Abstract
Human melanoma cells can be reprogrammed to terminally differentiate and irreversibly lose proliferative capacity by appropriate pharmacological manipulation. Subtraction hybridization identified melanoma differentiation-associated gene-5 (mda-5) as a gene induced during differentiation, cancer reversion, and programmed cell death (apoptosis). This gene contains both a caspase recruitment domain and putative DExH group RNA helicase domains. Atypical helicase motifs of MDA-5 deviate from consensus sequences but are well conserved in a potentially new group of cloned and hypothetical proteins. mda-5 is an early response gene inducible by IFN and tumor necrosis factor-α, responding predominantly to IFN-β. Protein kinase C activation by mezerein further augments mda-5 expression induced by IFN-β. Expression of mda-5 is controlled transcriptionally by IFN-β, and the MDA-5 protein localizes in the cytoplasm. mda-5 displays RNA-dependent ATPase activity, and ectopic expression of mda-5 in human melanoma cells inhibits colony formation. In these contexts, mda-5 may function as a mediator of IFN-induced growth inhibition and/or apoptosis. MDA-5 is a double-stranded RNA-dependent ATPase that contains both a caspase recruitment domain and RNA helicase motifs, with a confirmed association with growth and differentiation in human melanoma cells.
Terminal differentiation is essential for normal development and homeostasis (1). This process can go awry in cancer cells, resulting in uncontrolled proliferation and an inability to respond to normal growth-inhibitory signals (2, 3). A potentially less toxic form of therapy, differentiation therapy, is designed to reprogram cancer cells to a more differentiated state. This differentiation programming results in a loss of growth potential/terminal cell differentiation or programmed cell death (apoptosis; refs. 1–5). Moreover, by treatment with appropriate pharmacological agents it is possible to initiate a specific pathway, either differentiation or apoptosis, in cancer cells (1–5). To effectively use differentiation therapy it will be important to define the genes and pathways involved in determining cell fate.
In human melanoma cells, the combination of recombinant human IFN-β and the protein kinase C (PKC)-activating compound mezerein (MEZ) induces a rapid and irreversible loss in proliferation, an extinction of tumorigenic potential in nude mice, and terminal differentiation (1, 4, 6, 7). This process correlates with significant changes in cellular physiology including irreversible alterations in growth, enhanced melanogenesis, morphological changes, cell surface antigen modifications, and profound variations in gene expression (1, 4). When administered alone, IFN-β and MEZ can induce specific differentiation-associated changes in melanoma cells including growth suppression, melanogenesis, and morphology. However, these alterations in phenotype are fully reversible after removal of the inducer (1, 4, 6, 7). In these contexts, this differentiation system represents an ideal model to study growth control, differentiation (both reversible and terminal), and tumor suppression.
Phage-based cDNA-subtracted libraries and a recently developed rapid subtraction hybridization (RaSH) approach are being used to define the relevant gene changes associated with induction of terminal differentiation (1, 8). Initial screening of a melanoma differentiation-subtracted library identified a partial mda-5 expressed sequence tag (8). A full-length mda-5 cDNA has now been cloned by using a modified rapid amplification of cDNA ends approach. This gene is an early IFN- and tumor necrosis factor-α (TNF-α)-responsive gene, the transcription of which is increased by treatment with IFN-β and IFN-β + MEZ. Based on sequence analysis MDA-5 is a putative RNA helicase and as expected, it manifests strict RNA-dependent ATPase activity. An unanticipated feature of the MDA-5 protein is the presence of a caspase recruitment domain (CARD), which is unique among the DExH/D family of proteins. Ectopic expression of mda-5 suppresses colony formation in HO-1 human melanoma cells. In these contexts, mda-5 represents a putative RNA helicase with confirmed growth-suppressive properties. This gene may play a significant role in IFN-induced growth suppression and/or apoptosis.
Materials and Methods
Cell Cultures.
HO-1 human melanoma cells and 293 cells were grown as described (6). Sf9 cells were cultured in TNM-FH medium (Mediatech Laboratories, Cody, NY) supplemented with 10% FBS and penicillin/streptomycin (100 units/100 μg/ml) at 27°C in a humidified incubator.
Cloning and Sequencing of mda-5.
A partial mda-5 cDNA (3′, 1.8 kb) was cloned by screening a human placental cDNA library (CLONTECH; ref. 9). The remaining 5′ region of the mda-5 cDNA (1.5 kb) was obtained by using a modified rapid amplification of cDNA ends approach. This complete ORF approach involved an anchor primer instead of poly(A) or G tailing designed to anneal to the 5′ end of the mda-5 cDNA, followed by second-strand synthesis and subsequent PCR using an mda-5-specific reverse transcription primer and an anchor primer to generate a full-length mda-5 cDNA.
Northern Blot Analyses and Nuclear Run-On Assays.
RNA preparation and Northern blot hybridization were performed as described (9). Northern blots were probed with a 32P-labeled 2.5-kb EcoRI fragment of mda-5 cDNA and a 0.7-kb glyceraldehyde-3-phosphate dehydrogenase fragment. Nuclear run-on assays were performed as described (9). Probes used for nuclear run-on assays included: mda-5 5′, 9–837 bp; mda-5 3′, 2,531–3,365 bp; and a glyceraldehyde-3-phosphate dehydrogenase fragment (0.7 kb). Autoradiograms were quantitated by densitometry using a Molecular Dynamics densitometer.
mda-5 Expression Vectors.
A hemagglutinin (HA)-tagged mda-5 fragment was obtained by reverse transcription–PCR (GCCACCATGTACCCATACGACGTCCCAGACTACGCTATGTCGAATGGGTATTCCACAGACG/TCACTAATCCTCATCACTAAATAAACAGC) and was cloned into the EcoRV site of pcDEF3 with expression regulated by the EF-1α promoter. An antisense mda-5 expression vector was constructed by cloning the EagI/SpeI mda-5 genomic DNA (3.8-kbp) fragment from a bacterial artificial chromosome clone into the SpeI/NotI site of pcDEF3. The genomic DNA fragment consists of the first exon and part of the first intron. A green fluorescent protein (GFP)-mda-5 fusion expression vector was constructed by ligation of a reverse transcription–PCR-derived mda-5 cDNA product (ATGTCGAATGGGTATTCCACAGACG/TTTTTTTTTTTTCAGAGTAAAACAATC) into the SmaI site of pEGFP-C2 (CLONTECH).
Western Blot Analysis and Fluorescent Confocal Microscopy.
Transient transfections were performed by using SuperFect (Qiagen, Chatsworth, CA) as described in the manufacturer's protocol. Ten micrograms of supercoiled plasmid DNA (pcDEF3, pcDEF3/HA-mda-5, pEGFP-C2, and pEGFP-C2/mda-5) were transfected into ≈70% confluent 293 cells plated 1 day before transfection in a 10-cm plate, and cells were harvested 2 days after transfection. Protein sample preparation and Western blotting were performed as described (10). MDA-5 fusion proteins were probed with either α-HA antibody (Roche Molecular Biochemicals) or α-GFP antibody (CLONTECH) and with horseradish peroxidase-conjugated anti-Mouse IgG (Sigma) and detected by ECL (Amersham Pharmacia). For intracellular localization, 105 cells per well were seeded in 6-well plates containing a cover glass and transfected with the indicated plasmids (pEGFP-C2 and pEGFP-C2/mda-5, 2.5 μg per well). For fluorescence microscopy, the cover glass containing transfected cells was washed with PBS and mounted onto a glass slide with mounting medium. The cells were observed by fluorescent confocal microscopy.
Colony-Forming Assays.
HO-1 melanoma cells were plated at 8 × 105 in a 6-cm dish 1 day before transfection with 5 μg of supercoiled plasmid DNA. Two days after transfection, cells were trypsinized and replated at 105 cells per 6-cm dish with complete medium containing 750 μg of G418/ml. The G418-containing medium was replaced once a week for 3 weeks. Cells were fixed with methanol (−20°C) and stained with Giemsa (Sigma). Colonies with more than ≈50 cells were enumerated.
Purification of Glutathione S-Transferase (GST)-MDA-5 Fusion Protein.
A baculovirus transfer vector of GST-MDA-5 was constructed by ligation of the XhoI–BamHI mda-5 fragment from the pEGFP-C2/mda-5 and XhoI–BglII sites of pAcGHLT-C (PharMingen). The transfer vectors of GST only and GST-MDA-5 fusion (5 μg each) were transfected into Sf9 cells, and recombinant virus producer cells were selected as described by the manufacturer (PharMingen). GST and GST-MDA-5 fusion proteins were expressed and purified by affinity chromatography with glutathione-Sepharose 4B as described by the manufacturer (PharMingen).
ATPase Assay.
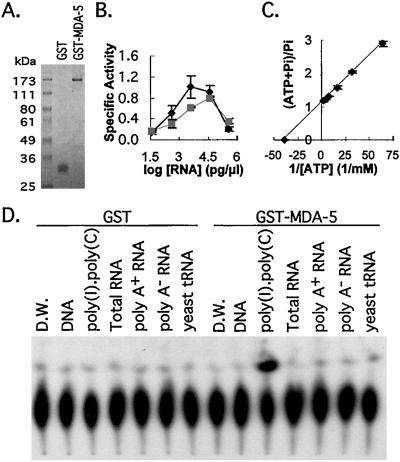
ATPase assays were performed in a 10-μl reaction mixture containing 50 mM MOPS, pH 6.5, 3 mM MgCl2, 2 mM DTT, 0.5 mM ATP, 5 μCi (1 Ci = 37 GBq) of [γ-32P]ATP (6,000 Ci/mmol; Amersham Pharmacia), DNA (sonicated salmon sperm) or RNA, and protein (0.2 μg) in a 37°C air incubator. Poly(I)⋅poly(C) (Amersham Pharmacia) and other RNA samples were reextracted with acid phenol/chloroform and precipitated. Specific assay conditions are described in the Fig. 5 legend. The reaction was started by addition of the ATP mixture and the dissociated Pi was resolved in polyethylene imine cellulose TLC plates as described (11).
Figure 5.
ATPase activity of MDA-5. (A) Electrophoretogram of purified proteins. GST and GST-MDA-5 were resolved on 9% SDS/PAGE and stained with Coomassie brilliant blue. (B) Effect of divalent metal ions and RNA concentration on MDA-5 ATPase activity. ATPase activity assay was performed with variable poly(I)⋅poly(C) concentrations (0.038, 0.375, 3.75, 37.5, and 375 ng/μl). MnCl2 (3 mM, ■) substituted for MgCl2 (3 mM, ♦). The results were quantitated by PhosphorImager. The data shown are the mean ± SD from two experiments. Specific activity is nmol/min/μg of protein. (C) Lineweaver–Burk plot of the effect of [ATP] on MDA-5 ATPase activity. ATPase activity was measured with various ATP concentrations (15.6, 31.3, 62.5, 125, 250, and 500 μM) for 20 min in a 37°C air incubator and quantitated by PhosphorImager. The result shown is the mean ± SD from three independent experiments. (D) Autoradiogram of MDA-5 ATPase assay with various RNA species. The indicated types of RNA (20 ng/μl) were added in standard reaction mixture, and the results presented were obtained by exposing a TLC plate to Kodak X-Omat film. D.W., distilled water.
Results and Discussion
IFN-β + MEZ Induces Terminal Differentiation in HO-1 Cells.
Treatment of HO-1 melanoma cells with IFN-β (2,000 units/ml) plus MEZ (10 ng/ml) results in rapid and irreversible growth suppression, induction of dendrite-like processes, and enhanced melanogenesis (6, 7). This combination treatment also results in a subset of detached cells that display Ao (hypodiploid) DNA content by fluorescence-activated cell sorter analysis, which is indicative of apoptosis. These floating cells are apparent first at 48 h (data not shown). In contrast, treatment with either agent alone inhibits growth and melanogenesis in a reversible manner, and treated cells do not terminally differentiate or undergo apoptosis (data not shown). In this context, induction of terminal differentiation in HO-1 cells by the combination treatment would be anticipated to involve the induction of genes that are relevant to the mode of action of the individual inducing agents, i.e., IFN-β and MEZ, related to both reversible and terminal differentiation, and associated with apoptosis (1, 7, 8).
Sequence Analysis of mda-5.
The cloned mda-5 cDNA [3,365 bp, excluding the poly(A) tail] contains a 1,025-amino acid-encoding ORF (nucleotides 169–3,246) with a predicted molecular mass of 116.7 kDa and a pI of 5.44. The first AUG at position 169 likely represents a genuine start codon caused by the presence of an in-frame stop codon at nucleotide 159 and an A−3 Kozak consensus sequence (AXXaugG; ref. 12). A polyadenylation signal (AATAAA) is located 23 bp upstream of the poly(A) tail. In vitro translation of the mda-5 cDNA resulted in a protein with apparent molecular mass of ≈120 kDa and additional smaller bands, most likely truncated translation products of MDA-5 (data not shown).
Sequence analysis of the MDA-5 protein in the Prosite database identifies two conserved domains, a DExH/D RNA helicase domain and a CARD. No nuclear localization signal or RNA binding domains were found in this protein. The C-terminal 102 amino acids (nucleotides 722–823) of MDA-5 show homology to the RNA helicase C-terminal conserved domain, a signature motif of helicase superfamily II, which consists of three subfamilies (DEAD, DEAH, and DExH) based on their ATPase B motif (13, 14). At least eight of nine motifs are well conserved among DExH group helicases (14). These eight DExH group helicase motifs display high conservation but show some divergence in MDA-5 (numbered in Fig. 1). The distance between consensus motifs, which also are well conserved in the DEAD group helicases (13), are extended further in MDA-5 (Fig. 1). In these contexts, MDA-5 seems to be a divergent form of DExH RNA helicase.
Figure 1.
Sequence alignment of putative RNA helicases. CLUSTALW alignment of helicase domains of putative RNA helicases that share the RNA helicase motifs with mda-5. Conserved residues in DExH group RNA helicase defined in Jankowsky and Jankowsky (14) are aligned with consensus sequence (uppercase Roman numeric). Those underlined and marked with lowercase Roman numeric are for conserved motifs in this subgroup. Asterisks (*), identical residues; colons (:), conserved substitutions; dots (.), semiconserved substitutions.
The ProDom database recognizes 10 cloned or hypothetical proteins closely related in their helicase domain to MDA-5 (dating back to early prokaryotes: Methanobacterium thermoautotrophicum, O27466; Schizosaccharomyces pombe, Q09884; Caenorhabditis elegans, Q17545, Q44165, and P34529; Arabidopsis thaliana, Q9SP32; Sus scrofa, RHIV-1; and Homo sapiens, RIG-1, Q9HAM6, and Q9UPY3). Proteins except RIG-1 (15), RHIV-1 (16), and Q9SP32 (17) contain hypothetical ORFs deduced from genomic sequence analysis of diverse species. CLUSTALW sequence alignment of representatives of these proteins (MDA-5, RIG-1, Q9HAM6, RHIV-1, P34529, Q9SP32, and Q09884) revealed that the DExH helicase motifs found in MDA-5 and 10 additional motifs/residues are well conserved within this group of proteins (Fig. 1). The greatest sequence homology to the MDA-5 helicase domain was observed with Q9HAM6, a hypothetical ORF from the human genome. The conserved pattern of helicase motifs among these molecules possibly signifies a newly identified subgroup of DExH RNA helicases. Among them, MDA-5 is the first protein with documented RNA-dependent ATPase activity (Fig. 5).
RNA helicases impinge on many biological phenomena including cell differentiation, proliferation, development, and viral life cycle (13, 14). Although RNA helicases are well conserved in their ATPase and helicase motifs, a direct correlation between sequence motifs and function is not clear-cut. In this context, it is not possible to deduce the potential function(s) of MDA-5 based on its primary structure. However, it is worth noting that four of the molecules (Q9SP32, Q09884, P34529, and Q9UPY3) of this group contain an RNase III motif as well as an RNA helicase domain. RNase III is an enzyme targeting double-stranded RNA and is involved in gene silencing by RNA interference (18). Thus, it is conceivable that MDA-5, through its ATP-dependent unwinding of RNA, most likely functions to promote message degradation by specific types of RNase.
The biological context of expression and function of the four cloned proteins (MDA-5, RIG-1, RHIV-1, and Q9SP32) are strikingly similar. RIG-1 is expressed during all-trans retinoic acid-induced promyelocytic differentiation (15) and also is induced by IFN-β in HO-1 cells (unpublished data). The expression of RHIV-1 is induced by viral infection suggesting IFN inducibility (16). In addition, Q9SP32 (also known as CAF) seems to suppress cell division in floral meristems (17). mda-5 is expressed during differentiation induced by IFN-β plus MEZ, which also involves growth inhibition. In this regard, it is possible that the putative helicases of this subgroup may participate in similar biochemical changes associated with growth inhibition and differentiation.
A distinctive attribute of MDA-5 involves the N-terminal one-third of this molecule. This region of the MDA-5 protein contains a CARD (amino acids 125–174), with significant but relatively low probability (P = 0.04). The CARD is a structural motif that consists of amphipathic α-helices and is present in various pro- and antiapoptotic molecules (19). The recruitment of caspase to apoptotic signaling receptor complexes through CARD–CARD interactions has been documented (20). Although the N-terminal 50 amino acids (125) of MDA-5 do not align with other CARD proteins, probably because CARD is a structural motif, secondary structure analysis of amino acids 101–200 of MDA-5 with secondary structural content prediction indicates 83.3% α-helical contents in this area. Based on these considerations, it is likely that the MDA-5 N terminus contains a CARD. If the CARD contained in MDA-5 influences apoptosis, this CARD containing RNA helicase would represent the first RNA-modulating molecule associated with programmed cell death.
mda-5 Expression Analysis.
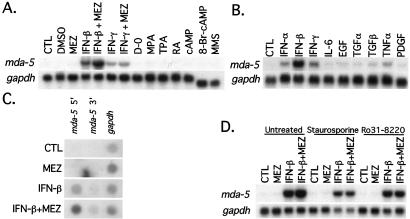
Because mda-5 was cloned in the context of induction of growth arrest and both reversible and terminal differentiation in HO-1 cells, experiments were performed to determine the effect on expression of agents affecting these parameters. Of the agents tested that influence melanocytic differentiation in human melanoma cells (ref. 21; Fig. 2A) and modulate melanoma growth (ref. 22; Fig. 2B), only IFNs (α, β, and γ) and TNF-α significantly increased steady-state mda-5 transcript levels within 24 h. However, the magnitude of induction by IFN-β was at least 3-fold greater than with IFN-γ, IFN-α, and TNF-α. Induction of DNA damage by exposure to the alkylating agent methyl methanesulfonate or growth in serum-free medium for 24 h did not induce mda-5 expression. Although MEZ treatment for 24 h by itself did not induce mda-5 expression, it augmented mda-5 expression by 1.5-fold when used in combination with IFN-β or IFN-γ (Fig. 2A). Although mda-5 is an IFN- and TNF-α-responsive gene, it responds primarily to IFN-β treatment. Because the other differentiation/growth-regulating reagents tested were not effective inducers of mda-5 expression, the primary role of mda-5 in the induction of terminal cell differentiation of HO-1 cells by IFN-β + MEZ seems to be restricted to IFN-β-mediated suppression of cell proliferation and/or induction of apoptosis (23).
Figure 2.
Expression of mda-5. Northern blot analyses of mda-5 after treatment with melanoma differentiation reagents (A) and growth factors (B) are shown. HO-1 cells were treated with the indicated reagents for 24 h. The concentrations in A are: control (CTL); 0.1% DMSO; 10 ng/ml MEZ; 2,000 units/ml IFN-β; 2,000 units/ml IFN-β + 10 ng/ml MEZ; 100 units/ml IFN-γ; 100 units/ml IFN-γ + 10 ng/ml MEZ; serum-free medium (D-O); 2.5 μM all-trans retinoic acid: 3 μM mycophenolic acid (MPA); 16 nM 12-O-tetradecanoylphorbol-13-acetate (TPA); 1 mM 3′-5′ cAMP; 1 mM 8-bromo-3′-5′ cyclic adenosine monophosphate (8-Br-cAMP); and 10 ng/ml methyl methanesulfonate (MMS). The reagents in B are: CTL, control; 1,000 units/ml IFN-α; 1,000 units/ml IFN-β; 1,000 units/ml IFN-γ; 1 ng/ml IL-6; 10 ng/ml epidermal growth factor (EGF); 10 ng/ml transforming growth factor-α (TGF-α); 2.5 ng/ml transforming growth factor-β (TGF-β); 1 ng/ml TNF-α; and 10 ng/ml platelet-derived growth factor (PDGF). (C) Nuclear run-on assays for mda-5. Nuclei were prepared from HO-1 melanoma cells treated with the indicated reagent(s) for 4 h. Blots were prepared and hybridized as described in Materials and Methods. gapdh, glyceraldehyde-3-phosphate dehydrogenase.(D) PKC inhibitors on mda-5 expression. RNA samples were extracted from HO-1 melanoma cells pretreated with 50 nM staurosporine or 0.2 μM Ro31-8220 (Calbiochem) for 30 min and treated with the indicated reagents for 8 h. RNA sample preparation and Northern hybridization were performed as described in Materials and Methods.
The timing of mda-5 expression was studied also by Northern blotting, indicating induction of mda-5 mRNA by 2 h of treatment with IFN-β or IFN-β + MEZ (data not shown). The mda-5 message level peaked between 6 and 8 h and remained elevated over a 96-h period. Although MEZ further increased mda-5 message level above that observed with IFN-β alone, it did not affect the timing of mda-5 expression. Pretreatment with the protein synthesis inhibitor cycloheximide (CHX) did not inhibit the induction of mda-5 by IFN-β or IFN-β + MEZ (data not shown). These results, which correspond with the early onset of mda-5 expression induced by these reagents, provide support for mda-5 being an early response gene induced without prior protein synthesis. They also indicate that mda-5 plays a critical role during an early stage of IFN-induced biological response. The organ-specific expression pattern of mda-5 was determined by hybridization with multiple tissue Northern blots (CLONTECH; data not shown). Most organs expressed mda-5 at low levels except the brain, testis, and lung, in which expression was barely detectable. Although mda-5 expression was ≈2-fold higher in placenta, pancreas, and spleen than other organs, no organ manifested significantly high enough expression to suggest a correlation between mda-5 expression and specific organ function. Northern blot analysis of mda-5 expression in cancer and normal cell lines of various sources again indicated low basal levels of expression with strong induction by IFN-β (data not shown).
Treatment of human skin fibroblasts with IFNs or TNF-α induces and MEZ augments mda-5 expression in a similar manner as in HO-1 cells (data not shown). These results suggest that induction of mda-5 by these agents may represent a general response of this gene occurring in different cellular contexts. Among the cytokines inducing mda-5 expression, IFN-β is the most potent. Gene expression profiles induced by type I IFNs (α/β), which bind to common receptors and share transcription factors, are quite similar, but the magnitude of differential gene expression and growth-inhibitory effects induced by IFN-β are more profound, as observed with mda-5 induction (21, 23, 24). Although differences have been reported in tyk2 requirement and specific coprecipitation of IFNAR1 with IFN-β but not with IFN-α, the mechanism underlying these differences in signal transduction between type I IFNs is not well understood (23). IFN-γ and IFN-γ + MEZ induce 3-fold lower levels of mda-5 mRNA than IFN-β and IFN-β + MEZ, but these treatments also directly induce mda-5 expression without prior protein synthesis (data not shown). It is possible that the mda-5 promoter contains an IFN-γ response sequence such as IFN-γ-activated sequence (23). Alternatively, the signal generated by IFN-γ may impact on mda-5 expression through shared signaling components with IFN-β but not through IFN-β production. In 3T3-L1 adipocytes, TNF-α activates the Janus kinase/signal transducer/activator of transcription signal transduction (Jak/STAT) pathway, which is stimulated by type I IFNs (25). TNF-α may induce mda-5 expression in HO-1 cells by activation of Jak/STAT signals as found in 3T3-L1 cells. Therefore, the Jak/STAT signal transduction pathway seems to be a major contributor to mda-5 expression.
IFNs were identified initially as molecules that provide immediate protection against viral infection by eliciting an antiviral state in exposed cells (23). IFN treatment evokes diverse responses depending on the target cell including growth inhibition, changes in differentiation, induction or inhibition of apoptosis, and changes in the expression of immune system modulating genes (1, 23, 26, 27). The highly inducible nature of mda-5 expression by IFNs regardless of cell type, especially IFN-β, the relatively low basal message level in various organs, and the rapid response to IFN treatment strongly suggest that mda-5 may play a critical role in responses that are specific for IFN signaling such as antiviral effect, growth inhibition, and apoptosis but may be less critical during normal physiological processes.
Although CARD interactions were not reported previously in IFN signaling, IFNs and their biological effectors do sensitize or induce apoptosis in specific cellular contexts, suggesting a link between IFN and apoptotic signaling (23, 28). Coincidentally, although not as effective as IFN-β, TNF-α, an apoptotic cytokine, also induces mda-5 expression in both HO-1 and human fibroblasts (ref. 29; Fig. 2B). Hence, MDA-5 may interact with death molecules through CARD and participate in the IFN- and/or TNF-α-induced apoptotic process by modulating RNA structure. MDA-5 also could have multiple functions with one or more of its potential enzymatic activities predominating, depending on the cellular context and presence of other coeffectors, e.g., CARD-containing proteins.
Nuclear run-on assays of HO-1 cells treated with IFN-β confirmed increases in mda-5 transcription in comparison with undetectable levels of transcription in untreated or MEZ-treated cells (Fig. 2C). The combination of IFN-β + MEZ further enhanced the transcription level of mda-5 as compared with IFN-β alone, an observation compatible with effects on steady-state message levels. These findings suggest that MEZ, in the combinatorial treatment protocol, augments mda-5 expression induced by IFN-β at a transcription level. Analysis of mda-5 mRNA stability by using actinomycin D to block transcription revealed that the half-life of the mda-5 transcript was ≈5 h and the decay rate of mda-5 message was indistinguishable between single- and combination-treated cells (data not shown). These data document that the increased steady-state levels of mda-5 message by IFN-β and IFN-β + MEZ treatment are primarily the result of increased mda-5 transcription.
The ability of IFN-β + MEZ to potentiate mda-5 mRNA levels suggests cooperativity between IFN and MEZ signaling in regulating mda-5 expression. Because MEZ is an activator of PKC, a potential role for PKC in enhancing mda-5 expression after IFN-β + MEZ treatment was tested by pretreatment of HO-1 cells with PKC-specific inhibitors, staurosporine or Ro31-8220 (refs. 30 and 31; Fig. 2D). Both reagents abolish the enhanced mda-5 induction by combination treatment. These findings indicate that both IFN and PKC signaling pathways are required for maximal induction of mda-5 expression in HO-1 cells. However, because MEZ does not always augment gene expression induced by IFN-β (e.g., human UBP43; ref. 21), crosstalk between PKC and IFN signaling pathways for all up-regulated genes seems unlikely. The higher level of induction seen in combination-treated cells seems to be through direct but independent action on the mda-5 promoter.
Intracellular Localization of MDA-5.
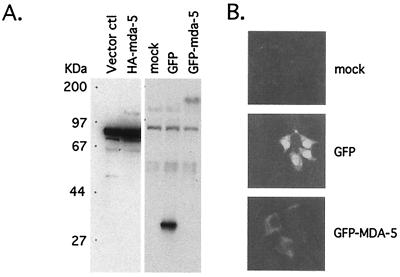
A GFP-MDA-5 fusion protein was transiently expressed in 293 cells and Western blot analysis of cell lysates detected a predicted ≈160-kDa protein (GFP-tagged) in mda-5 cDNA-transfected cells (Fig. 3A). Confocal fluorescence microscopy of 293 cells transiently transfected with GFP-MDA-5 fusion protein demonstrated that the protein localizes in the cytoplasm (Fig. 3B). The cytoplasmic localization of the MDA-5 protein corresponded with the absence of a nuclear localization signal in the MDA-5 protein. No specific localization pattern within the cytoplasm of the GFP-MDA-5 fusion protein was apparent. Cytoplasmic localization of MDA-5 suggests that MDA-5 plays a role in the translation, sequestration of specific mRNAs, or in regulating mRNA stability.
Figure 3.
Protein expression of mda-5 and intracellular localization. (A) Protein expression from mda-5 cDNA after transient transfection. Protein extracts were prepared from 293 cells transiently transfected with the indicated expression vector and resolved in 9% SDS/PAGE. Western blot analysis was performed with specified antibodies. ctl, control. (B) Intracellular localization of mda-5 protein. Transiently transfected 293 cells on cover slips with the indicated fusion protein constructs were mounted and photographed by using fluorescent confocal microscopy (×400).
mda-5 Inhibits the Colony-Forming Ability of HO-1 Cells.
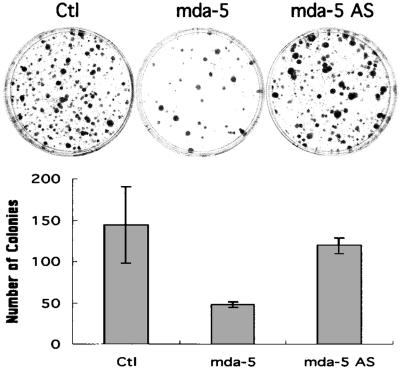
Treatment of HO-1 cells with 2,000 units/ml of IFN-β for 4 days results in ≈60% growth inhibition versus untreated controls (6, 7). Because mda-5 is induced primarily by IFN-β in HO-1 cells, ectopic expression of mda-5 could mimic the effect of IFN-β treatment and inhibit growth. To test this possibility, the mda-5 gene was expressed in HO-1 cells by transfection, and colony-forming ability was determined (Fig. 4 A and B). Expression of the mda-5 construct was confirmed in 293 cells, which are more efficient for transfection than HO-1 cells. Western blot analysis using α-HA antibody showed a protein band corresponding to the predicted size of MDA-5 (≈120 kDa) in HA-mda-5-transfected cell lysates (Fig. 3A). After verifying HA-MDA-5 expression, colony-forming efficiency of HO-1 cells was determined after transfection with parental vector, HA-mda-5 expression vector, or an antisense mda-5 expression vector. The number of G418-resistant HO-1 colonies developing after transfection with the mda-5 expression vector was reduced significantly (≈67%, P < 0.05) in comparison with cells transfected with parental vector (Fig. 4B). In contrast, transfection with the antisense mda-5 expression vector did not effect colony formation significantly, which supports a specific role of mda-5 in regulating the growth of HO-1 cells. Moreover, attempts to obtain stable HO-1 cell clones expressing mda-5 have proven unsuccessful thus far, providing substantiation for a growth-inhibitory effect of mda-5 in these cells.
Figure 4.
Effect of ectopic expression of mda-5 on HO-1 cells. Cells were transfected with the indicated expression vector, replated 2 days later, and selected with G418. (Upper) Representative colony-forming assays. G418-resistant colonies transfected with the indicated expression vectors are shown. Ctl, control; AS, antisense. (Lower) Quantitation of the effect of ectopic expression of mda-5 on colony formation. Giemsa-stained G418-resistant colonies containing more than ≈50 cells were counted. The results are the mean ± standard error from three independent transfections (three plates for each transfection) with two different plasmid batches.
The growth-inhibitory or apoptotic activity of type I IFNs are mediated by activation of IFN-inducible double-stranded RNA-dependent protein kinase (PKR) and 2′-5′ oligoadenylate synthase (OAS)/RNase L system (23). At present, it is not clear how ectopic expression of mda-5 inhibits HO-1 colony formation and which specific pathway(s) regulates this gene. Of note is the presence of CARD at the N terminus of MDA-5, which suggests possible interaction between MDA-5 and apoptotic signal molecules. However, unlike IFN-sensitive apoptotic cells, IFN-β suppresses the growth of HO-1 cells without causing apoptosis. A balance between death and survival signals regulates the apoptotic process (32). Apparently, treatment with only IFN-β or physiological expression of MDA-5 is insufficient to trigger apoptosis in HO-1 cells. It is possible that MDA-5 may be a component of death effector complexes or molecules but by itself lacks the capacity to trigger apoptosis. In this context, ectopic expression of MDA-5 may result in growth inhibition and not programmed cell death. Alternatively, the targets of MDA-5 may be growth-related messages, and this gene product may contribute to their metabolism when the death signal mediated by IFN impinges on CARD. It is possible also that the level of ectopic expression of mda-5 or the levels of MDA-5 induced by IFN-β in specific cell types may determine whether this molecule is growth-inhibitory or apoptotic. The fact that mda-5 expression is augmented by combined treatment with IFN-β plus MEZ, which induces terminal differentiation and apoptosis, agrees with this possibility.
ATPase Activity of MDA-5.
A GST-fusion protein of MDA-5 was purified by glutathione-Sepharose affinity chromatography and the purified GST-MDA-5 migrates as a single protein in SDS-PAGE with an apparent molecular mass of 170 kDa (Fig. 5A). GST as a control was expressed and purified in a manner similar to that for GST-MDA-5 (Fig. 5A). The ATPase activity of the MDA-5 fusion protein was 2-fold more active at pH 6.5 than at pH 7.5 but strictly depended on poly(I)⋅poly(C) (primarily exists as a double-stranded molecule) at both pH values (data not shown). Poly(I)⋅poly(C) has been shown to induce mda-5 in HO-1 and other cancer cells (data not shown). The requirement of divalent metal ions and effective RNA concentrations for MDA-5 ATPase activity were determined at various poly(I)⋅poly(C) concentrations by using either Mg2+ or Mn++ (Fig. 5B). The presence of Mn++ shifted the optimal poly(I)⋅poly(C) concentration for MDA-5 ATPase activity to a higher range. ATPase activity peaked at 3.75 ng of poly(I)⋅poly(C)/μl with Mg2+ and at 37.5 ng of poly(I)⋅poly(C)/μl with Mn++. The specific activity of MDA-5 at optimal conditions was calculated as 1.01 ± 0.22 nmol/min/μg of protein with Mg2+ and 0.80 ± 0.03 nmol/min/μg of protein with Mn++. These results suggest that Mg2+ is likely to be the preferred ion for MDA-5. Unexpectedly, increasing poly(I)⋅poly(C) concentrations did not increase MDA-5 ATPase activity. At the highest poly(I)⋅poly(C) concentration tested, ATPase activity of MDA-5 was reduced to 0.20, ≈0.25-fold of peak activity. Depletion of divalent metal ions by excessive poly(I)⋅poly(C) provides a possible explanation for this phenomenon. ATPase activity of MDA-5 also was determined at various ATP concentrations, and the calculated Km value for ATP as determined by double reciprocal plot between [ATP] and hydrolyzed Pi fraction was 26 μM (Fig. 5C). The Km value of MDA-5 for ATP is lower than other well characterized RNA helicases including eIF4A (DEAD helicase, 160 μM; ref. 33), NPH-II (DEAH helicase, 1.2 mM) (34), and hepatitis C virus NS3 protein (DECH helicase, 42 μM without RNA; ref. 35). Based on these results, MDA-5 is strictly an RNA-dependent ATPase, the activity of which may be limited more by the presence of double-stranded RNA than by [ATP]. These results also reinforce the suggestion that MDA-5 is an ATP-dependent RNA helicase.
To identify a potential target of MDA-5, the ATPase activity of MDA-5 was measured in the presence of cellular RNA samples. Of interest, no significant ATPase activity was observed with total, either poly(A) or poly(A)-negative RNA, from IFN-treated HO-1 cells and yeast tRNA (20 ng/μl in Fig. 5D; 375 ng/μl, data not shown). The lack of a stimulatory effect on MDA-5 ATPase activity by cellular RNA may reflect the heterogeneous nature of the cellular RNA populations or the extent of double-stranded RNA. However, although rRNA and tRNA are highly double-stranded and relatively homogeneous in comparison with poly(A) RNA, they did not activate MDA-5 ATPase activity, thereby ruling out these molecules as potential substrates for MDA-5. ATPase activity was apparent also with poly(A-U), which was significantly higher than with poly(I-U), suggesting no specific preference for inosine (data not shown). With regard to the biological role of RNA helicases and cytoplasmic localization of MDA-5, it is highly probable that MDA-5, by its ATP-dependent unwinding of RNA, may function to promote message degradation and/or inhibit translation during growth inhibition and/or apoptosis mediated by IFN or TNF-α treatment. This protein also may affect IFN-induced antiviral activity profoundly.
The present studies provide an initial glimpse into the biological function of MDA-5, a potentially important gene product in IFN-dependent growth inhibition. Sequence analysis of MDA-5 reveals the unique combination of two domains, CARD and RNA helicase, which is unprecedented for helicases. Moreover, the RNA-dependent ATPase activity of MDA-5 provides support for a role in ATP-dependent RNA-unwinding activity. The growth-inhibitory effect of ectopically expressed MDA-5 suggests a possible proapoptotic role of CARD. In this context, MDA-5 may represent the first apoptotic molecule identified to date that can modulate RNA directly. Unlike protein metabolism in apoptosis, RNA metabolism represents an understudied area. Understanding MDA-5 function could provide important insights into apoptotic processing of RNA. Additional experimentation is required to determine the role of mda-5 in normal cellular physiology and response to type I IFNs and TNF-α, as well as the relevance of the CARD in this putative RNA helicase to apoptosis. Important questions relate to the nature of the molecule(s) interacting with the CARD, the role of MDA-5 in RNA translation and/or degradation, the effector molecules regulated by MDA-5, and the identity of the potential target messages affected by MDA-5 helicase activity. Determination of the physiological role and molecular basis of mda-5 action should provide important insights into the mechanism of cellular defense conferred by IFN against viral attack and its role in growth control, differentiation, and apoptosis.
Acknowledgments
The present studies were supported in part by National Institutes of Health Grants CA35675 and CA74468, an award from the Samuel Waxman Cancer Research Foundation, and the Chernow Endowment. P.B.F. is the Michael and Stella Chernow Urological Cancer Research Scientist.
Abbreviations
- PKC
protein kinase C
- MEZ
mezerein
- mda
melanoma differentiation-associated
- TNF-α
tumor necrosis factor-α
- CARD
caspase recruitment domain
- HA
hemagglutinin
- GFP
green fluorescent protein
- GST
glutathione S-transferase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF095844).
References
- 1.Leszczyniecka M, Robert T, Dent P, Grant S, Fisher P B. Pharmacol Ther. 2001;90:105–156. doi: 10.1016/s0163-7258(01)00132-2. [DOI] [PubMed] [Google Scholar]
- 2.Waxman S. Differentiation Therapy. Vol. 10. Rome: Ares-Serono Symposia Publishers; 1996. pp. 1–531. [Google Scholar]
- 3.Freemerman A J, Vrana J A, Tombes R M, Jiang H, Chellappan S P, Fisher P B, Grant S. Leukemia. 1997;11:504–513. doi: 10.1038/sj.leu.2400625. [DOI] [PubMed] [Google Scholar]
- 4.Jiang H, Lin J, Fisher P B. Mol Cell Differ. 1994;2:221–239. [Google Scholar]
- 5.Wang Z, Su Z Z, Fisher P B, Wang S, VanTuyle G, Grant S. Exp Cell Res. 1998;244:105–116. doi: 10.1006/excr.1998.4191. [DOI] [PubMed] [Google Scholar]
- 6.Fisher P B, Prignoli D R, Hermo H, Jr, Weinstein I B, Pestka S. J Interferon Res. 1985;5:11–22. doi: 10.1089/jir.1985.5.11. [DOI] [PubMed] [Google Scholar]
- 7.Jiang H, Su Z-Z, Boyd J, Fisher P B. Mol Cell Differ. 1993;1:41–66. [Google Scholar]
- 8.Jiang H, Fisher P B. Mol Cell Differ. 1993;1:285–299. [Google Scholar]
- 9.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short Protocols in Molecular Biology. New York: Wiley; 1992. [Google Scholar]
- 10.Jiang H, Lin J J, Su Z Z, Goldstein N I, Fisher P B. Oncogene. 1995;11:2477–2486. [PubMed] [Google Scholar]
- 11.Wagner J D, Jankowsky E, Company M, Pyle A M, Abelson J N. EMBO J. 1998;17:2926–2937. doi: 10.1093/emboj/17.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozak M. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 13.Luking A, Stahl U, Schmidt U. Crit Rev Biochem Mol Biol. 1998;33:259–296. doi: 10.1080/10409239891204233. [DOI] [PubMed] [Google Scholar]
- 14.Jankowsky E, Jankowsky A. Nucleic Acids Res. 2000;28:333–334. doi: 10.1093/nar/28.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Y W. Ph.D. thesis. Sanghai, China: Sanghai Second Medical University; 1997. [Google Scholar]
- 16.Zhang X, Wang C, Schook L B, Hawken R J, Rutherford M S. Microb Pathog. 2000;28:267–278. doi: 10.1006/mpat.1999.0349. [DOI] [PubMed] [Google Scholar]
- 17.Jacobsen S E, Running M P, Meyerowitz E M. Development (Cambridge, UK) 1999;126:5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein E, Caudy A A, Hammond S M, Hannon G J. Nature (London) 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann K, Bucher P, Tschopp J. Trends Biochem Sci. 1997;22:155–156. doi: 10.1016/s0968-0004(97)01043-8. [DOI] [PubMed] [Google Scholar]
- 20.Chou J J, Matsuo H, Duan H, Wagner G. Cell. 1998;94:171–180. doi: 10.1016/s0092-8674(00)81417-8. [DOI] [PubMed] [Google Scholar]
- 21.Kang D, Jiang H, Wu Q, Pestka S, Fisher P B. Gene. 2001;267:233–242. doi: 10.1016/s0378-1119(01)00384-5. [DOI] [PubMed] [Google Scholar]
- 22.Garbe C, Krasagakis K. J Invest Dermatol. 1993;100:239S–244S. [PubMed] [Google Scholar]
- 23.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 24.Garbe C, Krasagakis K, Zouboulis C C, Schroder K, Kruger S, Stadler R, Orfanos C E. J Invest Dermatol. 1990;95:231S–237S. doi: 10.1111/1523-1747.ep12875837. [DOI] [PubMed] [Google Scholar]
- 25.Guo D, Dunbar J D, Yang C H, Pfeffer L M, Donner D B. J Immunol. 1998;160:2742–2750. [PubMed] [Google Scholar]
- 26.Grant S, Bhalla K, Weinstein B, Pestka S, Mileno M D, Fisher P B. Biochem Biophys Res Commun. 1985;130:379–388. doi: 10.1016/0006-291x(85)90428-0. [DOI] [PubMed] [Google Scholar]
- 27.Greiner J W, Schlom J, Pestka S, Langer J A, Giacomini P, Kusama M, Ferrone S, Fisher P B. Pharmacol Ther. 1985;31:209–236. doi: 10.1016/0163-7258(85)90023-3. [DOI] [PubMed] [Google Scholar]
- 28.Balachandran S, Roberts P C, Kipperman T, Bhalla K N, Compans R W, Archer D R, Barber G N. J Virol. 2000;74:1513–1523. doi: 10.1128/jvi.74.3.1513-1523.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallach D, Varfolomeev E E, Malinin N L, Goltsev Y V, Kovalenko A V, Boldin M P. Annu Rev Immunol. 1999;17:331–367. doi: 10.1146/annurev.immunol.17.1.331. [DOI] [PubMed] [Google Scholar]
- 30.Tamaoki T, Nomoto H, Takahashi I, Kato Y, Morimoto M, Tomita F. Biochem Biophys Res Commun. 1986;135:397–402. doi: 10.1016/0006-291x(86)90008-2. [DOI] [PubMed] [Google Scholar]
- 31.Morreale A, Mallon B, Beale G, Watson J, Rumsby M. FEBS Lett. 1997;417:38–42. doi: 10.1016/s0014-5793(97)01252-0. [DOI] [PubMed] [Google Scholar]
- 32.Strasser A, O'Connor L, Dixit V M. Annu Rev Biochem. 2000;69:217–245. doi: 10.1146/annurev.biochem.69.1.217. [DOI] [PubMed] [Google Scholar]
- 33.Pause A, Methot N, Sonenberg N. Mol Cell Biol. 1993;13:6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gross C H, Shuman S. J Virol. 1996;70:1706–1713. doi: 10.1128/jvi.70.3.1706-1713.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D W, Kim J, Gwack Y, Han J H, Choe J. J Virol. 1997;71:9400–9409. doi: 10.1128/jvi.71.12.9400-9409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]