the stearoyl metabolite receptor, in both GC and its liver metastasis, thereby exerting a significant influence on disease progression.
Looking ahead, there is considerable potential in enhancing treatment efficacy by targeting monocyte immune pathways to hinder the progression from GC to liver metastasis. This study not only advances our understanding of the dynamics involved in GC metastasis but also sets the stage for the development of more effective therapeutic approaches.
Conclusion
This study enhances our understanding of the relationship between stearoyl metabolism and ferroptosis in promoting liver metastasis of gastric cancer (LMGC) and their role in regulating tumor heterogeneity. These findings contribute to the advancement of clinical therapeutic strategies based on metabolic pathways, specifically the link between stearoyl metabolism and the essential ferroptosis gene NCOA4. In addition, the study provides deeper insights into the dynamics of the tumor microenvironment (TME) in gastric cancer, laying the groundwork for the development of more effective interventions to combat cancer metastasis.
Author contributions
ZY, HC, JF and LW conceived the study. ZY, YC, YM, HY, KC, YX, LS, LZ, YY, HC, JF and LW drafted the manuscript. ZY and LW performed the literature search and collected the data. ZY and LW completed in vitro experiments. LW analyzed and visualized the data. LW, HC and JF helped with the final revision of this manuscript. All authors reviewed and approved the final manuscript.
Funding
Sichuan Medical Association Project (S21048); Dazhou Science and Technology Bureau project (21ZDYF0025); Sichuan Medical Youth Innovation Research Project (No. Q23095).
Data availability
The datasets analyzed in this study can be found in GEO (https://www.ncbi.nlm.nih.gov/geo/), Xena (https://xena.ucsc.edu/) and GWAS (https://www.ebi.ac.uk/gwas/). However, the datasets used and/or analyzed in this study are available from the corresponding authors of this study upon reasonable request. All raw data are available at https://www.jianguoyun.com/p/DdwK6v4QhczHDBiIvdQFIAA.
Declarations
Competing interests
The authors affirm that the study was conducted without any commercial or financial relationships that could be perceived as a possible competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhongqiu Yang, Yuquan Chen, Yaping Miao and Haisheng Yan have contributed equally to this work.
Contributor Information
Hao Chi, Email: Chihao7511@163.com.
Jin Fu, Email: fujin7589@163.com.
Lexin Wang, Email: 13255859001@163.com.
References
- 1.Luo P, Chen G, Shi Z, Yang J, Wang X, Pan J, Zhu L. Comprehensive multi-omics analysis of tryptophan metabolism-related gene expression signature to predict prognosis in gastric cancer. Front Pharmacol. 2023;14:1267186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang J, Wei W, Xu Y, Chen K, Miao Y, Fan W, et al. CXC chemokine receptor 4-mediated immune modulation and tumor microenvironment heterogeneity in gastric cancer: Utilizing multi-omics approaches to identify potential therapeutic targets. BioFactors. 2024;2024:1. [DOI] [PubMed] [Google Scholar]
- 3.Wang L, Zhou X, Yan H, Miao Y, Wang B, Gu Y, et al. Deciphering the role of tryptophan metabolism-associated genes ECHS1 and ALDH2 in gastric cancer: implications for tumor immunity and personalized therapy. Front Immunol. 2024;15: 1460308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Chi H, Zhao X, Pan R, Wei Y, Han Y. Role of exosomes in immune microenvironment of hepatocellular carcinoma. J Oncol. 2022;2022: 2521025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seeneevassen L, Zaafour A, Sifré E, Genevois C, Nguyen TL, Pobiedonoscew Y, et al. Targeting metastasis-initiating cancer stem cells in gastric cancer with leukaemia inhibitory factor. Cell Death Discov. 2024;10:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su K, Shen Q, Tong J, Gu T, Xu K, Li H, et al. Construction and validation of a nomogram for HBV-related hepatocellular carcinoma: a large, multicenter study. Ann Hepatol. 2023;28: 101109. [DOI] [PubMed] [Google Scholar]
- 7.Bleve A, Durante B, Sica A, Consonni FM. Lipid metabolism and cancer immunotherapy: immunosuppressive myeloid cells at the crossroad. Int J Mol Sci. 2020;21:5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Guo L, Su K, Li C, Jiang Y, Wang P, et al. Construction and validation of TACE therapeutic efficacy by ALR score and nomogram: a large, multicenter study. J Hepatocell Carcinoma. 2023;10:1009–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su K, Liu Y, Wang P, He K, Wang F, Chi H, et al. Heat-shock protein 90α is a potential prognostic and predictive biomarker in hepatocellular carcinoma: a large-scale and multicenter study. Hepatol Int. 2022;16:1208–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang L, Zhang S, Jiang C, Chen H, Huang J, Yang J, et al. Integrative biomarker discovery and immune profiling for ulcerative colitis: a multi-methodological approach. Sci Rep. 2024;14:24290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu Y, Jiang L, Shui M, Luo H, Zhou X, Zhang S, Jiang C, Huang J, Chen H, Tang J, Fu Y, Luo H, Yang G, Xu K, Chi H, Liu J, Huang S. Revealing the association between East Asian oral microbiome and colorectal cancer through Mendelian randomization and multi-omics analysis. Front Cell Infect Microbiol. 2024;14:1452392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang L, Liu J, Zhang S, Jiang C, Huang J, Chen H, et al. Role of glycosylation-related gene MGAT1 in pancreatic ductal adenocarcinoma. Front Immunol. 2024;15:1438935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y, Cheng Y, Wang X, Fan J, Gao Q. Spatial omics: navigating to the golden era of cancer research. Clin Transl Med. 2022;12: e696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tu H, Hu Q, Ma Y, Huang J, Luo H, Jiang L, et al. Chen, Deciphering the tumour microenvironment of clear cell renal cell carcinoma: prognostic insights from programmed death genes using machine learning. J Cell Mol Med. 2024;28: e18524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang L, Ren X, Yang J, Chen H, Zhang S, Zhou X, et al. Mitophagy and clear cell renal cell carcinoma: insights from single-cell and spatial transcriptomics analysis. Front Immunol. 2024;15:1400431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang J, Zhang S, Jiang L, Liu J, Xu J, Jiang C, Chi H, et al. Causal relationship between immune cells and hepatocellular carcinoma: a Mendelian randomisation study. J Cancer. 2024;15:4219–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Zuo H, Huang J, Liu J, Jiang L, Jiang C, et al. Unravelling infiltrating T-cell heterogeneity in kidney renal clear cell carcinoma: integrative single-cell and spatial transcriptomic profiling. J Cell Mol Med. 2024;28: e18403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang C, Zhang S, Jiang L, Chen Z, Chen H, Huang J, et al. Precision unveiled: synergistic genomic landscapes in breast cancer-Integrating single-cell analysis and decoding drug toxicity for elite prognostication and tailored therapeutics. Environ Toxicol. 2024;39:3448–72. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh WC, Budiarto BR, Wang YF, Lin CY, Gwo MC, So DK, et al. Chen, Spatial multi-omics analyses of the tumor immune microenvironment. J Biomed Sci. 2022;29:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xing QR, Cipta NO, Hamashima K, Liou YC, Koh CG, Loh YH. Unraveling heterogeneity in transcriptome and its regulation through single-cell multi-omics technologies. Front Genet. 2020;11:662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng H, Wang M, Su J, Li Y, Long J, Chu J, Li Q, et al. Lipid metabolism and cancer. Life (Basel). 2022;12:784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He L, Ye Q, Zhu Y, Zhong W, Xu G, Wang L, et al. Lipid metabolism-related gene signature predicts prognosis and indicates immune microenvironment infiltration in advanced gastric cancer. Gastroenterol Res Pract. 2024;2024:6639205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kollara A, Brown TJ. Expression and function of nuclear receptor co-activator 4: evidence of a potential role independent of co-activator activity. Cell Mol Life Sci. 2012;69:3895–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mou Y, Wu J, Zhang Y, Abdihamid O, Duan C, Li B. Low expression of ferritinophagy-related NCOA4 gene in relation to unfavorable outcome and defective immune cells infiltration in clear cell renal carcinoma. BMC Cancer. 2021;21:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu C, Chang W, Wu J, Yao Y, Liu G, Yuan Y, et al. NCOA4: an immunomodulation-related prognostic biomarker in colon adenocarcinoma and pan-cancer. J Oncol. 2022;2022:5242437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christianson J, Oxford JT, Jorcyk CL. Emerging perspectives on leukemia inhibitory factor and its receptor in cancer. Front Oncol. 2021;11: 693724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang F, Wang Y, Li H, Li L, Yang X, You X, et al. Pan-cancer analysis identifies LIFR as a prognostic and immunological biomarker for uterine corpus endometrial carcinoma. Front Oncol. 2023;13:1118906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang S, Sun B, Li W, Yang H, Li N, Zhang X. Fatty acid metabolism is related to the immune microenvironment changes of gastric cancer and RGS2 is a new tumor biomarker. Front Immunol. 2022;13:1065927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Wang H, Tian Y, Li T, Zhang W, Ma L, et al. Discovery of a novel lipid metabolism-related gene signature to predict outcomes and the tumor immune microenvironment in gastric cancer by integrated analysis of single-cell and bulk RNA sequencing. Lipids Health Dis. 2023;22:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang H, Yu D, Yang P, Guo R, Kong M, Gao Y, et al. Revealing the transcriptional heterogeneity of organ-specific metastasis in human gastric cancer using single-cell RNA Sequencing. Clin Transl Med. 2022;12: e730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li D, Zhang X, Jiang L. Molecular mechanism and potential therapeutic targets of liver metastasis from gastric cancer. Front Oncol. 2022;12:1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen Y, Chen JX, Li M, Xiang Z, Wu J, Wang YJ. Role of tumor-associated macrophages in common digestive system malignant tumors. World J Gastrointest Oncol. 2023;15:596–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang N, Wang S, Wang X, Zheng Y, Yang B, Zhang J, et al. Research trends in pharmacological modulation of tumor-associated macrophages. Clin Transl Med. 2021;11: e288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shahait M, Dobbs R, Lee DI. Commentary RE: Balasubramanian S, Ronstrom C, Shiang A, Vetter JM, Sheets J, Palka J, Figenshau RS, Kim EH. Feasibility and safety of same-day discharge following single-port robotic-assisted laparoscopic prostatectomy. World J Urol. 2022 Nov 2:1–7. 10.1007/s00345-022-04204-y. Epub ahead of print. PMID: 36322183; PMCID: PMC9629187. World J Urol 41 (2023) 283–284. [DOI] [PMC free article] [PubMed]
- 35.Mamedov MR, Vedova S, Freimer JW, Sahu AD, Ramesh A, Arce MM, Kuball Z, Sebestyen EJ, Adams, Marson A, et al. CRISPR screens decode cancer cell pathways that trigger γδ T cell detection. Nature 2023;621:188–95. [DOI] [PMC free article] [PubMed]
- 36.Zhang S, Jiang C, Jiang L, Chen H, Huang J, Gao X, Zhang J, Chi H, Yang G, Tian G, et al. Construction of a diagnostic model for hepatitis B-related hepatocellular carcinoma using machine learning and artificial neural networks and revealing the correlation by immunoassay. Tumour Virus Res. 2023;16: 200271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vilbois S, Xu Y, Ho PC. Metabolic interplay: tumor macrophages and regulatory T cells. Trends Cancer. 2024;10:242–55. [DOI] [PubMed] [Google Scholar]
- 38.Ma J, Wu Y, Ma L, Yang X, Zhang T, Song G, et al. A blueprint for tumor-infiltrating B cells across human cancers. Science. 2024;384:eadj4857. [DOI] [PubMed] [Google Scholar]
- 39.Tellier J, Nutt SL. B cell trajectories influence cancer outcomes. Science. 2024;384:510–1. [DOI] [PubMed] [Google Scholar]
- 40.Tokunaga R, Naseem M, Lo JH, Battaglin F, Soni S, Puccini A, et al. Lenz, B cell and B cell-related pathways for novel cancer treatments. Cancer Treat Rev. 2019;73:10–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z, Wang H, Li Z, Dress RJ, Zhu Y, Zhang S, et al. Dendritic cell type 3 arises from Ly6C(+) monocyte-dendritic cell progenitors. Immunity. 2023;56:1761-1777.e6. [DOI] [PubMed] [Google Scholar]
- 42.Kwart D, He J, Srivatsan S, Lett C, Golubov J, Oswald EM, et al. Cancer cell-derived type I interferons instruct tumor monocyte polarization. Cell Rep. 2022;41: 111769. [DOI] [PubMed] [Google Scholar]
- 43.Bian X, Liu R, Meng Y, Xing D, Xu D, Lu Z. Lipid metabolism and cancer. J Exp Med. 2021;218:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hagihara M, Kato H, Yamashita M, Shibata Y, Umemura T, et al. Lung cancer progression alters lung and gut microbiomes and lipid metabolism. Heliyon. 2024;10: e23509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alannan M, Fayyad-Kazan H, Trézéguet V, Merched A. Targeting lipid metabolism in liver cancer. Biochemistry. 2020;59:3951–64. [DOI] [PubMed] [Google Scholar]
- 46.Pardo JC, Ruiz de Porras V, Gil J, Font A, Puig-Domingo M, Jordà M. Lipid metabolism and epigenetics crosstalk in prostate cancer. Nutrients. 2022;14:851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim SA, Su W, Chapman NM, Chi H. Lipid metabolism in T cell signaling and function. Nat Chem Biol. 2022;18:470–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bougarne N, Weyers B, Desmet SJ, Deckers J, Ray DW, Staels B, et al. Molecular actions of PPARα in lipid metabolism and inflammation. Endocr Rev. 2018;39:760–802. [DOI] [PubMed] [Google Scholar]
- 49.Pai JA, Hellmann MD, Sauter JL, Mattar M, Rizvi H, Woo HJ, Chow A, Satpathy AT, et al. Lineage tracing reveals clonal progenitors and long-term persistence of tumor-specific T cells during immune checkpoint blockade. Cancer Cell. 2023;41:776-790.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Topalian SL, Forde PM, Emens LA, Yarchoan M, Smith KN, Pardoll DM. Neoadjuvant immune checkpoint blockade: a window of opportunity to advance cancer immunotherapy. Cancer Cell. 2023;41:1551–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu RQ, Lao XM, Chen DP, Qin H, Mu M, Cao WJ, et al. Kuang, Immune checkpoint therapy-elicited sialylation of IgG antibodies impairs antitumorigenic type I interferon responses in hepatocellular carcinoma. Immunity. 2023;56:180-192.e11. [DOI] [PubMed] [Google Scholar]
- 52.Feng M, Wang F, Liu X, Hao T, Zhang N, Deng M, et al. Neutrophils as key regulators of tumor immunity that restrict immune checkpoint blockade in liver cancer. Cancer Biol Med. 2023;20:421–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chow A, Uddin FZ, Liu M, Dobrin A, Nabet BY, Mangarin L, Mansilla-Soto M, Sadelain CA, Klebanoff MD, Hellmann T, Sen E, de Stanchina JD, Wolchok T, Merghoub, Rudin CM, et al. The ectonucleotidase CD39 identifies tumor-reactive CD8(+) T cells predictive of immune checkpoint blockade efficacy in human lung cancer. Immunity 2023;56:93–106.e6. [DOI] [PMC free article] [PubMed]
- 54.Xu Q, Liu C, Wang H, Li S, Yan H, Liu Z, et al. Deciphering the impact of aggregated autophagy-related genes TUBA1B and HSP90AA1 on colorectal cancer evolution: a single-cell sequencing study of the tumor microenvironment. Discov Oncol. 2024;15:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Babl N, Decking SM, Voll F, Althammer M, Sala-Hojman A, Ferretti R, et al. MCT4 blockade increases the efficacy of immune checkpoint blockade. J Immunother Cancer. 2023;11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chakraborty C, Sharma AR, Bhattacharya M, Lee SS. A detailed overview of immune escape, antibody escape, partial vaccine escape of SARS-CoV-2 and their emerging variants with escape mutations. Front Immunol. 2022;13: 801522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mocellin S, Nitti D. Therapeutics targeting tumor immune escape: towards the development of new generation anticancer vaccines. Med Res Rev. 2008;28:413–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed in this study can be found in GEO (https://www.ncbi.nlm.nih.gov/geo/), Xena (https://xena.ucsc.edu/) and GWAS (https://www.ebi.ac.uk/gwas/). However, the datasets used and/or analyzed in this study are available from the corresponding authors of this study upon reasonable request. All raw data are available at https://www.jianguoyun.com/p/DdwK6v4QhczHDBiIvdQFIAA.