Abstract
Nucleotide excision repair (NER) of UV-induced cyclobutane pyrimidine dimers (CPDs) was measured in the individual strands of transcriptionally active and inactive ribosomal genes of yeast. Ribosomal genes (rDNA) are present in multiple copies, but only a fraction of them is actively transcribed. Restriction enzyme digestion was used to specifically release the transcriptionally active fraction from yeast nuclei, and selective psoralen crosslinking was used to distinguish between active and inactive rDNA chromatin. Removal of CPDs was followed in both rDNA populations, and the data clearly show that strand-specific repair occurs in transcriptionally active rDNA while being absent in the inactive rDNA fraction. Thus, transcription-coupled repair occurs in RNA polymerase I-transcribed genes in yeast. Moreover, the nontranscribed strand of active rDNA is repaired faster than either strand of inactive rDNA, implying that NER has preferred access to the active, non-nucleosomal rDNA chromatin. Finally, restriction enzyme accessibility to active rDNA varies during NER, suggesting that there is a change in ribosomal gene chromatin structure during or soon after CPD removal.
Nucleotide excision repair (NER) removes different types of lesions from DNA, including bulky adducts caused by chemicals, interstrand or intrastrand crosslinks, and the UV photoproducts cis-syn cyclobutane pyrimidine dimer (CPD) and pyrimidine (6–4) pyrimidone (1). If DNA lesions are not removed, mutations can occur after translesion replication (2). It is now well established that many transcriptionally active genes are repaired faster than inactive DNA (3, 4). Furthermore, preferential removal of CPDs from active genes is caused mainly by an increased rate of repair of the transcribed strand (TS). This transcription-coupled repair (TCR) was first discovered in mammalian cells (5), then in Escherichia coli (6) and yeast (7). Elongation by RNA polymerase II (pol II) is required for TCR (8, 9), and it is thought that only pol II-transcribed genes are subject to TCR (10).
RNA polymerase I (pol I) transcribes ribosomal genes (rDNA) at a very high rate. The rDNA is localized in the nucleolus, which is a dense chromatin region composed of rDNA, pol I, rRNA, assembling ribosomes, and proteins involved in cell-cycle regulation (11, 12). It is known that mammalian cells repair rDNA damaged by UV radiation (13) and chemicals (14–16). However, in rodent and human cells, CPDs are less efficiently repaired in rDNA than in either total genomic DNA or pol II-transcribed genes (16–20). Moreover, DNA repair does not exhibit strand bias in the rDNA of these cells (17, 19). To the contrary, CPDs are rapidly removed from both strands of total rDNA in yeast (21, 22). In addition, a strand bias during repair of total rDNA was observed in rad7 and rad16 mutants, even though strand-specific repair was not observed in total rDNA of wild-type cells (21).
Ribosomal genes are present in multiple copies organized in long tandem repeats (11), and in most cells only a fraction of rDNA is transcriptionally active (23). At the chromatin level, inactive rDNA is assembled in arrays of nucleosomes whereas canonical nucleosomes are not present on active rDNA (23). This coexistence of two distinct rDNA chromatin populations limits the interpretation of most biochemical assays used to study DNA processing in the rDNA locus. Indeed, determining strand-specific repair in rDNA by the standard Southern blot assay (5, 24) is compromised because results represent only an average of active and inactive rDNA copies.
Psoralen crosslinking has been used to separate active from inactive forms of rDNA (23, 25). With this technique, active rDNA chromatin binds more psoralen than the inactive chromatin fraction. Consequently, psoralen-crosslinked rDNA fragments from active genes have a slower migration on gels than fragments from inactive genes (26). Using this method, it was shown that the active rDNA fraction varies markedly in different cell types, as well as during the cell cycle (e.g., from ≈20% to ≈70%; refs. 23 and 27–29). Moreover, only the heavily psoralen crosslinked rDNA is actively transcribed, as the nascent rRNA transcripts crosslink only to this rDNA fraction (26, 28, 30).
For this article, we used differential psoralen crosslinking to distinguish between active and inactive rDNA genes in yeast and to confirm that active rDNA is preferentially released by digesting nuclei with EcoRI. Thus, active and inactive rDNA were separated and NER of CPDs was followed in both rDNA populations. The results clearly show that strand-specific repair occurs only in the transcriptionally active rDNA in wt yeast cells and, therefore, that TCR occurs in these pol I-transcribed genes. Furthermore, the nontranscribed strand (NTS) of active rDNA is repaired faster than either strand of inactive rDNA, suggesting that NER enzymes operate more efficiently in active rDNA chromatin. Finally, the accessibility of active rDNA to restriction enzyme digestion varies during NER, revealing changes in chromatin structure of ribosomal genes during DNA repair.
Materials and Methods
Yeast Cell Growth and UV Irradiation.
Yeast cells [Saccharomyces cerevisiae, strain JS311 (RAD+); ref. 31] were grown in complete medium (yeast extract/peptone/dextrose, YEPD) to early log-phase (OD600 = 0.4; ≈1.3 × 107 cells/ml). Cultures were harvested by centrifugation and resuspended in ice-cold PBS (137 mM NaCl, 2.5 mM KCl, 2 mM KH2PO4, 10 mM Na2HPO4, pH 7.0) to a final concentration of 2 × 107 cells/ml. Cell suspensions were poured into trays to a depth of ≈1 mm and irradiated (primary 254 nm) with a UV dose of 80 J/m2, measured with a Spectroline DM-254N short-wave UV meter (Spectronic, Westbury, NY). Cells were then harvested, resuspended in YEPD containing 100 mM hydroxyurea (Sigma) to prevent replicative DNA synthesis (32), and incubated in the dark at 30°C with continuous shaking for different repair times.
Nuclei Isolation and DNA Extraction.
Yeast cells (≈2 × 109) were collected, washed with ice-cold PBS, suspended in 1.5 ml of nuclei isolation buffer (NIB: 50 mM Mops, pH 8.0/150 mM potassium acetate/2 mM MgCl2/17% glycerol/0.5 mM spermine/0.15 mM spermidine) and transferred to 15-ml polypropylene tubes containing 1.5 ml of glass beads (425–600 μm, Sigma). Cells were disrupted by vortexing (12 × 30-s pulses with 30-s pauses on ice), the nuclear suspensions were collected, and the glass beads were rinsed four times with an equal volume of NIB. The combined suspensions (8 ml) were loaded onto 10 ml of 50% Percoll in NIB and centrifuged for 10 min at 4,000 rpm. Nuclei present at the interphase were collected in 2 ml NIB, pelleted, suspended in 0.5 ml of EcoRI digestion buffer, and digested with 50 units of EcoRI for 15 min at 37°C. DNA was extracted by adding SDS to 1.5% and incubating samples at 65°C for 3.5 h. Cell lysates were adjusted to 2.5 ml with TE buffer (10 mM Tris⋅HCl/1 mM EDTA, pH 8.0) and 2.5 ml of saturated NaCl. After centrifugation for 30 min at 8,000 rpm, supernatants were collected and DNA was precipitated in isopropanol. Pellets were dissolved in TE buffer, treated with RNase (Roche Diagnostics), phenol-extracted, and ethanol-precipitated.
Psoralen Crosslinking of Nuclei.
Crosslinking of nuclei was performed in 24-multiwell plates (Falcon, uncoated). Psoralen (4,5′,8-trimethylpsoralen, Sigma) stock solution (400 μg/ml in ethanol) was added at a volume equal to 0.025 × nuclei suspension volume. After 5 min on ice in the dark, the nuclear suspension was irradiated on ice for 10 min with a medium-pressure Hg lamp (450 W, Ace Glass), filtered to yield 320–380 nm light, and placed at a distance of 15 cm. The irradiation step was repeated twice.
Alkaline Gel Electrophoresis and Southern Blotting.
The DNA samples were cleaved specifically at CPDs by using T4 endonuclease V (T4 endo V; ref. 33), as described (e.g., ref. 7). After T4 endo V digestion, ≈5 μg of DNA per sample was separated on 1% alkaline agarose gels (34). DNA was transferred to Hybond N+ membranes (Amersham Pharmacia) in 0.4 M NaOH. Radioactive probes were generated by using random primers or strand-specific riboprobes (Promega). Hybridization and washing were performed at 70°C (26), and membranes were exposed to PhosphorImager screens (Molecular Dynamics).
Quantification of CPD Yield.
The number of CPDs present in genomic DNA was determined as described (35). DNA samples (5–10 μg) were digested with T4 endo V and resolved on 1% alkaline agarose gels. After electrophoresis, DNA was depurinated (0.25 M HCl for 30 min) and transferred to nylon membranes in 0.4 M NaOH. Radioactive probes were generated from EcoRI-digested yeast genomic DNA followed by random priming (Promega). Quantification of CPDs was performed on data from PhosphorImages of the Southern blots of these gels, using imagequant software (Molecular Dynamics) and number-average DNA length analysis (35). Measurement of CPDs in each strand of rDNA was performed as described (e.g., ref. 36).
Results
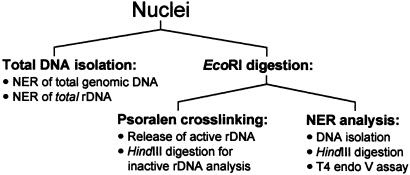
The strategy of these experiments is outlined in Fig. 1. Yeast cells were harvested at the specified repair times and nuclei were prepared. Total DNA was isolated from an aliquot of these nuclei to follow NER in both genomic DNA and total rDNA (Fig. 1, left branch). In parallel, aliquots of nuclei were digested with EcoRI to release the active fraction of rDNA chromatin (Fig. 1, right branch). Only the active rRNA genes are completely digested by EcoRI because nucleosomes are absent from these genes, making them accessible to the restriction endonuclease (23). To monitor the release of active ribosomal genes, half of each sample of EcoRI-digested nuclei was photo-crosslinked with psoralen. The DNA was purified and analyzed on native agarose gels. The inactive rRNA genes were analyzed by HindIII digestion of DNA isolated from EcoRI-treated nuclei. Because EcoRI sites are located within the two HindIII sites (Fig. 2), the complete HindIII band (≈6.4 kb) can originate only from rDNA that was not cleaved by EcoRI. Thus, the complete HindIII fragment is derived from inactive rDNA where EcoRI accessibility is inhibited by the presence of nucleosomes. To follow NER in the active and inactive ribosomal genes, DNA was isolated from the remaining EcoRI-digested nuclei, digested with HindIII, and treated with T4 endo V to generate strand breaks specifically at CPD sites. This double digestion allows a direct analysis of active (EcoRI band) and inactive (HindIII band) rDNA from the same cells.
Figure 1.
Experimental design.
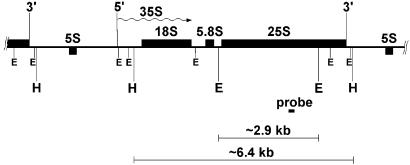
Figure 2.
Map of the yeast 35S rRNA gene. The rRNA gene, 5′ and 3′ ends, and direction of transcription (wavy arrow) are shown. The short black box represents the probe (≈140 bp) used in this work. The EcoRI (E) and HindIII (H) restriction sites are indicated, together with the sizes of the restriction fragments (solid bars).
Repair of Total Genomic DNA.
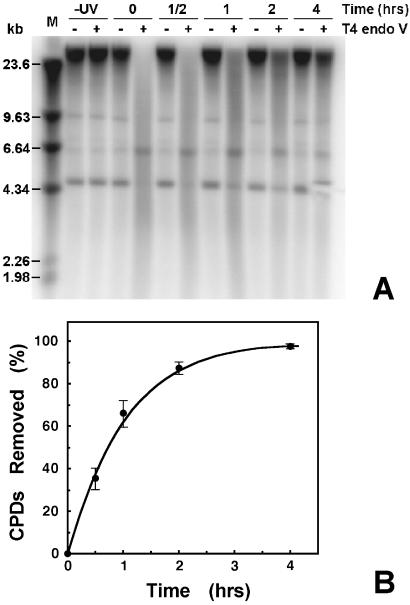
To determine whether the time course of repair in rDNA is similar to that of the genome overall, total DNA was isolated after different repair time incubations. Samples were treated with (or without) T4 endo V, separated on alkaline agarose gels, transferred to membranes, and hybridized with random primer-labeled probes to genomic DNA. A typical gel image from these experiments is shown in Fig. 3A, where the change in DNA mobility after T4 endo V treatment (compare − and + lanes) reflects the number of strand breaks at CPD sites. The shift in number-average length of the DNA molecules was used to calculate the average number of CPDs/kb at each repair time (35, 37). Immediately after irradiation, 0.25 ± 0.01 CPDs/kb were detected in genomic DNA and the decrease in this number with increased incubation time was used to monitor repair. As shown in Fig. 3B, almost all of the CPDs (≈97%) are removed from genomic DNA within 4 h. We note that the faint bands in the 4- to 10-kb region are also visible on ethidium bromide-stained gels.
Figure 3.
Repair of CPDs from total genomic DNA. Yeast cells were irradiated with 80 J/m2 UV and harvested at the times indicated. Total DNA was purified, treated with T4 endo V, and separated on 1% alkaline agarose gels. After blotting, filters were hybridized with random primer-labeled total genomic DNA. (A) Representative Southern blot. Repair times (in h) after UV irradiation are indicated above the lanes. Other labels are: −UV, DNA from unirradiated cells; − and +, samples mock-treated or treated with T4 endo V, respectively; and M, λ DNA digested with HindIII. (B) Percent of CPDs removed as a function of repair time. The number of CPDs present in genomic DNA at each time was determined as described (35). Data are the mean ± 1 SD of three independent experiments.
Repair of Individual Strands of Total rDNA.
Aliquots of the DNA used to follow NER of genomic DNA (Fig. 3) were digested with either EcoRI or HindIII, before treatment with T4 endo V. Digestion with EcoRI releases a ≈2.9-kb fragment from the central part of the rDNA transcription unit, whereas the HindIII fragment (≈6.4 kb) contains most of the transcribed region (Fig. 2). Blots from alkaline agarose gels were hybridized to strand-specific “riboprobes” of rDNA (Fig. 2).
The induction of CPDs in each strand of the EcoRI and HindIII fragments is shown in Fig. 4 A and B, respectively (lanes 3 and 4). After 80 J/m2, the values obtained for the TS of total rDNA are 0.68 ± 0.09 and 1.52 ± 0.23 CPDs for the EcoRI and HindIII fragments, respectively (mean ± 1 SD of four experiments). The values obtained for the NTS were 0.51 ± 0.08 and 1.38 ± 0.28 CPDs for the EcoRI and HindIII fragments, respectively. These values correspond to an average yield of ≈0.23 CPDs/kb in each strand of the HindIII fragment and are similar to the value obtained for genomic DNA (≈0.25 CPDs/kb).
Figure 4.
Repair of CPDs from total rDNA. DNA from cells, irradiated as in Fig. 3, was digested with either EcoRI or HindIII before treatment with T4 endo V. Filters were hybridized with strand-specific riboprobes. (A) PhosphorImage for the TS and NTS of EcoRI-digested total rDNA. (B) PhosphorImage for the TS and NTS of HindIII-digested total rDNA. Labeling of the gel is the same as in Fig. 3. (C) Quantification of PhosphorImages. DNA repair is expressed as percent of CPDs removed vs. repair time. Filled and empty symbols represent data for the TS and the NTS, respectively. Circles and dashed lines denote HindIII digests, and triangles and solid lines denote EcoRI digests. Data are the mean ± 1 SD of four independent experiments.
Repair of total rDNA in these fragments is shown in Fig. 4 A and B (lanes 5–12) and analysis of the data indicates that over 90% of the CPDs are removed during the 4-h repair time (Fig. 4C). The time course of rDNA repair is similar to that obtained for total genomic DNA (compare Figs. 3B and 4C). In addition, the time course for CPD removal is the same for both the 2.9-kb EcoRI fragment and the 6.4-kb HindIII fragment (Fig. 4C, compare triangles with circles).
Analysis of rDNA Chromatin by Psoralen Crosslinking.
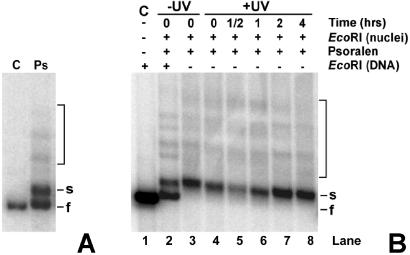
Transcriptionally active and inactive rDNA chromatin was analyzed by psoralen photo-crosslinking in whole yeast cells and isolated nuclei. DNA isolated from psoralen-crosslinked cells was digested with EcoRI and separated by gel electrophoresis. As shown in Fig. 5A (lane Ps), the two bands corresponding to inactive and active rDNA populations (designated f and s, respectively) are well resolved. The relative intensity of these bands is about 60% and 40%, respectively. Conversely, control DNA isolated from uncrosslinked cells shows only the expected EcoRI DNA fragment (lane C). The faint bands above the EcoRI bands (Fig. 5A, bracket) correspond to incomplete digestion of some rDNA sequences, because of the presence of psoralen crosslinks close to the restriction sites (27, 38).
Figure 5.
EcoRI digestion of nuclei. (A) DNA was extracted from cells treated (lane Ps) or untreated (lane C) with psoralen. After EcoRI digestion, DNA was separated on 1% native agarose gels, blotted, and hybridized with labeled rDNA probe. The s- (slow) and f- (fast) bands correspond to the active and inactive rDNA, respectively. (B) Nuclei were isolated from unirradiated (lane 3) or irradiated (lanes 4–8) cells, before (lane 4) and during NER (lanes 5–8). Nuclei were digested with EcoRI and then treated with psoralen (lanes 2–8). DNA was purified from the nuclei, and the DNA samples were separated by gel electrophoresis (lanes 3–8) or redigested with EcoRI before gel electrophoresis (lane 2). As control (C), DNA was isolated from uncrosslinked nuclei and digested with EcoRI (lane 1). The bracket indicates partial EcoRI digest bands.
Release of Active Ribosomal Genes by EcoRI Digestion of Nuclei.
The selective release of active rDNA chromatin by EcoRI digestion was followed during repair. For each repair time, nuclei were isolated, digested with EcoRI (Fig. 1), and then photo-reacted with psoralen to separate the active and inactive fractions (Fig. 5B). Because nucleosomes are an impediment to restriction enzyme accessibility (39, 40), only the active rDNA (s-band) is released (Fig. 5B, lanes 3–8). The inactive rDNA copies migrate in native gels as very high molecular weight DNA (not shown). As a control, an aliquot of the same DNA separated in Fig. 5B, lane 3 was redigested with EcoRI to release the inactive rDNA fragments from the high molecular weight DNA (Fig. 5B, lane 2). It is clear that EcoRI releases only the active rDNA fraction from nuclei, whereas the inactive fraction is released after redigestion of the isolated DNA. In addition, EcoRI accessibility to rDNA chromatin changes during NER, being low at early repair times and increasing at late repair times (Fig. 5B).
Release of Inactive Ribosomal Genes by Redigestion of DNA with HindIII.
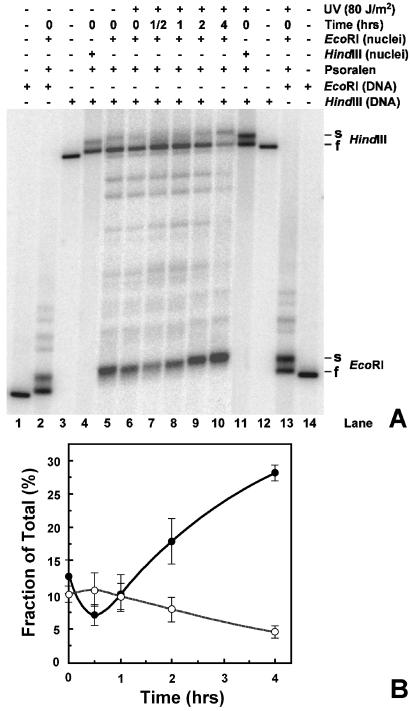
After different repair times, nuclei were isolated and active rDNA was released by EcoRI digestion as described above (Fig. 5B). Because the two EcoRI sites are within the HindIII fragment (Fig. 2), a full-length fragment (≈6.4 kb) obtained after HindIII digestion of DNA isolated from EcoRI-treated nuclei contains primarily inactive rDNA. As shown in Fig. 5B, EcoRI digests of nuclei, isolated from cells incubated for varying repair times, contain only active rDNA (also see Fig. 6A, lanes 5–10, EcoRI s-bands). When the isolated DNA is redigested with HindIII, intact HindIII fragments contain primarily inactive rDNA (Fig. 6A, lanes 5–10, HindIII f-bands). As controls, single digests by EcoRI and HindIII of DNA from psoralen-crosslinked nuclei show the migration of both active and inactive rDNA (Fig. 6A, lanes 2, 13 and 4, 11, respectively). The faint bands present between the EcoRI and HindIII fragments represent partial EcoRI digests of rDNA chromatin in nuclei (Fig. 2, small E). These data demonstrate that most of the active rDNA is released from nuclei by EcoRI, whereas there is a marked enrichment for inactive rDNA in the HindIII fraction.
Figure 6.
Separation of active and inactive ribosomal chromatin. (A) Nuclei were isolated from unirradiated (lane 5) and irradiated (lanes 6–10) cells that were harvested after different repair times. These nuclei were digested with EcoRI before psoralen crosslinking (lanes 5–10). The isolated DNA was then digested with HindIII, separated on a 1% native agarose gel, blotted, and hybridized with labeled rDNA probe (see Fig. 2). As controls, genomic DNA was isolated from uncrosslinked cells and digested with either EcoRI (lanes 1 and 14) or HindIII (lanes 3 and 12). The presence of active and inactive rDNA chromatin was monitored by digesting nuclei with EcoRI or HindIII before psoralen crosslinking and by redigesting the isolated DNA with EcoRI and HindIII, respectively (lanes 2 and 13, and lanes 4 and 11). Labels on the right denote active rDNA, s- (slow) band, and inactive rDNA, f- (fast) band. (B) Signals of the EcoRI and HindIII bands in each lanes were quantified and expressed as percent of the total signal measured in the corresponding lanes. Data are the mean ± 1 SD of three independent experiments.
Quantification of active (EcoRI band) and inactive (HindIII band) populations (Fig. 6A, lanes 6–10) at each repair time are presented in Fig. 6B. Signals of each population are expressed as the percent of total signal measured in each lane.
Repair of Individual Strands of Active and Inactive rDNA.
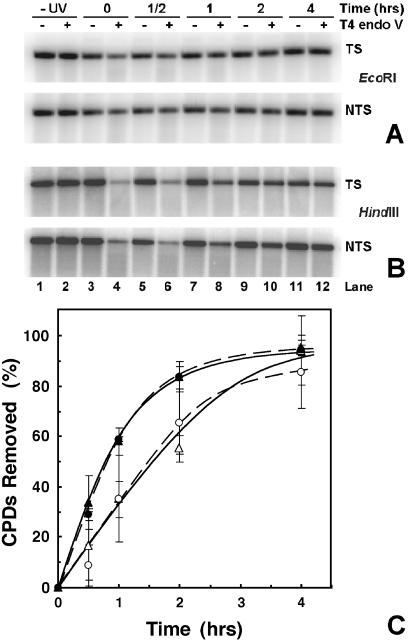
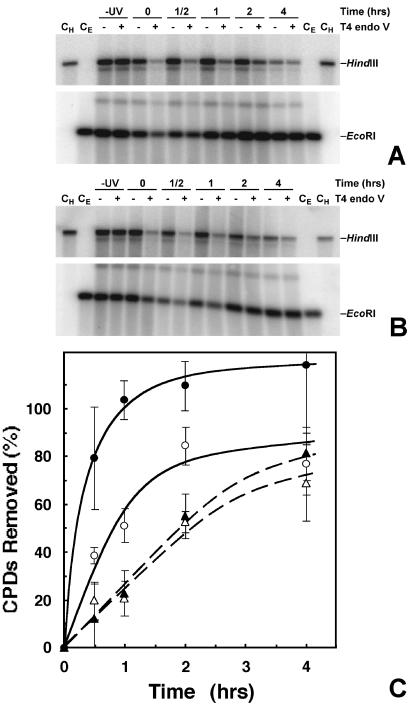
To follow NER, DNA was isolated from aliquots of EcoRI-digested nuclei (Fig. 1). The DNA samples were redigested with HindIII to release inactive rDNA and then treated with T4 endo V. Representative gels for repair of the TS and NTS of active (EcoRI band) and primarily inactive (HindIII band) rDNA are shown in Fig. 7 A and B, respectively.
Figure 7.
Repair of individual strands of active and inactive rDNA. After different repair times, DNA was isolated from EcoRI-treated nuclei and digested with HindIII. DNA samples, mock-treated or treated with T4 endo V, are denoted by − and +, respectively. −UV denotes nuclei from unirradiated cells, and +UV denotes nuclei from irradiated cells harvested after the indicated repair times. Samples were separated on a 1% alkaline agarose gel, blotted, and hybridized with strand-specific riboprobes (see Fig. 2). As controls, genomic DNA was isolated from nontreated cells and digested with either HindIII or EcoRI, respectively (CH and CE). (A) TS and (B) NTS. To conserve space, the central portion of each gel is not shown. (C) Quantification of PhosphorImages. DNA repair is expressed as percent of CPDs removed as a function of repair time. Data are from active rDNA (EcoRI, circles) and inactive rDNA (HindIII, triangles). Solid and open symbols represent data from the TS and NTS, respectively. Data are the mean ± 1 SD of three independent experiments.
The yield of CPDs in each strand of active and inactive rDNA was similar. For the active fraction 0.20 ± 0.05 and 0.18 ± 0.05 CPDs/kb were obtained for the TS and NTS, respectively. For the inactive fraction 0.20 ± 0.07 and 0.20 ± 0.03 CPDs/kb were obtained for the TS and NTS, respectively (mean ± 1 SD of three experiments). Therefore, the average yield of CPDs in each strand of active and inactive rDNA is not statistically different from the values determined for total rDNA (see above).
Importantly, DNA repair is significantly faster in the TS of active rDNA than in the NTS of that fraction (Fig. 7C, compare solid and open circles). Indeed, over twice as many CPDs are removed from the TS, compared with the NTS, in 1 h. Conversely, there is no difference in DNA repair between the TS and NTS of inactive rDNA (Fig. 7C, compare solid and open triangles). These data demonstrate that TCR occurs in pol I-transcribed genes of yeast and is essentially complete in the TS after 60 min of repair.
Discussion
We used psoralen crosslinking in yeast to separate active from inactive rDNA, and CPD removal was followed in the two rDNA populations. Our results show that TCR occurs in active rDNA and is absent in the inactive rDNA (Fig. 7). Previously, the existence of strand-specific repair was shown in total rDNA of rad7, rad16, and rad4 yeast strains (21). However, those results did not confirm the existence of TCR because samples contained both active and inactive rDNA (Fig. 5A; ref. 28), leaving open the possibility that strand-specific repair occurs in both fractions, independent of transcription (21).
This work extends our current knowledge on TCR, the process that repairs DNA lesions that arrest transcription (4). In contrast to the TCR observed in many pol II genes of mammalian cells (10), no evidence for TCR was found in the rDNA of these cells (17, 19). Because each class of RNA polymerase uses a distinct set of transcription factors, which are assembled into transcription initiation complexes at specific promoters (41), it is possible that there is no TCR in rDNA because of the differences between pol II and pol I complexes. However, an intrinsic problem in those studies (17, 19) is that either the active and inactive rDNA copies were not separated for NER measurements (17) or little repair was observed in the active and total rDNA fractions (19). In the present article, experiments where active and inactive rDNA are not separated show only a small bias for strand-specific repair in total rDNA (Fig. 4).
At present, it remains to be determined whether TCR of pol I genes is a unique feature of yeast cells. Indeed, yeast and higher eukaryotes present some differences in both NER (42) and chromatin (43, 44). This question could be addressed by measuring NER in both rDNA fractions of exponentially growing mammalian cells.
In a current model, Hanawalt (45) suggests that TCR may involve an “obstacle-recognition” step that is needed before repair of damage to transcribing genes, in which pol II is stalled at a lesion. In this model, Cockayne Syndrome proteins CSA and CSB could be involved in removing the arrested pol II, whereas transcription factor TFIIH and xeroderma pigmentosum (group G) protein XPG would “assess the nature of the obstruction” and recruit NER enzymes (45). Because we observed TCR in rDNA (Fig. 7), this process could also be true for arrested pol I. However, the helicases XPB and XPD, required for both TCR and global genomic repair, are subunits of TFIIH (46), which is essential only for pol II transcription (41). Therefore, considering the fast removal of CPDs from rDNA in yeast (compare Figs. 3B and 4C), TFIIH would have to rapidly penetrate the nucleolus.
Another model suggests that TCR is a subpathway of NER (46). In this case, the TS is preferentially repaired just upstream of where pol II clears the promoter and releases TFIIH. Beyond that point, TCR requires CSA and CSB (or Rad26p in yeast) to re-recruit TFIIH to the repair complex when pol II is arrested at a lesion (4, 10). Potentially, Rad26p also could recruit TFIIH to pol I stalled at a lesion. Christians and Hanawalt (47) examined removal of CPDs from rDNA in CSA and CSB cells, which display normal levels of NER in the genome overall (48) but lack TCR in pol II genes (49). Interestingly, lower than normal repair of CPDs was observed in the rDNA of both cell types (47). Conversely, the RAD26 gene in yeast was not found to be involved in removal of CPDs from rDNA (21). Thus, the question of whether CSA and CSB (or Rad26p) are involved in TCR of pol I genes remains unresolved.
Very few studies have analyzed NER in RNA polymerase III (pol III)-transcribed genes. In human fibroblasts, CPDs present in tRNA genes were repaired by NER but TCR was not observed (50). Different results were found in yeast, where the NTS was actually repaired faster than the TS of the SNR6 gene (51). In yeast, the pol III-transcribed 5S rRNA gene is located between rDNA transcription units (Fig. 2). It would be interesting to investigate whether TCR extends to the 5S rRNA gene.
The data in Fig. 4C indicate there may be a small bias for repair of the TS in total rDNA. Additionally, repair of the TS of total rDNA is similar to that of genomic DNA, whereas repair of the NTS appears to be slightly lower (compare Figs. 3B and 4C). This can be explained because the data are an average of active and inactive rDNA copies. Compared with repair of genomic DNA, repair of the TS of active rDNA is faster, repair of the NTS of active rDNA is similar, and repair of both strands of inactive rDNA is slower (compare Figs. 3B and 7C). Because the NTS of active rDNA is repaired faster than either strand of inactive rDNA (Fig. 7C), nucleosomes may be an impediment for NER in rDNA. Moreover, NER is faster in genomic DNA than in the inactive rDNA (compare Figs. 3B and 7C), possibly because most of the genome in yeast is actively transcribed (52) whereas the silent rDNA is folded into inactive chromatin (28).
Because chromatin rearrangements occur during DNA repair (53, 54), the accessibility of EcoRI to rDNA chromatin was followed during NER (Figs. 5 and 6). Generally, DNA is more accessible to restriction enzymes in unfolded (open) chromatin (39, 40). EcoRI digestion of nuclei, combined with psoralen as a probe for chromatin structure, indicates that rDNA becomes less accessible to EcoRI during early repair times (up to 1 h) and more accessible at later repair times (2–4 h) (Fig. 6). These results suggest that during NER chromatin rearrangements occur in the rDNA locus. The rearrangements could be a direct result of NER or possibly the result of arrest and reinitiation of transcription after CPD removal.
In conclusion, we examined the induction and removal of CPDs from individual strands of active and inactive rDNA chromatin in yeast and compared these with CPD removal from total rDNA and bulk chromatin. Our results show that TCR occurs in actively transcribing rDNA as strand-specific repair and is not found in the inactive rDNA copies. In addition, changes in chromatin structure of the rDNA locus occur during NER.
Acknowledgments
We thank Drs. J. S. Smith and J. D. Boeke for the JS311 yeast cells, Dr. R. S. Lloyd for supplying T4 endo V, and Ms. D. Fahy for critical reading of the manuscript. This study was supported by Grant ES04106 from the National Institute of Environmental Health Sciences.
Abbreviations
- NER
nucleotide excision repair
- CPD
cyclobutane pyrimidine dimer
- TS
transcribed strand
- TCR
transcription-coupled repair
- pol II
polymerase II
- pol I
polymerase I
- NTS
nontranscribed strand
- endo V
endonuclease V
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Freidberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol. Press; 1995. [Google Scholar]
- 2.Woodgate R. Genes Dev. 1999;13:2191–2195. doi: 10.1101/gad.13.17.2191. [DOI] [PubMed] [Google Scholar]
- 3.Vrieling H, van Zeeland A A, Mullenders L H. Mutat Res. 1998;400:135–142. doi: 10.1016/s0027-5107(98)00064-5. [DOI] [PubMed] [Google Scholar]
- 4.Tornaletti S, Hanawalt P C. Biochimie. 1999;81:139–146. doi: 10.1016/s0300-9084(99)80046-7. [DOI] [PubMed] [Google Scholar]
- 5.Mellon I, Spivak G, Hanawalt P C. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 6.Mellon I, Hanawalt P C. Nature (London) 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 7.Smerdon M J, Thoma F. Cell. 1990;61:675–684. doi: 10.1016/0092-8674(90)90479-x. [DOI] [PubMed] [Google Scholar]
- 8.Leadon S A, Lawrence D A. Mutat Res. 1991;255:67–78. doi: 10.1016/0921-8777(91)90019-l. [DOI] [PubMed] [Google Scholar]
- 9.Christians F C, Hanawalt P C. Mutat Res. 1992;274:93–101. doi: 10.1016/0921-8777(92)90056-9. [DOI] [PubMed] [Google Scholar]
- 10.Hanawalt P C, Spivak G. In: Advances in DNA Damage and Repair. Dizdaroglu M, Karakaya A E, editors. New York: Plenum; 1999. pp. 169–179. [Google Scholar]
- 11.Sollner-Webb B, Mougey E B. Trends Biochem Sci. 1991;16:58–62. doi: 10.1016/0968-0004(91)90025-q. [DOI] [PubMed] [Google Scholar]
- 12.Visintin R, Amon A. Curr Opin Cell Biol. 2000;12:372–377. doi: 10.1016/s0955-0674(00)00102-2. [DOI] [PubMed] [Google Scholar]
- 13.Cohn S M, Lieberman M W. J Biol Chem. 1984;259:12456–12462. [PubMed] [Google Scholar]
- 14.Matsumoto A, Vos J M, Hanawalt P C. Mutat Res. 1989;217:185–192. doi: 10.1016/0921-8777(89)90070-0. [DOI] [PubMed] [Google Scholar]
- 15.Vos J M H, Wauthier E L. Mol Cell Biol. 1991;11:2245–2252. doi: 10.1128/mcb.11.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fritz L K, Suquet C, Smerdon M J. J Biol Chem. 1996;271:12972–12976. doi: 10.1074/jbc.271.22.12972. [DOI] [PubMed] [Google Scholar]
- 17.Christians F C, Hanawalt P C. Biochemistry. 1993;32:10512–10518. doi: 10.1021/bi00090a030. [DOI] [PubMed] [Google Scholar]
- 18.Stevnsner T, May A, Peterson L N, Larminat F, Pirsel M, Bohr V A. Carcinogenesis. 1993;14:1591–1596. doi: 10.1093/carcin/14.8.1591. [DOI] [PubMed] [Google Scholar]
- 19.Fritz L K, Smerdon M J. Biochemistry. 1995;34:13117–13124. doi: 10.1021/bi00040a024. [DOI] [PubMed] [Google Scholar]
- 20.Balajee A S, May A, Bohr V A. Nucleic Acids Res. 1999;27:2511–2520. doi: 10.1093/nar/27.12.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verhage R A, van de Putte P, Brouwer J. Nucleic Acids Res. 1996;24:1020–1025. doi: 10.1093/nar/24.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conconi A, Jager-Vottero P, Zhang X, Beard B C, Smerdon M J. Mutat Res. 2000;459:55–64. doi: 10.1016/s0921-8777(99)00057-9. [DOI] [PubMed] [Google Scholar]
- 23.Lucchini R, Sogo J M. In: Transcription of Ribosomal RNA Genes by Eukaryotic RNA Polymerase I. Paule M R, editor. Austin, TX: Landes Bioscience; 1998. pp. 254–276. [Google Scholar]
- 24.Bohr V A, Smith C A, Okumoto D S, Hanawalt P C. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 25.Sogo J M, Conconi A, Widmer R M. In: Photochemical Probes in Biochemistry. Nielsen P E, editor. New York: Kluwer; 1989. pp. 179–194. [Google Scholar]
- 26.Conconi A, Widmer R M, Koller T, Sogo J M. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- 27.Conconi A, Sogo J M, Ryan C A. Proc Natl Acad Sci USA. 1992;89:5256–5260. doi: 10.1073/pnas.89.12.5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dammann R, Lucchini R, Koller T, Sogo J M. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dammann R, Lucchini R, Koller T, Sogo J M. Mol Cell Biol. 1995;15:5294–5303. doi: 10.1128/mcb.15.10.5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lucchini R, Sogo J M. Mol Cell Biol. 1992;12:4288–4296. doi: 10.1128/mcb.12.10.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith J S, Caputo E, Boeke J D. Mol Cell Biol. 1999;19:3184–3197. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slater M L. J Bacteriol. 1973;113:263–270. doi: 10.1128/jb.113.1.263-270.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dodson M L, Lloyd R S. Mutat Res. 1989;218:49–65. doi: 10.1016/0921-8777(89)90011-6. [DOI] [PubMed] [Google Scholar]
- 34.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab Press; 1982. [Google Scholar]
- 35.Bespalov V A, Conconi A, Zhang X, Fahy D, Smerdon M J. Environ Mol Mutagen. 2001;38:166–174. doi: 10.1002/em.1068. [DOI] [PubMed] [Google Scholar]
- 36.Mueller J P, Smerdon M J. Nucleic Acids Res. 1995;23:3457–3464. doi: 10.1093/nar/23.17.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutherland B M, Shih A G. Biochemistry. 1983;22:745–749. doi: 10.1021/bi00273a006. [DOI] [PubMed] [Google Scholar]
- 38.Lucchini R, Pauli U, Braun R, Koller T, Sogo J M. J Mol Biol. 1987;196:829–843. doi: 10.1016/0022-2836(87)90408-6. [DOI] [PubMed] [Google Scholar]
- 39.Ness P J, Labhart P, Banz E, Koller T, Parish R W. J Mol Biol. 1983;166:361–381. doi: 10.1016/s0022-2836(83)80090-4. [DOI] [PubMed] [Google Scholar]
- 40.Anderson J D, Lowary P T, Widom J. J Mol Biol. 2001;307:977–985. doi: 10.1006/jmbi.2001.4528. [DOI] [PubMed] [Google Scholar]
- 41.Grummt I. Prog Nucleic Acid Res Mol Biol. 1999;62:109–154. doi: 10.1016/s0079-6603(08)60506-1. [DOI] [PubMed] [Google Scholar]
- 42.Prakash S, Prakash L. Mutat Res. 2000;451:13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- 43.Vogelauer M, Wu J, Suka N, Grunstein M. Nature (London) 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- 44.Grunstein M. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 45.Hanawalt P C. Nature (London) 2000;405:415–416. doi: 10.1038/35013197. [DOI] [PubMed] [Google Scholar]
- 46.Hanawalt P C. Mutat Res. 2001;485:3–13. doi: 10.1016/s0921-8777(00)00071-9. [DOI] [PubMed] [Google Scholar]
- 47.Christians F C, Hanawalt P C. Mutat Res. 1994;323:179–187. doi: 10.1016/0165-7992(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 48.Mayne L V, Lehmann A R, Waters R. Mutat Res. 1982;106:179–189. doi: 10.1016/0027-5107(82)90200-7. [DOI] [PubMed] [Google Scholar]
- 49.Venema J, Mullenders L H, Natarajan A T, van Zeeland A A, Mayne L V. Proc Natl Acad Sci USA. 1990;87:4707–4711. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dammann R, Pfeifer G P. Mol Cell Biol. 1997;17:219–229. doi: 10.1128/mcb.17.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aboussekhra A, Thoma F. Genes Dev. 1998;12:411–421. doi: 10.1101/gad.12.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fangman W L, Zakian V A. In: The Molecular Biology of the Yeast Saccharomyces. Broach J, Jones E, Strathern J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1981. pp. 27–58. [Google Scholar]
- 53.Smerdon M J, Conconi A. Prog Nucleic Acid Res Mol Biol. 1999;62:227–255. doi: 10.1016/s0079-6603(08)60509-7. [DOI] [PubMed] [Google Scholar]
- 54.Thoma F. EMBO J. 1999;18:6585–6598. doi: 10.1093/emboj/18.23.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]