Abstract
Presentation of metabolites by the major histocompatibility complex class I–related protein 1 (MR1) molecule to mucosal-associated invariant T cells is impaired during herpes simplex virus type 1 (HSV-1) and type 2 (HSV-2) infections. This is surprising given these viruses do not directly synthesise MR1 ligands. We have previously identified several HSV proteins responsible for rapidly downregulating the intracellular pool of immature MR1, effectively inhibiting new surface antigen presentation, while preexisting ligand-bound mature MR1 is unexpectedly upregulated by HSV-1. Using flow cytometry, immunoblotting, and high-throughput fluorescence microscopy, we demonstrate that the endocytosis of surface MR1 is impaired during HSV infection and that internalized molecules accumulate in EEA1-labeled early endosomes, avoiding degradation. We establish that the short MR1 cytoplasmic tail is not required for HSV-1–mediated downregulation of immature molecules; however it may play a role in the retention of mature molecules on the surface and in early endosomes. We also determine that the HSV-1 US3 protein, the shorter US3.5 kinase and the full-length HSV-2 homolog, all predominantly target mature surface rather than total MR1 levels. We propose that the downregulation of intracellular and cell surface MR1 molecules by US3 and other HSV proteins is an immune-evasive countermeasure to minimize the effect of impaired MR1 endocytosis, which might otherwise render infected cells susceptible to MR1-mediated killing by mucosal-associated invariant T cells.
Keywords: herpesvirus, immunosuppression, viral immunology, receptor endocytosis, MR1, HSV
The major histocompatibility complex (MHC) class I (MHC-I)-related protein 1 (MR1) molecule presents a variety of small metabolites (1, 2) to mucosal-associated invariant T (MAIT) cells (3) and other MR1-restricted T cells (4). The unique structure and intracellular life cycle of this nonclassical antigen presentation molecule complements the characteristics of its metabolic ligand. MR1 is highly conserved in mammals (5). The canonical ligand 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil condenses from unstable intermediaries in the biosynthesis pathway of vitamin B2, synthesized by many commensal and pathogenic bacterial and fungal species (6), but not by mammalian host cells, nor viruses. Unlike classical MHC-I molecules, MR1 molecules lacking bound ligand can still accumulate in the endoplasmic reticulum (ER). This ready store of molecules supports a rapid response to ligand, the binding of which triggers a conformational change in MR1, stabilization of the otherwise unstable cargo, association with beta-2 microglobulin (β2m), and trafficking through the secretory pathway, with the resulting mature MR1–β2m–antigen complexes displayed at the cell surface (7). Whereas classical MHC-I peptide binding stability affects the half-life of complexes on the plasma membrane (PM) (8, 9), the presence or characteristics of the bound ligand does not impact longevity of MR1 on the cell surface (7). The relatively slow rate of clathrin-mediated endocytosis is regulated by binding of a noncanonical tyrosine motif in the short MR1 cytoplasmic tail to the endocytic adaptor protein 2 (AP2) complex (7, 10). This interaction also controls transit through endosomal compartments, which supports limited exchange of cargo with that supplied by intracellular pathogens (10, 11, 12). Unlike another nonclassical antigen presentation molecule CD1d, which relies on endosomal recycling to present lipids to invariant natural killer T cells (13), most mature MR1 molecules are degraded in lysosomal compartments, with little recycling back to the PM for detection by MR1-restricted T cells (7, 10).
Establishment of the immunological synapse between the MAIT T cell receptor (TCR) and antigen-bound MR1 drives a tissue repair signature in MAIT cells (14, 15). This establishes a specific role sensing and responding to the integrity of barrier sites, according to fluctuations in exogenous microbial metabolic by-products. Signs of infection detected by pattern recognition and cytokine receptors, in combination with TCR-mediated signaling, are required for maximal MAIT cell proliferation, production of proinflammatory molecules, and cytolytic killing across the immunological synapse (16, 17). In the absence of MR1-mediated activation, MAIT cells can still respond to these signs of infection, which allows them to mediate antiviral activity (18, 19, 20).
Herpes simplex viruses (HSVs) encode many mechanisms to hide their presence and inhibit antiviral responses. This enables their rapid transmission across several different cell types in the skin and mucosa to access sensory neurons, where they can establish life-long latency (21, 22). Surprisingly, we have established that these viruses, as well as human cytomegalovirus, murine cytomegalovirus, and varicella zoster virus (VZV), also downregulate MR1 antigen presentation in epithelial cells, even though, to the best of our knowledge, they do not directly contribute to MR1 ligand production (23, 24, 25). HSV type 1 (HSV-1) and type 2 (HSV-2) both primarily target the pool of immature ER-resident MR1 molecules through expression of viral proteins ICP22 and the RNase vhs (23, 26). This depletion of the MR1 reservoir blocks ligand-induced MR1 maturation and trafficking to the PM and consequent TCR-mediated T cell activation (23). However, there is evidence of an additional point of interference in the MR1 trafficking pathway during HSV infection, which may not be advantageous to the virus. Specifically, molecules that escape the ER prior to viral infection are instead up regulated on the cell surface during infection (23, 26). Even with a strong depletion of the MR1 reservoir by HSV, this unexpected increase in surface MR1 might expose infected cells to TCR-mediated killing, potentially explaining the biological imperative to inhibit MR1 synthesis and accumulation.
In this study, we reveal the opposing forces that affect the level of surface MR1 during HSV infection by focusing on the fate of endocytosed molecules at late stages of HSV-1 and HSV-2 infection. Using high-throughput quantitative fluorescence microscopy, we examine the modulation of MR1 at key locations in its trafficking pathway in cells pretreated with MR1 ligand, confirming that MR1 molecules are not only protected on the PM but also accumulate in early endosomes. Furthermore, we identify that impaired internalization contributes to the extended retention of surface MR1 during HSV-1 infection.
Previously, we identified that ectopic expression of HSV-1 US3 serine-threonine kinase downregulates endogenous and overexpressed surface MR1 (23). Here, we extend this finding to include both HSV-1 US3.5, a shortened, naturally expressed, kinase-active form of the protein, and the HSV-2 US3 homolog, and establish that these viral proteins primarily target surface rather than total cellular MR1. Inactivation of the kinase domain only partially rescues the loss of surface MR1, confirming that US3 enzymatic activity contributes to the modulation of mature MR1.
Furthermore, we identify that impaired internalization contributes to the extended retention of surface MR1 during HSV-1 infection. We propose that this is the by-product of viral infection and demonstrate that it is effectively counteracted by the depletion of MR1 by US3 and other viral proteins, under conditions of continuous ligand availability that mimic homeostasis in an intact mucosa.
Here, we provide the first evidence explaining why herpes viruses encode a multipronged and strong downregulation of MR1, by identifying an unexpected impact of viral infection. We propose that the downregulation of MR1 molecules during HSV infection is an immune-evasive countermeasure offsetting the increased retention of surface MR1, which would otherwise render infected cells vulnerable to rapid MAIT cell detection and cytolytic killing.
Results
Mature MR1 accumulates in EEA1-labeled endosomes
Immature MR1 accumulates in an open conformation in the ER, with little closed conformation, mature MR1 detectable on the PM, unless the molecule is artificially overexpressed (7, 27). This intracellular pool allows the cell to respond rapidly and sensitively to exogenous ligand, which binds to ER-resident MR1 and triggers its maturation and trafficking through the Golgi and trans Golgi network to the PM, for detection by the MAIT TCR. During HSV-1 and HSV-2 infection of epithelial cells, a substantial loss of the immature MR1 is evident by 6 h post infection (h p.i.), however detecting the downstream effect of this loss on mature surface MR1 is usually only significant at later timepoints (23, 26). Furthermore, loss of surface MR1 is dependent on ligand conditions. In particular, maximizing the amount of preexisting mature surface MR1, by inducing strong trafficking to the PM in the period prior to infection, instead reveals an upregulation of endogenous and overexpressed surface MR1 (23). This phenomenon is most evident when cells are pretreated with ligand, which is then washed from the cells at the time of infection.
To investigate the impact of HSV on mature ligand-bound MR1, the relative amounts of MR1 molecules were quantified at five subcellular locations within ARPE-19 MR1 cells, by high-throughput fluorescence microscopy (Figs. 1 and S1). Cells were pretreated with acetyl-6-formylpterin (Ac-6-FP), a synthetic MR1 ligand that has been used to characterize MR1 localization due to its stability and strong promotion of MR1 maturation and trafficking from the ER to the PM (7, 28). After 6 h the cells were washed and infected with either HSV-1 (strain F, multiplicity of infection (MOI) 5) or HSV-2 (strain 186, MOI 3). Based on the expected location of MR1 in both the secretory and endocytic pathways (7, 11), cells were then stained at 15 h p.i. for the PM (wheat germ agglutinin [WGA]), then MR1 and the ER (calreticulin), GA (GM130), early endosomes (EEA1) or late endosomes/lysosomes (LAMP1), and finally for the nucleus (4′,6-diamidino-2-phenylindole). MR1 was detected by intracellular staining with the 26.5 clone antibody, which labels only the closed conformations of the MR1 antigenic cleft mostly associated with mature glycosylated MR1 (7, 29), rather than molecules in the open conformation that dominate the MR1 pool in the ER (27).
Figure 1.
Upregulation of surface and early endosomal MR1 during HSV-1 and HSV-2 infection. ARPE-19 MR1 cells were pretreated with Ac-6-FP (5 μM) for 6 h and then infected with HSV-1 (strain F, MOI 5), HSV-2 (strain 186, MOI 3), or mock-infected in parallel. Cells were stained at 15 h p.i. with WGA (plasma membrane) and then permeabilized and stained for MR1 (clone 26.5), and calreticulin (ER), GM130 (Golgi apparatus), EEA1 (early endosomes), or LAMP1 (late endosomes/lysosomes). Cells were then stained with fluorophore-conjugated secondary antibodies and DAPI (nucleus). A, representative images of DAPI + WGA, MR1, EEA1, and merged images, with white arrows highlighting examples of colocation in enlarged image (box) on right. The scale bar represents 50 μm or 20 μm in the enlarged images on the right. Cell segmentation map for each field of view (FOV) was completed with ilastik software after training with a representative image set. ImageJ software was then used for quantification of fold change (FC) in MR1 MFI intensity relative to mock (B, C) or median cellular MR1 (D, E) within each FOV after overlaying the Ilastik-generated map. Violin plots depict fold change in MFI in mock (white), HSV-1 (red), and HSV-2 (blue) infected cells to either mock (C) or median cellular MR1 (E), with median and quartiles shown as horizontal lines. Statistical significance was calculated with one-way ANOVA with multiple comparisons, with p values < 0.05 annotated. Analysis of at least 14 FOV in two independent experiments. Ac-6-FP, acetyl-6-formylpterin; DAPI, 4′,6-diamidino-2-phenylindole; ER, endoplasmic reticulum; h p.i., hour post infection; HSV-1, herpes simplex virus type 1; HSV-2, herpes simplex virus type 2; MOI, multiplicity of infection; MR1, major histocompatibility complex class I–related protein 1; WGA, wheat germ agglutinin.
Infected cells demonstrated general morphological changes consistent with the expression of viral proteins, including punctate nuclear staining associated with the formation of nuclear bodies and virus-enriched chaperone-enhanced domains (30), increased WGA staining of membranes, cell rounding from cytoskeleton disruption (31), and formation of multinucleated cells associated with this HSV-2 strain (32) (Figs. 1A and S1). There was clear loss of MR1 from within the infected cells (Fig. 1A), with residual MR1 primarily located in cytoplasmic accumulations or on the PM. The MR1 intensity colocated with these markers was quantified within multiple fields of view (FOVs) per sample and the ilastik machine learning software (https://www.ilastik.org/) (33) trained to create a segmentation mask of the nucleus, subcellular marker, cytoplasm, and PM for each FOV. The median MR1 fluorescence signal at each of these locations, and within the whole cell, was quantified using ImageJ software (https://imagej.net/ij/).
Firstly, we analyzed the effect of HSV infection on the MR1 signal intensity in each subcellular compartment by calculating the fold change of the fluorescent signal in infected to mock infected samples. Both HSV-1 and HSV-2 caused a significant downregulation of total MR1 within the cell and a selective downregulation in the ER, Golgi, and late endocytic/recycling compartments (Fig. 1, B, C and S1). Mature MR1 was preferentially retained on the PM in HSV-1–infected cells but significantly upregulated by HSV-2 (Fig. 1, B and C) and accumulated in early endosomal compartments with both viral infections (Fig. 1, A, B and C) compared to the uninfected samples.
The accumulation in early endosomes may have impacted transition to late endosomes/lysosomes, thus explaining the reduction of MR1 in this compartment. Alternately, HSV infection may actively promote lysosomal degradation of MR1. To determine if the latter is true, infected ARPE-19 MR1-GFP and MR1 cells were treated with lysosomal inhibitors folimycin or leupeptin after the 1-h period of viral adsorption. Total MR1 protein was analyzed at 6 h p.i. by flow cytometry (MR1-GFP, Fig. S2A) and immunoblot (MR1) (Fig. S2B). Infected treated cells did not demonstrate any rescue of the loss of MR1-GFP or MR1, establishing that HSV-1 infection does not promote lysosomal degradation of MR1.
A direct comparison of the relative amount of MR1 in one subcellular compartment to the others was not possible as the EEA1 staining required permeabilization with a stronger detergent (Triton X-100) than the other probes (Saponin) to minimize background binding of the EEA1 antibody, resulting in reduced staining with the MR1 antibody (Fig. S3). Nevertheless, by comparing the median MR1 signal in each subcellular compartment to the median cell signal within the FOV with the same permeabilization agent, the relative modulation of MR1 at different locations was assessed. The relative strength of MR1 mean fluorescence intensity (MFI) signal colocated with calreticulin, compared to the average across the cell, was only modestly downregulated in the ER in HSV-2 infected cells; however there was a stronger relative loss in the GA with both viral infections. The relative amount of MR1 colocating with EEA1 was increased only in HSV-1–infected cells (Fig. 1C), however treatment with the stronger detergent reduced staining with the MR1 antibody (Fig. S3), making comparison to the median cellular MFI within the FOV less sensitive. Nevertheless, there was a clear localization with EEA1 staining in the infected cells that was not evident in the mock-infected samples (Fig. 1A, white arrows) or for any of the other intracellular markers (Fig. S1). Finally, both HSV-1 and HSV-2–infected cells demonstrated a strong relative increase in MR1 detected on the PM (Fig. 1E), compared to the median MR1 signal within the cell.
In summary, these results demonstrate that the loss of immature MR1 from the ER (23) results in a comparable loss of mature MR1 in the secretory pathway, but that this does not flow through to the PM or endocytic compartments. Instead, we observe an upregulation on the PM and retention in early endocytic endosomes that may prevent migration to late endosomal/lysosomal compartments for degradation. This establishes that additional factors present during viral infection impact the localization of mature MR1 molecules.
Downregulation of MR1 is not dependent on the MR1 cytoplasmic tail
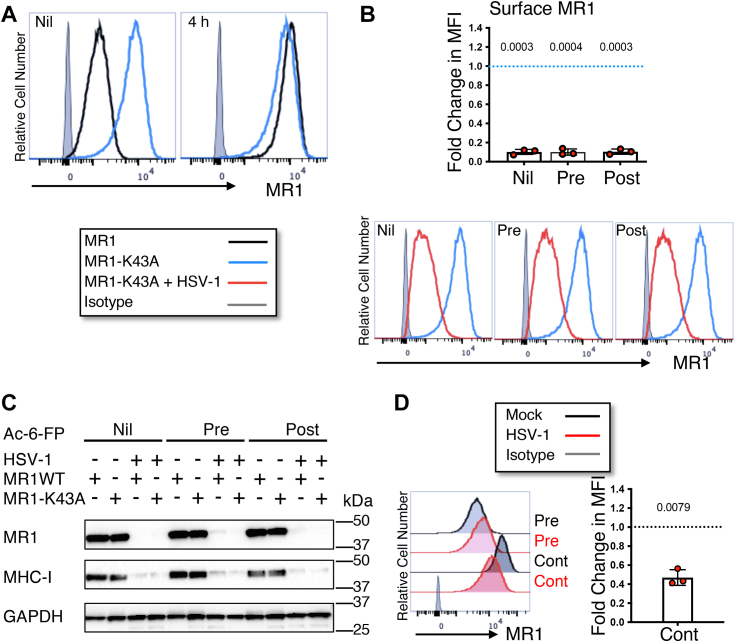
Herpes viruses target the localization of other antigen presentation molecules by association with and modification of their cytoplasmic domain. For example, HSV-2 ICP47 protein associates with the human leukocyte antigen (HLA)-C cytoplasmic domain to downregulate its cell surface presentation (34) and Kaposi sarcoma–associated herpes virus protein modulator of immune recognition ubiquitinates the CD1d tail to promote endocytosis (35). MR1 encodes a short cytoplasmic tail (36), deletion of which does not affect pooling of the molecule in the ER, maturation, or regulation of trafficking through the secretory pathway (10). To determine whether the cytoplasmic tail is critical for the modulation of MR1 by HSV-1, ARPE-19 cells were transduced by retrovirus (10) to overexpress an MR1 mutant lacking the final 15 amino acids of the C-terminal domain (ARPE-19 ΔP305). Given that manipulating the timing of ligand addition, relative to the time of infection, affects the ability of HSV-1 to modulate surface WT MR1 (23), three different ligand conditions were evaluated. Cells were optionally treated with Ac-6-FP ligand for 24 h prior to infection with HSV-1 (pre) and washed from the assay at the time of infection as previously described or added for the final 4 h (post). Cells were harvested at 18 h p.i. and MR1 analyzed by flow cytometry and immunoblotting. No difference was detected in the surface expression of WT or ΔP305 MR1 both in the absence of ligand (nil), and with 4 h ligand treatment, in mock-infected cells (Fig. 2A), corroborating that the cytoplasmic tail is not required for trafficking to the PM in this cell line. Modulation of surface ΔP305 during HSV-1 infection mirrored that of WT MR1 (23), demonstrating a significant loss of ΔP305 in the absence of ligand, and with post infection treatment, while pretreatment resulted in a small but significant upregulation (Fig. 2B).
Figure 2.
HSV-1 mediated loss of total and surface MR1 is not dependent on the cytoplasmic tail.A, ARPE-19 MR1 (black) or ARPE-19 ΔP305 (blue) cells transduced to express MR1 lacking the final C-terminal 15 amino acids were left untreated (nil) or treated with Ac-6-FP (5 μM) for 4 h prior to harvesting. Cells were stained for surface MR1 or isotype control (gray) and analyzed by flow cytometry. Histograms representative of two independent experiments. B and C, ARPE-19 ΔP305 cells were mock (blue) or HSV-1 (strain F, MOI 5, red) infected in parallel. Cells were left untreated (nil) or treated with Ac-6-FP (5 μM) 24 h prior to infection (pre) or 14 h p.i. (post). B, cells were harvested at 18 h p.i. when they were stained for surface MR1 or isotype control (gray) and analyzed by flow cytometry. Fold change of mean fluorescence intensity in infected cells relative to mock-infected cells (dotted blue line) with matching ligand treatment is graphed with SD. Statistical significance was calculated by paired Student’s t test, with p values < 0.05 annotated. Analysis of four independent experiments. C, cell lysates were harvested at 18 h p.i. and separated by gel electrophoresis before immunoblotting for MR1, MHC-I, and GAPDH. Ac-6-FP, acetyl-6-formylpterin; h p.i., hour post infection; HSV-1, herpes simplex virus type 1; MHC-I, MHC class I; MOI, multiplicity of infection; MR1, major histocompatibility complex class I–related protein 1.
Total ΔP305 during HSV-1 infection was detected by immunoblotting lysed cells at 18 h p.i. with the same ligand treatments (Fig. 2C). ΔP305 could not be detected in the infected cells in the absence of ligand (nil) and in those treated 4 h prior to harvest (post); however, a band was detected in the infected cells pretreated with (pre) (Fig. 2C) consistent with HSV-1 modulation of the WT molecule (23). Together, these results demonstrate that the MR1 cytoplasmic tail is not required for either the loss of cellular or surface MR1 during HSV-1 infection or for the protection and upregulation of surface MR1 with ligand treatment prior to infection.
Continuous MR1 trafficking effectively masks the HSV-1–mediated upregulation of surface MR1 associated with ligand pretreatment
Very little MR1 traffics to the PM in the absence of ligand, although this stochastic event is amplified by overexpression of MR1 (27). Formation of the Schiff base between the charged K43 lysine residue in the A′ pocket and bound ligand triggers the conformational change that initiates MR1 egress from the ER (7, 29). Controlling the availability of ligand to either before or after the time of infection is a useful means of highlighting the differential capacity of HSV to modulate points in MR1 trafficking. However this does not represent biologically relevant conditions, as prior to HSV infection of a healthy mucosal barrier site such as the skin or the anogenital area, ligand would be continuously available from commensal bacteria. Consequently, two different models were used to examine the impact of HSV-1 infection on surface MR1 under conditions of continuous ligand availability.
Firstly, ARPE-19 cells were transduced to overexpress the MR1-K43A mutant (2, 37), which automatically adopts the closed conformation of the ligand-bound WT molecule, and consequently traffics independent of ligand availability. Although this mutant cannot stably bind ligand due to its inability to form a Schiff bond (37), it may have some capacity to capture ligand, given the increased half maximum melting temperatures of MR1-K43A molecules folded in the presence, versus the absence, of ligand (28). With 4 h ligand treatment prior to harvest, surface MR1 WT in mock-infected cells increased in response to ligand, however surface expression of MR1-K43A was unchanged (Fig. 3A). The ARPE-19 MR1-K43A cells were then infected with HSV-1 for 18 h with the three different controlled ligand conditions (nil, pre, and post). HSV-1 strongly downregulated MR1-K43A surface expression regardless of ligand treatment (Fig. 3B), failing to replicate the protection or upregulation of surface MR1 WT seen with controlled preinfection ligand treatment (Fig. 1). As detected by immunoblot, HSV-1 also downregulated total MR1 WT and MR1-K43A under all three ligand conditions (Fig. 3C), with a weak band of high molecular weight remaining in the MR1 WT–expressing cells pretreated with ligand, consistent with previous findings (23).
Figure 3.
Surface MR1 is downregulated during HSV-1 infection under conditions of continuous trafficking.A, ARPE-19 MR1 (black) or ARPE-19 MR1-K43A (blue) cells transduced to express MR1 auto folding mutant were left untreated (nil) or treated with Ac-6-FP (5 μM) for 4 h prior to harvesting. Cells were stained for surface MR1 or isotype control (gray) and analyzed by flow cytometry. Histograms representative of two independent experiments. B, ARPE-19 MR1-K43A cells were mock (blue) or HSV-1 (strain F, MOI 5, red) infected in parallel. Cells were left untreated (nil) or treated with Ac-6-FP (5 μM) 24 h prior to infection (pre) or 14 h p.i. (post). Cells were harvested at 18 h p.i. when they were stained for surface MR1 or isotype control (gray) and analyzed by flow cytometry. Fold change in mean fluorescence intensity of infected cells relative to mock-infected cells (dotted blue line) with matching ligand treatment is graphed with SD. Statistical significance was calculated by paired Student’s t test, with p values < 0.05 annotated. Analysis of three independent experiments. C, ARPE-19 MR1 or MR1-K43A cells were mock or HSV-1 (strain F) infected in parallel. Cells were left untreated or treated with Ac-6-FP (5 μM) 24 h prior to infection (pre) or 14 h p.i. (post) before harvesting at 18 h p.i. Cell lysates were separated by gel electrophoresis and before immunoblotting for MR1, MHC-I, GAPDH, and GFP. D, ARPE-19 MR1 cells were mock or HSV-1 (strain F) infected in parallel. Cells were left treated with Ac-6-FP (5 μM) 24 h prior to infection (pre) and optionally treated again immediately post infection (cont) before harvesting at 18 h p.i., when they were stained for surface MR1 or isotype control (gray) and analyzed by flow cytometry. Fold change of infected cells (red) relative to mock-infected cells (dotted line) is graphed with SD. Statistical significance was calculated by paired Student’s t test with p values < 0.05 annotated. Analysis of three independent experiments. Ac-6-FP, acetyl-6-formylpterin; HSV-1, herpes simplex virus type 1; MOI, multiplicity of infection; MR1, major histocompatibility complex class I–related protein 1; p.i., hour post infection.
Secondly, ARPE-19 MR1 cells pretreated with ligand were HSV-1–infected, and then optionally retreated with ligand after the period of viral adsorption to provide continuous ligand availability. HSV-1 downregulated surface MR1 under these conditions (Fig. 3D), though not as strongly as seen with the MR1-K43A mutant (Fig. 3B). Nevertheless, neither of these models replicated the extent of loss of surface MR1 in HSV-1– or HSV-2–infected cells treated with ligand post infection (23, 26), instead demonstrating an intermediate phenotype. Together these results demonstrate that when there is a constant flow of MR1 to the PM, surface MR1 expression is effectively inhibited by HSV-1.
US3 viral protein predominantly modulates surface MR1
Our findings establish that MR1 accumulates at the surface and in early endosomes during HSV infection (Fig. 1). We previously demonstrated that single gene expression of HSV-1 viral kinase US3 results in the downregulation of surface MR1 (23), suggesting that HSV-1 employs US3 to at least partially counteract this surface-accumulated MR1. However, the ability of ectopically expressed US3 to target total MR1 has not been evaluated. Furthermore, infection of ARPE-19 MR1 cells with a US3-deficient HSV-1 mutant resulted in a partial recovery of surface, but not total MR1, suggesting US3 may preferentially target mature molecules in the trafficking pathway. The ability of the HSV-2 US3 homolog or the shorter, kinase-active US3.5 protein, to modulate total and surface MR1, has not been investigated.
The HSV family of US3 proteins play diverse roles during infection including modulation of cell morphology through cytoskeleton remodeling (31, 38, 39), increased neurovirulence (40), inhibition of apoptosis (41, 42), promotion of viral genome replication and transcription (43, 44, 45), suppression of antiviral responses (46, 47, 48, 49), egress of nucleocapsids through the nuclear lamina (50, 51) and regulating functionality of viral glycoproteins (52, 53). US3 downregulates surface expression of other antigen presentation molecules including MHC-I, in part, through inhibition of antiviral interferon signaling (54), and CD1d, through modulation of cellular protein KIF3A to impair CD1d recycling (55, 56). During HSV-1 infection US3.5 expression is less than that of the full-length protein (57). Furthermore, US3.5 lacks some of the functionality of the full-length protein, including inhibition of apoptosis and optimal facilitation of nuclear capsids across the nuclear membrane (57, 58).
To examine the ability of US3 and US3.5 to modulate both total and surface MR1, HSV-1 (strain F) and HSV-2 (strain 186) US3 and HSV-1 US3.5 from codon 77 were cloned with an N-terminal FLAG tag, into the replication incompetent pAdZ5-C5 adenovirus vector (59). ARPE-19 MR1 cells were infected with parental (RAd-Ctrl), HSV-1 US3 (RAd-H1-US3, MOI 100), US3.5 (RAd-H1-US3.5, MOI 25), or HSV-2 US3 (RAd-H2-US3, MOI 25) expressing adenovirus or mock-infected. Cell lysates were harvested at 44 h p.i. to confirm the size of the expressed protein through FLAG probe in a Western blot (Fig. 4A). Total MR1 was substantially reduced by US3 (HSV-1, HSV-2) and US3.5 (HSV-1) ectopic expression. MR1 depletion by HSV-2 US3 was slightly weaker, potentially due to lower expression of the viral kinase as detected by the FLAG probe, rather than functional differences between the homologs.
Figure 4.
HSV-1 and HSV-2 US3 protein expression modulation of surface MR1 precedes loss of total MR1.A, ARPE-19 MR1 cells were infected with RAd-Ctrl, RAd-H1-US3, RAd-H1-US3.5, RAd-H2-US3, or mock-infected. Cells were harvested at 44 h p.i. and lysates were separated by gel electrophoresis before immunoblotting for FLAG (to detect US3 expression), MR1, MHC-I, and GAPDH. B, ARPE-19 MR1-GFP cells were infected with RAd-Ctrl adenovirus or RAd-Ctrl modified to encode HSV-1 US3 (RAd-H1-US3), US3.5 gene (RAd-H1-US3.5), or HSV-2 US3 gene (RAd-H2-US3). Cells were treated with Ac-6-FP (5 μM) for 4 h prior to harvesting at 30 or 44 h p.i. Cells were stained for surface MR1 and analyzed by flow cytometry. C, ARPE-19 MR1-GFP cells were infected with RAd-Ctrl adenovirus (black), RAd-H1-US3 (red), RAd-H1-US3.5 (orange), or RAd-H2-US3 (blue). Cells were left untreated (nil) or treated with Ac-6-FP (5 μM) for 4 h (post) prior to harvesting at 44 h p.i. Cells were stained for surface MR1, MHC-I, or matching isotype control (gray) and analyzed by flow cytometry. Fold change in MFI of live-infected cells relative to RAd-Ctrl with matching ligand treatment is graphed with SD. Statistical significance was calculated by paired Student’s t test, with p values < 0.05 annotated. Analysis of four independent experiments. D, 293T cells were transfected with pSY10 (parental plasmid), pSY10-US3 (HSV-1 US3), or pSY10-US3kd (kinase dead US3, K220A). After adding ligand (Ac-6-FP) at 24 h, cells were harvested at 28 h and stained for surface MR1 and MHC-I and analyzed by flow cytometry. Fold change in MFI of GFP+ cells relative to mean pSY10 MFI is graphed with SD. Statistical significance was calculated by one-way ANOVA with multiple comparisons, with p values < 0.05 annotated. Analysis of four independent transfections for each plasmid. Ac-6-FP, acetyl-6-formylpterin; h p.i., hour post infection; HSV-1, herpes simplex virus type 1; HSV-2, herpes simplex virus type 2; MFI, mean fluorescence intensity; MHC-I, MHC class I; MR1, major histocompatibility complex class I–related protein 1.
ARPE-19 MR1-GFP cells were similarly infected with the US3- and US3.5-expressing constructs for quantitative flow cytometry analysis of total cellular MR1 (GFP signal) and surface MR1 and MHC-I (antibody). Cells were optionally treated with Ac-6-FP ligand 4 h (nil, post) prior to harvesting at either 30 or 44 h p.i. By 30 h p.i. the percentage of ligand-treated cells expressing high total and surface MR1 was reduced from 95.6% (RAd-Ctrl) to as low as 62.7% with RAd-H1-US3.5 infection (Fig. 4B). Correspondingly, cells expressing low surface MR1 increased from a low of 0.58% in the control adenovirus-infected cells to as high as 26.4% in the RAd-H1-US3.5–infected cells. This trend continued at 44 h p.i., at which point 63.9% of the RAd-H1-US3–infected cells were gated in the high total, low surface MR1 quadrant. Together, these results suggest that US3 and US3.5 predominantly downregulate surface MR1 and that this occurs prior to the loss of total MR1.
The downregulation of surface and total MR1-GFP was quantified at the later time point (44 h p.i.), in both the ligand-free and postinfection-treated cells. There was a small but significant, MOI-dependent, increase in total but not surface MR1 by the RAd-Ctrl (Fig. S4). Consequently, all statistical analysis of the modulation of MR1-GFP, surface MR1, and MHC-I, by the viral gene products, was calculated relative to the parental virus with matching MOI, rather than mock-infected cells. Total MR1-GFP was downregulated independent of ligand treatment with HSV-1 US3 and US3.5 expression. The reduction of MR1 associated with HSV-2 US3 expression was somewhat variable, but less than that seen with the other viral kinases (Fig. 4C), consistent with the immunoblot result (Fig. 4A). Surface MR1, however, was strongly downregulated by each of the three viral proteins to levels approaching isotype. Furthermore, surface, and to a lesser extent, total MHC-I molecules were also reduced (Fig. 4, A and C), although this phenotype is not always detected with ectopic US3 expression (60).
Kinase functionality of the US3 protein can be disabled by replacing one lysine residue with alanine (K220A) (46, 61). To determine whether the kinase domain encoded by both US3 and US3.5 is required for the modulation of MR1, a plasmid-expressing HSV-1 US3 and GFP separated by a T2A sequence that facilitates translation of two separate proteins from the one transcript (23), was modified to express a kinase dead US3 (US3kd). 293T cells transfected with either the parental plasmid (pSY10), US3 (pSY10-US3), or kinase dead US3 (pSY10-US3kd), were treated with ligand after 24 h. The surface expression of MR1 and MHC-I in the GFP+ cells was analyzed by flow cytometry 4 h later (Fig. 4D). Inactivation of the kinase domain partially rescued the loss of both surface MR1 and MHC-I, indicating that while kinase activity contributes to the modulation of MR1, it is not absolutely required for the modulation of these antigen presentation molecules.
Together these findings demonstrate that US3 protein encoded by both HSV viruses, as well as US3.5 of HSV-1, modulate surface MR1 prior to the loss of total MR1, and that the kinase activity contributes to this phenotype. This contrasts to the modulation of MR1 during HSV infection, which is led by the loss of total protein. These results establish that US3 primarily acts against mature MR1 in a way that reduces surface antigen presentation, although it can also drive a reduction in total protein.
Increased retention of surface MR1 during HSV-1 infection
While the C-terminal cytoplasmic domain is not required for MR1 trafficking to the PM, it does play a role controlling the rate of MR1 endocytosis from the PM into EEA1-labeled, early endosomes (10). MR1 contains a noncanonical tyrosine motif encoded in the cytoplasmic tail that mediates a suboptimal interaction with the endocytic AP2 complex (7, 10). This causes MR1 to remain on the PM longer than other typical AP2 cargo. Although variation between cell types has been observed in the rate of this clathrin-mediated process, ∼50% of the molecule can be expected to internalize within several hours (7, 10). This is in stark contrast to the transferrin receptor CD71 molecule, which encodes a canonical AP2 binding motif and internalizes within minutes (62, 63). Furthermore, only a small percentage of endocytosed MR1 molecules recycle back to the PM, although again the proportion varies according to cell type (10).
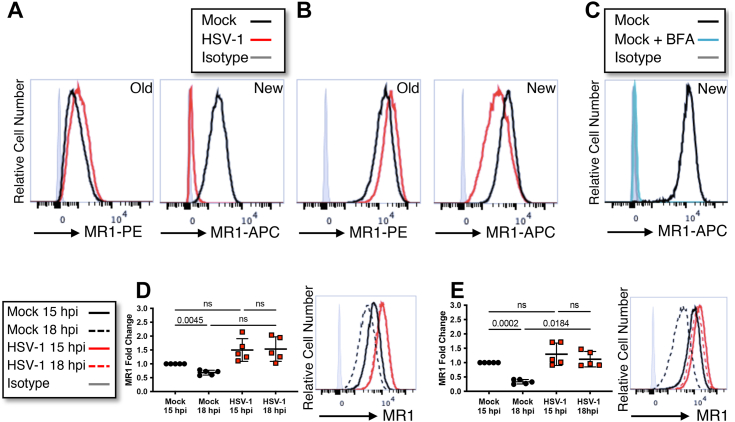
Through manipulating availability of ligand, increased MR1 can be detected on the PM during HSV infection. It is unclear, however, whether this is due to an accumulation of newly trafficked molecules, increased recycling from endosomal compartments or retention of preexisting surface molecules. Several assays were designed to identify the main driver of this phenotype. To examine the relative contribution of newly trafficked and preexisting molecules, ARPE-19 MR1 cells were optionally pretreated with ligand. Surface MR1 was labeled with anti-MR1-phycoerythrin (PE) for the final 30 min of the period of viral adsorption, to label preexisting (old) surface MR1. At 16 h p.i. surface MR1 was stained again with the same MR1 antibody clone conjugated to a different fluorophore (allophycocyanin [APC]), to detect molecules that arrived post infection (new) (Fig. 5, A and B). To validate that the assay only detected new surface MR1 and not preexisting stained MR1 from which the antibody had dissociated, cells were treated with brefeldin A to inhibit new MR1 trafficking from the ER, and no staining was detected at 16 h (Fig. 5C). Both in the absence of ligand (Fig. 5A), where only a small proportion of MR1 progresses to the cell surface, and with ligand pretreatment, which drives MR1 to the cell surface (Fig. 5B), there was a reduction in new MR1 molecules. This is most likely due to depletion of the MR1 pool available to traffic to the PM. The downregulation was particularly striking in the absence of ligand, where new surface molecules approached isotype levels in the HSV-1–infected cells. In this assay, the old molecules include those that remain on the PM, are located in intracellular endocytic vesicles, and those that have recycled back to the cell surface. Both in the absence of ligand, and with treatment prior to infection, there was an increase in the amount of Old MR1 in the infected cells (Fig. 5, A and B). This is consistent with the MR1 accumulation on the PM, and in EEA1 labelled compartments, detected by fluorescence microscopy (Fig. 1).
Figure 5.
Endocytosis of MR1 is reduced during HSV-1 infection.A and B, ARPE-19 MR1 cells were mock or HSV-1 (strain F) infected in parallel. Cells were treated with Ac-6-FP (5 μM) 24 h prior to infection (B) or left untreated (A). Existing surface MR1 (old) was captured by staining for surface MR1 (anti-MR1-PE or isotype control in gray) during the final 30 min of the 1-h period of viral adsorption. Newly trafficked surface MR1 (new) was detected at 16 h p.i., when cells were again stained for surface MR1 (anti-MR1-APC or isotype control in gray). C, ARPE-19 MR1 cells were stained for existing surface MR1 (anti-MR1-PE), treated with Ac-6-FP (5 μM), and optionally treated with brefeldin A. After 16 h, cells were harvested and stained for surface MR1 (new, anti-MR1-APC, or isotype control in gray). Histogram representative of two independent experiments. D, ARPE-19 MR1 cells were mock or HSV-1 (strain F) infected in parallel. Cells were treated with Ac-6-FP (5 μM) 6 h prior to infection. At 15 h p.i., cells were stained with anti-MR1-APC and then harvested immediately or after a further 3 h. E, ARPE-19 MR1 cells were mock or HSV-1 (strain F) infected in parallel. Cells were treated with Ac-6-FP (5 μM) 6 h prior to infection. At 15 h p.i., cells were stained with anti-MR1-biotin and then harvested immediately or after a further 3 h. Cells were stained with streptavidin-APC at time of harvest. D and E, Cells were analyzed by flow cytometry with analysis based on mean fluorescence intensity fold change to mock infected at 15 h p.i. Statistical significance was calculated by one-way ANOVA with multiple comparisons, with p values < 0.05 annotated. Analysis of five independent experiments. Ac-6-FP, acetyl-6-formylpterin; APC, allophycocyanin; h p.i., hour post infection; HSV-1, herpes simplex virus type 1; MR1, major histocompatibility complex class I–related protein 1.
To further explore the retention of MR1 molecules after their arrival at the PM, but over a shorter timeframe, ARPE-19 MR1 cells were again pretreated with ligand and then infected with HSV-1. Cells were stained with anti-MR1-APC at 15 h p.i. on ice and optionally incubated a further 3 h at 37 °C. There was a significant loss of the signal between the two time points in the mock infected cells, which was not evident in the HSV-1–infected cells (Fig. 5D). Furthermore, there was relatively more MR1-APC detected at both 15 h p.i. and 18 h p.i. in the infected cells, compared to mock controls, although this trend did not reach statistical significance.
To examine whether this result was due to increased retention on the PM, or intracellular accumulations, the internalization rate of infected cells was compared to the mock-infected controls. Cells were pretreated with ligand and infected with HSV-1, with the surface MR1 labeled on ice with biotin-conjugated MR1 antibodies at 15 h p.i. Cells were stained with streptavidin-APC immediately, or after a further 3 h incubation, to detect only those molecules remaining on the PM. There was a significant loss of surface MR1 over the 3 h in the mock-infected cells, consistent with the expected rate of endocytosis (7, 10), which was not observed in the infected cells (Fig. 5E). The difference in the relative amount of surface MR1 between mock and infected cells reached statistical significance at 18 h p.i. Given the low rates of MR1 recycling, the remaining signal likely reflects surface retention, although it is not possible to exclude infection-induced upregulation of recycling as contributing to this result. Together, this data suggests that during HSV-1 infection MR1 endocytosis is impaired, resulting in extended periods of antigen presentation, and increasing the likelihood of detection by MAIT cells.
Discussion
Our findings define the fate of residual MR1 molecules, during both HSV-1 and HSV-2 infection (Fig. 6), after the significant depletion of ligand-free, ER-resident, immature molecules (Fig. 1). We observe the downstream reduction in molecules trafficking through the secretory pathway but establish that this is not necessarily reflected on the PM, due to impaired MR1 endocytosis (Fig. 5). Furthermore, HSV infection drives MR1 accumulation in early endosomes, but not late endosomes/lysosomes. We confirm that the MR1 cytoplasmic tail is not required for depletion of immature molecules (Fig. 2), but its role in the surface retention and accumulation in early endosomes remains unclear. Viral proteins US3 and US3.5 primarily modulate surface, rather than total MR1 (Fig. 4), in part through their kinase activity. However this, together with the ICP22- and vhs-mediated loss of immature MR1 (26), are sufficient to downregulate surface MR1, under the homeostatic conditions of continuous ligand availability present at healthy intact mucosal sites (Fig. 3). We propose that the targeting of intracellular and cell surface MR1 molecules by HSV is an immune-evasive countermeasure to minimize the increased retention of surface MR1, which may be a by-product of viral infection. Whether the modulation of MR1 has any impact on HSV infection and/or the anti-HSV immune response awaits the development of an animal model to assess HSV pathogenesis in this context.
Figure 6.
The fate of MR1 molecules during HSV infection. HSV infection depletes the pool of immature MR1 molecules in the endoplasmic reticulum, limiting trafficking through the Golgi apparatus in response to ligand availability. Infection impairs endocytosis of ligand-bound mature molecules on the plasma membrane, although this is offset by HSV targeting of intracellular and surface MR1 molecules for degradation. After endocytosis MR1 preferentially accumulates in early endosomal compartments, but not late endosomes/lysosomes. HSV, herpes simplex virus; MR1, major histocompatibility complex class I–related protein 1.
The reduced rate of MR1 endocytosis exhibited by HSV-1–infected ARPE-19 MR1 cells (Fig. 5), that is effectively masked under conditions of continuous ligand availability (Fig. 3) consistent with homeostatic bacterial synthesis of riboflavin metabolites, is the first data that helps to directly explain the immuno-evasive advantage of the early loss of immature MR1 molecules (23). HSV infection could be associated with increased availability of ligand, with enhanced MAIT cell cytolytic capacity and proinflammatory effector functions. MAIT cells represent up to 7% of the resident T cell population in orofacial and anogenital mucosa (64, 65, 66), where HSV-1 and HSV-2 primary infections predominantly occur (67, 68). Entry to permissive keratinocytes, fibroblasts, and epithelial cells is enhanced by microabrasions, and pathology-induced barrier damage, and inflammation (69, 70), all of which could be associated with bacterial translocation and altered MR1 ligand availability. Eczema herpeticum, a widespread HSV skin infection present in up to 24% of people with atopic dermatitis (71), is associated with increased risk of bacterial skin infection, particularly with the riboflavin synthesising commensal Staphylococcus aureus (3, 72, 73). Furthermore, HSV continuously reactivates from its reservoir site of latency in peripheral sensory neurons resulting in recurrent herpetic lesions (74, 75). In the female genital tract, this can be associated with chronic alterations to the composition of the microbiome, including reductions in Lactobacilli, and increased bacterial infections (76). Interestingly, Lactobacilli-derived factors help dampen MAIT cell responses to S. aureus. HSV-induced dysbiosis may be removing critical regulation that prevents MAIT cell over-responsiveness to essential riboflavin synthesis (77). This could render HSV-infected cells more susceptible to detection and cytolytic killing by resident MAIT cells, which would be effectively offset by viral downregulation of MR1 antigen presentation.
Cellular responses to viral infection also prime the MR1–MAIT cell axis (78). TLR agonists and NF-κB signaling, which are features of HSV infections, are associated with increased MR1 antigen presentation in some cell types (79, 80, 81, 82). At the same time, antiviral type I interferons heighten the cytolytic capacity of MAIT cells through increased production of granzyme B, potentially amplifying the response of MAIT cells to MR1 antigen presentation during viral infections (83). Consequently, there could be multiple factors in addition to delayed MR1 antigen internalisation during HSV infection that, in the absence of viral-mediated rapid depletion of MR1 intracellular reserves, would otherwise expose infected cells to TCR-mediated MAIT cell detection.
Further research is required to characterize the endocytosis of surface MR1 to examine endogenous MR1 expression in primary human cells, as well as to confirm whether HSV-2 exhibits the same phenotype. Surface levels of other antigen presentation molecules, including CD1d and MHC-I are reduced during HSV infection (84, 85, 86, 87), while other surface molecules, including the CD71 transferrin receptor (55, 88, 89), remain steady. Given that there is no evidence of widespread surface retention of cellular glycoproteins during infection, a global effect is unlikely, although an AP2-specific impact is plausible. HSV glycoproteins are synthesized in the ER, glycosylated during transit through the secretory pathway, and then group together on the PM where they form lipid rafts. AP2 consensus sequences encoded in the cytoplasmic tails of some of the glycoproteins supports endocytosis of the clusters, after which they are transported to virus assembly sites such as the trans Golgi network and endocytic tubules (90, 91, 92). Although there is some evidence suggesting increased expression of AP2 subunits during HSV-1 infection (93), this may be insufficient to prevent competitive inhibition, allowing the highly expressed HSV proteins to monopolise the AP2 endocytic machinery. Interestingly, there are some other examples of upregulation of AP2-endocytosed surface molecules during HSV-1 infection, including CD63 (LAMP3) and LAMP1, both of which cycle between the PM and late endosomal or lysosomal compartments (55, 87, 94). However, CD1d, which presents glycolipid antigens to invariant natural killer T cells (95, 96), is inhibited during HSV-1 infection through US3 and glycoprotein B–mediated blocking of endocytic recycling (85, 87). This variability could be due to differential binding affinities or other cellular factors involved in the regulation of endocytosis (97, 98, 99). Furthermore, given that HSV commonly modulates several steps in antigen presentation pathways, downregulation does not exclude the possibility this masks an underlying retention of CD1d on the PM.
A second explanation is that direct interactions between viral proteins and MR1 impact longevity on the PM and perhaps also drive the observed accumulation in early endosomes. Interestingly, HSV glycoprotein B binds to CD1d molecules not only on the PM but also between immature ER-resident molecules, however US3 phosphorylation of type II kinesin motor protein KIF3A is responsible for the reduction in endocytic recycling that decreases surface CD1d expression (55, 85). Data examining the direct interactions between MR1 and viral proteins may prove useful in resolving this question.
The short MR1 cytoplasmic tail is not required for the downregulation of immature molecules during HSV infection (Fig. 2). The noncanonical AP2 motif in the MR1 tail is required for optimal antigen presentation (10), and while mutations in the sequence impact the rate of internalization, deletion of the short cytoplasmic domain surprisingly does not alter endocytosis. The role of the MR1 tail and the AP2 binding sequence in the preferential retention of mature molecules is currently unclear, as we show that WT and MR1 molecules lacking expression of the C-terminal domain are both upregulated on the PM during HSV infection.
We have previously demonstrated that single gene expression of HSV-1 US3 and the VZV homolog ORF66 both downregulate surface MR1. Infection with viruses lacking their expression, followed by ligand treatment, only partially rescues this loss with HSV-1 but not VZV infection (23, 25). This reflects the contribution by other viral proteins to the inhibition of MR1 antigen presentation. Here, we demonstrate that both the shorter US3.5 kinase and the full-length HSV-2 homolog also impact surface MR1 expression, and that modulation of surface, rather than total expression, is the dominant phenotype (Fig. 4). Furthermore, we establish that the kinase domain contributes to this modulation but may not be the only mechanism by which US3 impacts presentation of antigen by both MR1 and MHC-I. Given that MR1 has no known phosphorylation sites and does not include the requisite serine/threonine residues in the cytoplasmic tail, an indirect mechanism of action is likely. The US3 family of kinases share substrate similarities to cellular PKA and protein kinase B (Akt) (43, 100). Although a few putative US3 viral and cellular targets have been identified, only a small proportion has been confirmed, leaving open additional possible mechanisms of modulation.
Interestingly, the phosphorylation of motor protein KIF3A with transient US3 expression in HeLa cells also results in reduced surface expression of CD1d, CD63, and LAMP1 (87). It is possible that phosphorylation of KIF3A is also responsible for the downregulation of surface MR1, however several points do not support this theory. Firstly, unlike CD1d, CD63, and LAMP1, MR1 does not predominantly traffic between late endosomes and the PM; moreover the β2m–MR1 complex is unstable at the low pH found in late endosomes/lysosomes (10). Secondly, this kinesin is primarily associated with late and not early endosomes (101); however, we detected increased accumulation in early endosome, and relatively stronger loss in LAMP1-associated compartments, with no recovery of MR1 in cells treated with two different lysosomal inhibitors (Fig. S2). Indeed CD1d accumulates in multivesicular LAMP1 vesicles during HSV-1 infection, where its relocation from internal vesicles to the lysosome limiting membrane may be responsible for the inhibition of recycling (87). Additional examination of ectopic US3 expression is required to confirm whether it is involved in the EEA1 accumulation of MR1 and evaluate the effect of KIF3A phosphorylation on MR1 localization. Finally, given that regulation of US3 location and enzymatic activity is controlled during infection by autophosphorylation and the expression of both cellular and viral factors, including viral UL13, UL47, and UL34 proteins (57, 102, 103, 104), it will be important to examine these findings in US3 mutant viruses.
In conclusion, we have identified two additional points in the life-cycle of MR1 antigen presentation that are modulated by both HSV-1 and HSV-2. We observe striking similarities in the phenotype exhibited by these two closely related alpha herpes viruses, which suggests a strong biological imperative, given these viral species have independently evolved from different cross-species transmissions over millions of years (105, 106, 107). We propose that HSV encodes multiple mechanisms to downregulate surface MR1, at least in part to offset an increased retention of mature molecules on the PM, thus avoiding cytolytic killing by the most populous clonal population of T cells at sites of primary infection and reactivation.
Experimental procedures
Cells
Human retinal pigment epithelial (ARPE-19) cell lines expressing MR1 lacking the final 15 amino acids of C-terminal domain (ARPE-19 ΔP305) (10) or the MR1-K43A auto folding mutant (ARPE-19 MR1-K43A) were generated as described previously by retroviral transduction (7). The MR1 constructs were encoded in the parental pMIG-II vector, a gift from Dario Vignali (University of Pittsburgh; plasmid #52107), under the control of the Murine stem cell virus LTR, with EGFP coexpressed from the same promoter via a downstream internal ribosomal entry site. ARPE-19 MR1, ΔP305, and MR1-K43A and ARPE-19 MR1-GFP cell lines, 293T (American Type Culture Collection, CRL-3216), Vero (American Type Culture Collection, CCL-81), and T-REx-293 cells were grown at 37 °C and 5% CO2 in Dulbecco’s Modified Eagle’s Medium (Lonza, #12-604F) supplemented with 10% fetal calf serum (Cytiva, #SH30084.02). All cells are periodically tested for mycoplasma and are free from contamination.
Viruses
HSV-1 and HSV-2 strains used in this study were HSV-1 strain F (courtesy Dr Russell Diefenbach, Macquarie University) and HSV-2 strain 186 (courtesy Dr Naomi Truong, The Westmead Institute for Medical Research) (108). After first replacing the media, cells were infected for the 1-h period of adsorption (with gentle rocking at 37 °C) and then washed again before continuing with the assay. All HSV strains were grown and titrated on Vero cells.
Replication-deficient adenovirus constructs were generated from the pAdZ5-C5 vector (59). Adenoviruses used in this study were: RAd-Ctrl (no exogenous protein-coding region) (59), RAd-H1-US3 and RAd-H1-US3.5 expressing the corresponding gene encoded by HSV-1 (strain F), or RAd-H2-US3 from HSV-2. Two-step PCR using Q5 High-Fidelity polymerase (NEB) was used to amplify the viral genes adding an N-terminal FLAG tag, with the second round of amplification adding approximately 80 bp of homology to allow recombination with the adenovirus construct. All primers are listed in Table 1, with the gene-specific sequence underlined. Purified PCR products were incorporated into the vector by homologous recombination as previously described (26, 59) with the sequence confirmed by Sanger sequencing.
Table 1.
Primers used to amplify HSV genes for recombination into pAdZ5-C5 vector
| Target | Forward | Reverse |
|---|---|---|
| HSV-1F US3 | 5′-GACCGATCCAGCCTGGATCCATGGACTACAAAGACGATGACGACAAGGGCGCCGCCTGTCGTAAGTTTTGTCG-3′a | 5′-ACAATAGTGACGTGGGATCCTCATTTCTGTTGAAACAGCGGCAAAC-3′ |
| HSV-1F US3.5 | 5′-GACCGATCCAGCCTGGATCCATGGACTACAAAGACGATGACGACAAGGGCGCCTACGGAAACCAGGACTACCC-3′ | 5′-ACAATAGTGACGTGGGATCCTCATTTCTGTTGAAACAGCGGCAAA-3′ |
| HSV-2186 US3 | 5′-GACCGATCCAGCCTGGATCCATGGACTACAAAGACGATGACGACAAGGGCGCCGCCTGTCGTAAGTTCTGTGG-3′ | 5′-ACAATAGTGACGTGGGATCCTCACTTAGGGTGAAATAGCGGCAGGC-3′ |
| pAdZ5 Round 2 | 5′-GAGAGTTTAGTGAACCGTCAGATCGCCTGGAGACGCCATCCACGCTGTTTTGACCTCCATAGAAGACACCGGGACCGATCCAGCCTGGATC-3′ | 5′-GGCGTGACACGTTTATTGAGTAGGATTACAGAGTATAACATAGAGTATAATATAGAGTATACAATAGTGACGTGGGATCC-3′ |
HSV, herpes simplex virus.
Gene-specific sequence is bold.
Confirmed constructs were purified using the NucleoBond Xtra Midi kit (Macherey-Nagel, #740410) and transfected into T-REx-293 using FuGene HD (Promega, #E2231). After several days the infected cells were collected, and the pellet lysed by resuspending in equal volumes of PBS and tetrachloroethylene. After centrifugation, the upper layer of PBS containing the adenovirus was removed and stored at −80 °C. Virus was titrated after 48 h p.i. in T-REx-293 cells, using goat anti-adenovirus primary antibody (Sigma-Aldrich, #AB1056), anti-goat HRP secondary antibodies, and 3,3′-diaminobenzidine substrate (Pierce, #34065) for quantification of infected cells.
Plasmids and transfections
Two primer sets (Forward: 5′-GTAATCGTGGCAGCGGGGTG-3′ and Reverse: 5′-TCCTTGAGAGTTTTCGCCCC-3′; Forward 5′-GGGGCGAAAACTCTCAAGGA-3′ and Reverse: 5′-CACCCCGCTGCCACGATTAC-3′) and were used to amplify two halves of the pSY10-US3 plasmid (23), replacing lysine (K220) with alanine (bold in primers) to create the pSY10-US3kd kinase dead plasmid. The two fragments were recombined using NEB HiFi builder (E2621L) and competent Escherichia coli (NEB, #C2987H) transformed with the plasmid were selected on Luria-Bertani agar with ampicillin (50 mg/ml), grown in LB broth with ampicillin overnight and purified (Bioline #740490). The full US3 sequence was confirmed by Sanger sequencing.
293T cells were transfected with the pSY10 parental plasmid (pCDH_EF1-MCS-T2A-copGFP vector, Systems Bioscience, #CD526A-1), pSY10-US3, or pSY10-US3kd using Fugene HD (Promega).
Immunoblotting
Cells were harvested in cell lysis buffer (150 mM NaCl, 50 mM Tris pH8, 1% IGEPAL, 1% Triton X-100) supplemented with protease inhibitor cocktail (1% V/V, Sigma) and allowed to incubate on ice for 20 min. Lysates were then centrifuged (16,000g for 20 min at 4 °C) and the supernatant collected. Lysates were denatured by heating at 95 °C for 5 min in reducing sample buffer (Bio-Rad) and resolved by SDS-PAGE on precast polyacrylamide gels (Bio-Rad) before immunoblotting onto polyvinylidene difluoride membranes. Membranes were probed with the designated primary antibodies in 3% bovine serum albumin in phosphate buffered solution with 0.5% (V/V) Tween20 (Sigma-Aldrich P9416 (PBS-T)), followed by incubation with an appropriate horseradish peroxidase–conjugated secondary antibody (Santa Cruz Biotechnology) and visualized using Clarity Western ECL Substrate (Bio-Rad, #1705061). The following primary antibodies were utilized: anti-MR1 CT or clone 8G3 (27), anti-MR1 (Abcam #ab55164) and anti-HLA-A, B, C (Abcam, clone EMR8-5, #ab70328), anti-DDDDK (Abcam, clone EPR20018-251, #Ab205606), and anti-GAPDH (Santa Cruz Biotechnology, clone FL-335, #sc-257778 or Thermo Fisher Scientific #TAB1001). Immunoblots in Figure 4A depict the same mock and RAd-Ctrl lanes from a previous manuscript as the experiments were conducted concurrently (26).
MR1 ligand and cellular inhibitors
The MR1 ligand Ac-6-FP (5 μM, Schircks Laboratories, #11.418) was added as indicated to the culture medium. To inhibit lysosomal degradation of proteins, cells were treated with folimycin (concanamycin A, 50 nM Sigma-Aldrich #C9705) or leupeptin (100 μM, Pierce, #78435), following the 1-h period of viral adsorption. To inhibit trafficking of MR1 to the PM, cells were treated with brefeldin A for 16 h (5 μg/ml, BioLegend #420601).
Flow cytometry
The following antibodies were used to detect surface molecules: MR1 was detected by anti-MR1-biotin (clone 26.5) (7), followed by streptavidin-PE (eBioscience, #12-4317-87) or streptavidin-APC (eBioscience, #17-4317-82), or using anti-MR1 directly conjugated to PE or APC (BioLegend, clone 26.5, #361105, #361107); MHC-I by anti-HLA-A,B,C-PE (Miltenyi Biotec, clone REA230, #130-120-055), anti-HLA-A,B,C-APC (BD Pharmingen, clone G46-26, #555555), or anti-HLA,B,C-SB436 (Invitrogen, clone W6/32, #62998342). All cells were stained at 4 °C to minimize internalization, unless otherwise stated. Live cells were identified using Zombie NIR Fixable Viability Kit (Biolegend) or Live/Dead Fixable blue (Invitrogen, #L23105). After staining, cells were treated with BD Cytofix Fixation buffer (BD Biosciences, #554655). Flow cytometry was performed using a LSR Fortessa X-20 or BD-LSR-II (BD Biosciences) and fluorescence intensity (MFI) analyzed using FlowJo software (Treestar Inc., https://www.flowjo.com, version 10.8).
Fluorescence imaging
ARPE-19 MR1 cells were seeded in duplicate on 96-well PhenoPlates (PerkinElmer, 6055302) precoated with Geltrex basement membrane matrix (Gibco, #A1413301) and then infected as described above with either HSV-1 or HSV-2. Cells were stained at 4 °C for delineation of PM (WGA, CF405M Conjugate Biotium #29028) in Hanks Balanced Salt Solution (Gibco, #14170) with 2% normal donkey serum (NDS, Sigma-Aldrich, #D9663). They were then fixed (BD Cytofix Fixation buffer, BD Biosciences, #554655), permeabilized, and blocked with the staining buffer (PBS, 1 mM CaCl2/MgCl2, 2% NDS) containing either 0.01% saponin (Sigma-Aldrich, #S7900) or for EEA1 staining 0.01% Triton X-100 (Sigma-Aldrich). The following primary rabbit anti-human antibodies were used to label intracellular compartments: calreticulin for the ER (Sigma-Aldrich, #C4606), GM130 for the GA (Cell Signalling Technology, clone D6B1, #12480), EEA1 for early endosomes (Thermo Fisher Scientific, clone F.43.1, #MA514794) or LAMP1 for late endosomes/lysosomes (Abcam, #ab24170). This was followed by donkey anti-rabbit IgG conjugated to Alexa Fluor 546 (Thermo Fisher Scientific, #A10040) and 4′,6-diamidino-2-phenylindole nuclear staining (Sigma-Aldrich, #MBD0015). Cells were also stained for MR1 (BioLegend, clone 26.5, #361102), followed by donkey anti-mouse IgG conjugated to Alexa Fluor 647 (Thermo Fisher Scientific, #A31571). Cells were washed several times between each staining step and then covered in degassed imaging buffer (5% (v/v) glycerol, 2.5% (w/v) DABCO Powder in PBS, pH 8.5). Two biological repeats were completed for each set of assay conditions.
Cells were imaged using the Opera Phenix Plus (PerkinElmer) with the 40X water objective under the control of the Harmony software. A minimum of 4 FOVs were imaged per well, with five planes, each 0.5 μm apart, acquired in each channel. Maximum Z-stack projections were created for each FOV, and flat field correction was applied through ImageJ software. A segmentation map of the cells within each FOV defining the subcellular compartments was created with ilastik software (33). Given that the nucleus and PM are morphologically distinct, they were acquired in the same fluorescence channel and machine learning was used to identify their separate locations in the segmentation map. After normalizing the median MR1 FOV signal to the control samples within the corresponding experimental condition, the median MR1 signal at each subcellular location was calculated with ImageJ by overlaying the segmentation map. The MR1 signal fold change in the FOV at each subcellular compartment in infected cells compared to corresponding mock-infected samples was calculated. In addition, the median MR1 signal within each FOV for the same permeabilization agent was used to calculate the relative strength of MR1 signal in the subcellular compartment compared to the median cell signal. Statistical significance was evaluated with a minimum of 14 FOV.
Quantification and statistical analysis
Paired Student’s t tests or one-way with multiple comparisons analysis were performed as indicated using GraphPad Prism software (GraphPad Software, LLC., https://www.graphpad.com/scientific-software/prism/version 9. Data are presented as dot plots (flow cytometry and reverse transcriptase quantitative polymerase chain reaction with the mean and SD or violin plots (fluorescence microscopy) with the median and quartiles.
Data availability
Further information and requests for resources, data, and reagents should be directed to and will be fulfilled by the Lead Contact, Barry Slobedman (barry.slobedman@sydney.edu.au). Cells, viral mutants, and constructs are available upon request, subject to our institutional and Material Transfer Agreements, and those with the host institution at which these reagents were generated.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors wish to thank members of the Herpes virus Pathogenesis and Viral Immunology research groups (School of Medical Sciences, The University of Sydney) for helpful discussions and acknowledge the Sydney Cytometry Core Research Facility, a joint initiative of Centenary Institute and the University of Sydney, for assistance with flow and imaging cytometry experiments.
Author contributions
C. S. conceptualization; C. S., H. E. G. M., and J. G. B. formal analysis; C. S. investigation; C. S., H. E. G. M., B. P. M., J. G. S., and B. S. methodology; C. S. writing–original draft; C. S., H. E. G. M., J. G. B., R. J. S., J. R., and J. A. V. resources; C. S., H. E. G. M., B. P. M., J. G. B., R. J. S., J. R., J. A. V., A. A., and B. S. writing–review and editing; A. A. and B. S. funding acquisition; A. A. and B. S. supervision.
Funding and additional information
C. S. was supported by an Australian Postgraduate Award/Australian Government Research Training Program Scholarship. We acknowledge grant support from the National Health and Medical Research Council (NHMRC) of Australia, 198704 (A. A. and B. S.), 2003192 (H. E. G. M.), 2013621 (J. G. B.), and 1113293 and 1154502 (J. A. V.), the US National Institutes of Health RO1 R01AI148407 (J. R. and J. A. V.), NHMRC Leadership Investigator grants 2008913 (J. R.) and 2016969 (J. A. V.), Australian Research Council (ARC) DP170102471 (J. A. V.), Wellcome Trust 226615/Z/22/Z (R. J. S.), and UK Medical Research Council (MRC) MR/S00971X/1 (R. J. S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Craig Cameron
Supporting information
References
- 1.Keller A.N., Eckle S.B.G., Xu W., Liu L., Hughes V.A., Mak J.Y.W., et al. Drugs and drug-like molecules can modulate the function of mucosal-associated invariant T cells. Nat. Immunol. 2017;18:402–411. doi: 10.1038/ni.3679. [DOI] [PubMed] [Google Scholar]
- 2.Kjer-Nielsen L., Patel O., Corbett A.J., Le Nours J., Meehan B., Liu L.G., et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–723. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- 3.Gold M.C., Cerri S., Smyk-Pearson S., Cansler M.E., Vogt T.M., Delepine J., et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gherardin N.A., Keller A.N., Woolley R.E., Le Nours J., Ritchie D.S., Neeson P.J., et al. Diversity of T Cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity. 2016;44:32–45. doi: 10.1016/j.immuni.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Tsukamoto K., Deakin J.E., Graves J.A.M., Hashimoto K. Exceptionally high conservation of the MHC class I-related gene, MR1, among mammals. Immunogenetics. 2013;65:115–124. doi: 10.1007/s00251-012-0666-5. [DOI] [PubMed] [Google Scholar]
- 6.Corbett A.J., Eckle S.B.G., Birkinshaw R.W., Liu L.G., Patel O., Mahony J., et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361–365. doi: 10.1038/nature13160. [DOI] [PubMed] [Google Scholar]
- 7.McWilliam H.E.G., Eckle S.B.G., Theodossis A., Liu L.G., Chen Z.J., Wubben J.M., et al. The intracellular pathway for the presentation of vitamin B-related antigens by the antigen-presenting molecule MR1. Nat. Immunol. 2016;17:531–537. doi: 10.1038/ni.3416. [DOI] [PubMed] [Google Scholar]
- 8.Howarth M., Williams A., Tolstrup A.B., Elliott T. Tapasin enhances MHC class I peptide presentation according to peptide half-life. Proc. Natl. Acad. Sci. 2004;101:11737–11742. doi: 10.1073/pnas.0306294101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montealegre S., Venugopalan V., Fritzsche S., Kulicke C., Hein Z., Springer S. Dissociation of β2-microglobulin determines the surface quality control of major histocompatibility complex class I molecules. FASEB J. 2015;29:2780–2788. doi: 10.1096/fj.14-268094. [DOI] [PubMed] [Google Scholar]
- 10.Lim H.J., Wubben J.M., Garcia C.P., Cruz-Gomez S., Deng J., Mak J.Y.W., et al. A specialized tyrosine-based endocytosis signal in MR1 controls antigen presentation to MAIT cells. J. Cell Biol. 2022;221 doi: 10.1083/jcb.202110125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harriff M.J., Karamooz E., Burr A., Grant W.F., Canfield E.T., Sorensen M.L., et al. Endosomal MR1 trafficking plays a key role in presentation of Mycobacterium tuberculosis ligands to MAIT cells. PLOS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karamooz E., Harriff M.J., Narayanan G.A., Worley A., Lewinsohn D.M. MR1 recycling and blockade of endosomal trafficking reveal distinguishable antigen presentation pathways between Mycobacterium tuberculosis infection and exogenously delivered antigens. Sci. Rep. 2019;9:4797. doi: 10.1038/s41598-019-41402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barral D.C., Brenner M.B. CD1 antigen presentation: how it works. Nat. Rev. Immunol. 2007;7:929–941. doi: 10.1038/nri2191. [DOI] [PubMed] [Google Scholar]
- 14.Lu B., Liu M., Wang J., Fan H., Yang D., Zhang L., et al. IL-17 production by tissue-resident MAIT cells is locally induced in children with pneumonia. Mucosal Immunol. 2020;13:824–835. doi: 10.1038/s41385-020-0273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salou M., Lantz O. A TCR-dependent tissue repair potential of MAIT cells. Trends Immunol. 2019;40:975–977. doi: 10.1016/j.it.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Chen Z., Wang H., D'Souza C., Sun S., Kostenko L., Eckle S.B.G., et al. Mucosal-associated invariant T-cell activation and accumulation after in vivo infection depends on microbial riboflavin synthesis and co-stimulatory signals. Mucosal Immunol. 2017;10:58–68. doi: 10.1038/mi.2016.39. [DOI] [PubMed] [Google Scholar]
- 17.Slichter C.K., McDavid A., Miller H.W., Finak G., Seymour B.J., McNevin J.P., et al. Distinct activation thresholds of human conventional and innate-like memory T cells. JCI Insight. 2016;1 doi: 10.1172/jci.insight.86292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Wilgenburg B., Scherwitzl I., Hutchinson E.C., Leng T.Q., Kurioka A., Kulicke C., et al. MAIT cells are activated during human viral infections. Nat. Commun. 2016;7 doi: 10.1038/ncomms11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dias J., Hengst J., Parrot T., Leeansyah E., Lunemann S., Malone D.F.G., et al. Chronic hepatitis delta virus infection leads to functional impairment and severe loss of MAIT cells. J. Hepatol. 2019;71:301–312. doi: 10.1016/j.jhep.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jouan Y., Guillon A., Gonzalez L., Perez Y., Boisseau C., Ehrmann S., et al. Phenotypical and functional alteration of unconventional T cells in severe COVID-19 patients. J. Exp. Med. 2020;217 doi: 10.1084/jem.20200872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flowerdew S.E., Wick D., Himmelein S., Horn A.K.E., Sinicina I., Strupp M., et al. Characterization of neuronal populations in the human trigeminal ganglion and their association with latent herpes simplex virus-1 infection. PloS one. 2013;8:9. doi: 10.1371/journal.pone.0083603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim M., Truong N.R., James V., Bosnjak L., Sandgren K.J., Harman A.N., et al. Relay of herpes simplex virus between langerhans cells and dermal dendritic cells in human skin. PLOS Pathog. 2015;11:19. doi: 10.1371/journal.ppat.1004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McSharry B.P., Samer C., McWilliam H.E.G., Ashley C.L., Yee M.B., Steain M., et al. Virus-mediated suppression of the antigen presentation molecule MR1. Cell Rep. 2020;30:2948–2962. doi: 10.1016/j.celrep.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashley C.L., McSharry B.P., McWilliam H.E.G., Stanton R.J., Fielding C.A., Mathias R.A., et al. Suppression of MR1 by human cytomegalovirus inhibits MAIT cell activation. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1107497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purohit S.K., Samer C., McWilliam H.E.G., Traves R., Steain M., McSharry B.P., et al. Varicella zoster virus impairs expression of the non-classical major histocompatibility complex class I-related gene protein (MR1) J. Infect. Dis. 2021;227:391–401. doi: 10.1093/infdis/jiab526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samer C., McWilliam H.E.G., McSharry B.P., Velusamy T., Burchfield J.G., Stanton R.J., et al. Multi-targeted loss of the antigen presentation molecule MR1 during HSV-1 and HSV-2 infection. iScience. 2024;27 doi: 10.1016/j.isci.2024.108801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McWilliam H.E.G., Mak J.Y.W., Awad W., Zorkau M., Cruz-Gomez S., Lim H.J., et al. Endoplasmic reticulum chaperones stabilize ligand-receptive MR1 molecules for efficient presentation of metabolite antigens. Proc. Natl. Acad. Sci. U. S. A. 2020;117:24974–24985. doi: 10.1073/pnas.2011260117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckle S.B.G., Birkinshaw R.W., Kostenko L., Corbett A.J., McWilliam H.E.G., Reantragoon R., et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J. Exp. Med. 2014;211:1585–1600. doi: 10.1084/jem.20140484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang S.X., Gilfillan S., Cella M., Miley M.J., Lantz O., Lybarger L., et al. Evidence for MR1 antigen presentation to mucosal-associated invariant T cells. J. Biol. Chem. 2005;280:21183–21193. doi: 10.1074/jbc.M501087200. [DOI] [PubMed] [Google Scholar]
- 30.Livingston C.M., Ifrim M.F., Cowan A.E., Weller S.K. Virus-induced chaperone-enriched (VICE) domains function as nuclear protein quality control centers during HSV-1 infection. PLOS Pathog. 2009;5:15. doi: 10.1371/journal.ppat.1000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naghavi M.H., Gundersen G.G., Walsh D. Plus-end tracking proteins, CLASPs, and a viral Akt mimic regulate herpesvirus-induced stable microtubule formation and virus spread. Proc. Natl. Acad. Sci. U. S. A. 2013;110:18268–18273. doi: 10.1073/pnas.1310760110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spang A.E., Godowski P.J., Knipe D.M. Characterization of herpes simplex virus 2 temperature-sensitive mutants whose lesions map in or near the coding sequences for the major DNA-binding protein. J. Virol. 1983;45:332–342. doi: 10.1128/jvi.45.1.332-342.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berg S., Kutra D., Kroeger T., Straehle C.N., Kausler B.X., Haubold C., et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods. 2019;16:1226–1232. doi: 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- 34.Elboim M., Grodzovski I., Djian E., Wolf D.G., Mandelboim O. HSV-2 specifically down regulates HLA-C expression to render HSV-2-Infected DCs susceptible to NK cell killing. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez D.J., Gumperz J.E., Ganem D. Regulation of CD1d expression and function by a herpesvirus infection. J. Clin. Invest. 2005;115:1369–1378. doi: 10.1172/JCI24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geraghty D.E., Koller B.H., Orr H.T. A human major Histocompatibility Complex Class-I gene that encodes a protein with a shortened cytoplasmic sement. Proc. Natl. Acad. Sci. U. S. A. 1987;84:9145–9149. doi: 10.1073/pnas.84.24.9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reantragoon R., Corbett A.J., Sakala I.G., Gherardin N.A., Furness J.B., Chen Z.J., et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 2013;210:2305–2320. doi: 10.1084/jem.20130958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mou F., Forest T., Baines J.D. U(S)3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 2007;81:6459–6470. doi: 10.1128/JVI.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leach N., Bjerke S.L., Christensen D.K., Bouchard J.M., Mou F., Park R., et al. Emerin is hyperphosphorylated and redistributed in herpes simplex virus type 1-infected cells in a manner dependent on both UL34 and US3. J. Virol. 2007;81:10792–10803. doi: 10.1128/JVI.00196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato A., Shindo K., Maruzuru Y., Kawaguchi Y. Phosphorylation of a herpes simplex virus 1 dUTPase by a viral protein kinase, Us3, dictates viral pathogenicity in the central nervous system but not at the periphery. J. Virol. 2014;88:2775. doi: 10.1128/JVI.03300-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benetti L., Munger J., Roizman B. The herpes simplex virus 1 US3 protein kinase blocks caspase-dependent double cleavage and activation of the proapoptotic protein BAD. J. Virol. 2003;77:6567–6573. doi: 10.1128/JVI.77.11.6567-6573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogg P.D., McDonell P.J., Ryckman B.J., Knudson C.M., Roller R.J. The HSV-1 Us3 protein kinase is sufficient to block apoptosis induced by overexpression of a variety of Bcl-2 family members. Virology. 2004;319:212–224. doi: 10.1016/j.virol.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 43.Chuluunbaatar U., Roller R., Feldman M.E., Brown S., Shokat K.M., Mohr I. Constitutive mTORC1 activation by a herpesvirus Akt surrogate stimulates mRNA translation and viral replication. Genes Dev. 2010;24:2627–2639. doi: 10.1101/gad.1978310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato A., Tsuda S., Liu Z., Kozuka-Hata H., Oyama M., Kawaguchi Y. Herpes simplex virus 1 protein kinase Us3 phosphorylates viral dUTPase and regulates its catalytic activity in infected cells. J. Virol. 2014;88:655. doi: 10.1128/JVI.02710-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poon A.P., Gu H., Roizman B. ICP0 and the US3 protein kinase of herpes simplex virus 1 independently block histone deacetylation to enable gene expression. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9993–9998. doi: 10.1073/pnas.0604142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chuluunbaatar U., Roller R., Mohr I. Suppression of extracellular signal-regulated kinase activity in herpes simplex virus 1-infected cells by the Us3 protein kinase. J. Virol. 2012;86:7771–7776. doi: 10.1128/JVI.00622-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang K., Ni L., Wang S., Zheng C. Herpes simplex virus 1 protein kinase US3 hyperphosphorylates p65/RelA and dampens NF-κB activation. J. Virol. 2014;88:7941. doi: 10.1128/JVI.03394-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.You H.J., Lin Y.Y., Lin F., Yang M.Y., Li J.H., Zhang R.Z., et al. Beta-catenin is required for the cGAS/STING signaling pathway but antagonized by the herpes simplex virus 1 US3 protein. J. Virol. 2020;94:13. doi: 10.1128/JVI.01847-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Gent M., Chiang J.J., Muppala S., Chiang C., Azab W., Kattenhorn L., et al. The US3 kinase of herpes simplex virus phosphorylates the RNA sensor RIG-I to suppress innate immunity. J. Virol. 2022;96:e0151021. doi: 10.1128/jvi.01510-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wisner T.W., Wright C.C., Kato A., Kawaguchi Y., Mou F., Baines J.D., et al. Herpesvirus gB-induced fusion between the virion envelope and outer nuclear membrane during virus egress is regulated by the viral US3 kinase. J. Virol. 2009;83:3115–3126. doi: 10.1128/JVI.01462-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mou F., Wills E., Baines J.D. Phosphorylation of the U(L)31 protein of herpes simplex virus 1 by the U(S)3-encoded kinase regulates localization of the nuclear envelopment complex and egress of nucleocapsids. J. Virol. 2009;83:5181–5191. doi: 10.1128/JVI.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kato A., Arii J., Shiratori I., Akashi H., Arase H., Kawaguchi Y. Herpes simplex virus 1 protein kinase Us3 phosphorylates viral envelope glycoprotein B and regulates its expression on the cell surface. J. Virol. 2009;83:250–261. doi: 10.1128/JVI.01451-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kato A., Ando T., Oda S., Watanabe M., Koyanagi N., Arii J., et al. Roles of Us8A and its phosphorylation mediated by Us3 in herpes simplex virus 1 pathogenesis. J. Virol. 2016;90:5622. doi: 10.1128/JVI.00446-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang L., Roizman B. Expression of gamma interferon-dependent genes is blocked independently by virion host shutoff RNase and by U(S)3 protein kinase. J. Virol. 2008;82:4688–4696. doi: 10.1128/JVI.02763-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiong R., Rao P., Kim S., Li M., Wen X.S., Yuan W.M. Herpes simplex virus 1 US3 phosphorylates cellular KIF3A to downregulate CD1d expression. J. Virol. 2015;89:6646–6655. doi: 10.1128/JVI.00214-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rao P., Wen X.S., Lo J.H., Kim S., Li X., Chen S.Y., et al. Herpes simplex virus 1 specifically targets human CD1d antigen presentation to enhance its pathogenicity. J. Virol. 2018;92:12. doi: 10.1128/JVI.01490-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poon A.P., Roizman B. Herpes simplex virus 1 ICP22 regulates the accumulation of a shorter mRNA and of a truncated US3 protein kinase that exhibits altered functions. J. Virol. 2005;79:8470–8479. doi: 10.1128/JVI.79.13.8470-8479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poon A.P., Benetti L., Roizman B. U(S)3 and U(S)3.5 protein kinases of herpes simplex virus 1 differ with respect to their functions in blocking apoptosis and in virion maturation and egress. J. Virol. 2006;80:3752–3764. doi: 10.1128/JVI.80.8.3752-3764.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stanton R.J., McSharry B.P., Armstrong M., Tomasec P., Wilkinson G.W.G. Re-engineering adenovirus vector systems to enable high-throughput analyses of gene function. BioTechniques. 2008;45:659–668. doi: 10.2144/000112993. [DOI] [PubMed] [Google Scholar]
- 60.Imai T., Koyanagi N., Ogawa R., Shindo K., Suenaga T., Sato A., et al. Us3 kinase encoded by herpes simplex virus 1 mediates downregulation of cell surface major histocompatibility complex class I and evasion of CD8+ T cells. PloS one. 2013;8 doi: 10.1371/journal.pone.0072050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ryckman B.J., Roller R.J. Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 2004;78:399–412. doi: 10.1128/JVI.78.1.399-412.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dumont C., Czuba E., Chen M., Villadangos J.A., Johnston A.P.R., Mintern J.D. DNA-based probes for flow cytometry analysis of endocytosis and recycling. Traffic. 2017;18:242–249. doi: 10.1111/tra.12466. [DOI] [PubMed] [Google Scholar]
- 63.Mayle K.M., Le A.M., Kamei D.T. The intracellular trafficking pathway of transferrin. Biochim. Biophys. Acta Gen. Subj. 2012;1820:264–281. doi: 10.1016/j.bbagen.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sobkowiak M.J., Davanian H., Heymann R., Gibbs A., Emgard J., Dias J., et al. Tissue-resident MAIT cell populations in human oral mucosa exhibit an activated profile and produce IL-17. Eur. J. Immunol. 2019;49:133–143. doi: 10.1002/eji.201847759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gibbs A., Leeansyah E., Introini A., Paquin-Proulx D., Hasselrot K., Andersson E., et al. MAIT cells reside in the female genital mucosa and are biased towards IL-17 and IL-22 production in response to bacterial stimulation. Mucosal Immunol. 2017;10:35–45. doi: 10.1038/mi.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Constantinides M.G., Linehan J.L., Sen S., Shaik J., Roy S., LeGrand J.L., et al. Mucosal-associated invariant T cells respond to cutaneous microbiota. J. Immunol. 2017;198:218.15. [Google Scholar]
- 67.Durukan D., Fairley C.K., Bradshaw C.S., Read T.R.H., Druce J., Catton M., et al. Increasing proportion of herpes simplex virus type 1 among women and men diagnosed with first-episode anogenital herpes: a retrospective observational study over 14 years in Melbourne, Australia. Sex. Transm. Infections. 2019;95:307–313. doi: 10.1136/sextrans-2018-053830. [DOI] [PubMed] [Google Scholar]
- 68.Kaufman H.E., Azcuy A.M., Varnell E.D., Sloop G.D., Thompson H.W., Hill J.M. HSV-1 DNA in tears and saliva of normal adults. Invest. Ophthalmol. Vis. Sci. 2005;46:241–247. doi: 10.1167/iovs.04-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cunningham A.L., Diefenbach R.J., Miranda-Saksena M., Bosnjak L., Kim M., Jones C., et al. The cycle of human herpes simplex virus infection: virus transport and immune control. J. Infect. Dis. 2006;194:S11–S18. doi: 10.1086/505359. [DOI] [PubMed] [Google Scholar]
- 70.Möckel M., De La Cruz N.C., Rübsam M., Wirtz L., Tantcheva-Poor I., Malter W., et al. Herpes simplex virus 1 can bypass impaired epidermal barriers upon ex vivo infection of skin from atopic dermatitis patients. J. Virol. 2022;96:e0086422. doi: 10.1128/jvi.00864-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hemani S.A., Edmond M.B., Jaggi P., Cooley A. Frequency and clinical features associated with eczema herpeticum in hospitalized children with presumed atopic dermatitis skin infection. Pediatr. Infect. Dis. J. 2020;39:263–266. doi: 10.1097/INF.0000000000002542. [DOI] [PubMed] [Google Scholar]
- 72.Beck L.A., Boguniewicz M., Hata T., Schneider L.C., Hanifin J., Gallo R., et al. Phenotype of atopic dermatitis subjects with a history of eczema herpeticum. J. Allergy Clin. Immunol. 2009;124:260–269. doi: 10.1016/j.jaci.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vitreschak A.G., Rodionov D.A., Mironov A.A., Gelfand M.S. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 2002;30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramchandani M., Kong M., Tronstein E., Selke S., Mikhaylova A., Magaret A., et al. Herpes simplex virus type 1 shedding in tears and nasal and oral mucosa of healthy adults. Sex. Transm. Dis. 2016;43:756–760. doi: 10.1097/OLQ.0000000000000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cook M.L., Stevens J.G. Pathogenesis of herpetic neuritis and ganglionitis in mice: evidence for intra-axonal transport of infection. Infect. Immun. 1973;7:272–288. doi: 10.1128/iai.7.2.272-288.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Esber A., Vicetti Miguel R.D., Cherpes T.L., Klebanoff M.A., Gallo M.F., Turner A.N. Risk of bacterial vaginosis among women with herpes simplex virus type 2 infection: a systematic review and meta-analysis. J. Infect. Dis. 2015;212:8–17. doi: 10.1093/infdis/jiv017. [DOI] [PubMed] [Google Scholar]
- 77.Johansson M.A., Björkander S., Forsberg M.M., Qazi K.R., Celades M.S., Bittmann J., et al. Probiotic lactobacilli modulate Staphylococcus aureus-induced activation of conventional and unconventional T cells and NK cells. Front. Immunol. 2016;7:273. doi: 10.3389/fimmu.2016.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Samer C., Traves R., Purohit S.K., Abendroth A., McWilliam H.E.G., Slobedman B. Viral impacts on MR1 antigen presentation to MAIT cells. Crit. Rev. Immunol. 2021;41:49–67. doi: 10.1615/CritRevImmunol.2022041981. [DOI] [PubMed] [Google Scholar]
- 79.Ussher J.E., van Wilgenburg B., Hannaway R.F., Ruustal K., Phalora P., Kurioka A., et al. TLR signaling in human antigen-presenting cells regulates MR1-dependent activation of MAIT cells. Eur. J. Immunol. 2016;46:1600–1614. doi: 10.1002/eji.201545969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holm C.K., Jensen S.B., Jakobsen M.R., Cheshenko N., Horan K.A., Moeller H.B., et al. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat. Immunol. 2012;13:737–743. doi: 10.1038/ni.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aravalli R.N., Hu S., Rowen T.N., Palmquist J.M., Lokensgard J.R. Cutting edge: TLR2-mediated proinflammatory cytokine and chemokine production by microglial cells in response to herpes simplex virus. J. Immunol. 2005;175:4189–4193. doi: 10.4049/jimmunol.175.7.4189. [DOI] [PubMed] [Google Scholar]
- 82.Leoni V., Gianni T., Salvioli S., Campadelli-Fiume G. Herpes simplex virus glycoproteins gH/gL and gB bind toll-like receptor 2, and soluble gH/gL is sufficient to activate NF-kappa B. J. Virol. 2012;86:6555–6562. doi: 10.1128/JVI.00295-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lamichhane R., Galvin H., Hannaway R.F., de la Harpe S.M., Munro F., Tyndall J.D., et al. Type I interferons are important co-stimulatory signals during T cell receptor mediated human MAIT cell activation. Eur. J. Immunol. 2019;50:178–191. doi: 10.1002/eji.201948279. [DOI] [PubMed] [Google Scholar]
- 84.Fruh K., Ahn K., Djaballah H., Sempe P., Vanendert P.M., Tampe R., et al. A viral inhibitor of peptide transporters for antigen presentation. Nature. 1995;375:415–418. doi: 10.1038/375415a0. [DOI] [PubMed] [Google Scholar]
- 85.Rao P., Pham H.T., Kulkarni A., Yang Y., Liu X., Knipe D.M., et al. Herpes simplex virus 1 glycoprotein B and US3 collaborate to inhibit CD1d antigen presentation and NKT cell function. J. Virol. 2011;85:8093–8104. doi: 10.1128/JVI.02689-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.York I.A., Roop C., Andrews D.W., Riddell S.R., Graham F.L., Johnson D.C. A cytosolic herpes-simplex virus protein inhibits antigen presentation to CD8(+) T-lymphocytes. Cell. 1994;77:525–535. doi: 10.1016/0092-8674(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 87.Yuan W., Dasgupta A., Cresswell P. Herpes simplex virus evades natural killer T cell recognition by suppressing CD1d recycling. Nat. Immunol. 2006;7:835–842. doi: 10.1038/ni1364. [DOI] [PubMed] [Google Scholar]
- 88.Campbell T.M., McSharry B.P., Steain M., Slobedman B., Abendroth A. Varicella-zoster virus and herpes simplex virus 1 differentially modulate NKG2D ligand expression during productive infection. J. Virol. 2015;89:7932–7943. doi: 10.1128/JVI.00292-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moody D.B., Porcelli S.A. Intracellular pathways of CD1 antigen presentation. Nat. Rev. Immunol. 2003;3:11–22. doi: 10.1038/nri979. [DOI] [PubMed] [Google Scholar]
- 90.Albecka A., Laine R.F., Janssen A.F., Kaminski C.F., Crump C.M. HSV-1 glycoproteins are delivered to virus assembly sites through dynamin-dependent endocytosis. Traffic. 2016;17:21–39. doi: 10.1111/tra.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hollinshead M., Johns H.L., Sayers C.L., Gonzalez-Lopez C., Smith G.L., Elliott G. Endocytic tubules regulated by Rab GTPases 5 and 11 are used for envelopment of herpes simplex virus. Embo J. 2012;31:4204–4220. doi: 10.1038/emboj.2012.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sugimoto K., Uema M., Sagara H., Tanaka M., Sata T., Hashimoto Y., et al. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J. Virol. 2008;82:5198–5211. doi: 10.1128/JVI.02681-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kulej K., Avgousti D.C., Sidoli S., Herrmann C., Della Fera A.N., Kim E.T., et al. Time-resolved global and chromatin proteomics during herpes simplex virus type 1 (HSV-1) infection. Mol. Cell Proteomics. 2017;16:S92–S107. doi: 10.1074/mcp.M116.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jung K.K., Liu X.W., Chirco R., Fridman R., Kim H.R. Identification of CD63 as a tissue inhibitor of metalloproteinase-1 interacting cell surface protein. Embo J. 2006;25:3934–3942. doi: 10.1038/sj.emboj.7601281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bendelac A., Mattner J., DeBord K.L., Ismail N., Goff R.D., Cantu C., et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 96.Fischer K., Scotet E., Niemeyer M., Koebernick H., Zerrahn J., Maillet S., et al. Mycobacterial phosphatidylinositol mannoside is a natural antigen for CD1d-restricted T cells. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10685–10690. doi: 10.1073/pnas.0403787101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Conner S.D., Schmid S.L. Differential requirements for AP-2 in clathrin-mediated endocytosis. J. Cel. Biol. 2003;162:773–779. doi: 10.1083/jcb.200304069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kelly B.T., McCoy A.J., Späte K., Miller S.E., Evans P.R., Höning S., et al. A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature. 2008;456:976–979. doi: 10.1038/nature07422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jayawardena-Wolf J., Benlagha K., Chiu Y.H., Mehr R., Bendelac A. CD1d endosomal trafficking is independently regulated by an intrinsic CD1d-encoded tyrosine motif and by the invariant chain. Immunity. 2001;15:897–908. doi: 10.1016/s1074-7613(01)00240-0. [DOI] [PubMed] [Google Scholar]
- 100.Benetti L., Roizman B. Herpes simplex virus protein kinase US3 activates and functionally overlaps protein kinase A to block apoptosis. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9411–9416. doi: 10.1073/pnas.0403160101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bananis E., Nath S., Gordon K., Satir P., Stockert R.J., Murray J.W., et al. Microtubule-dependent movement of late endocytic vesicles in vitro: requirements for dynein and kinesin. Mol. Biol. Cell. 2004;15:3688–3697. doi: 10.1091/mbc.E04-04-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Benedyk T.H., Muenzner J., Connor V., Han Y., Brown K., Wijesinghe K.J., et al. pUL21 is a viral phosphatase adaptor that promotes herpes simplex virus replication and spread. PLOS Pathog. 2021;17:33. doi: 10.1371/journal.ppat.1009824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kato A., Liu Z., Minowa A., Imai T., Tanaka M., Sugimoto K., et al. Herpes simplex virus 1 protein kinase Us3 and major tegument protein UL47 reciprocally regulate their subcellular localization in infected cells. J. Virol. 2011;85:9599. doi: 10.1128/JVI.00845-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bjerke S.L., Roller R.J. Roles for herpes simplex virus type 1 U(L)34 and U(S)3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology. 2006;347:261–276. doi: 10.1016/j.virol.2005.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Norberg P., Tyler S., Severini A., Whitley R., Liljeqvist J.-Å., Bergström T. A genome-wide comparative evolutionary analysis of herpes simplex virus type 1 and varicella zoster virus. PloS One. 2011;6:e22527. doi: 10.1371/journal.pone.0022527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Severini A., Tyler S.D., Peters G.A., Black D., Eberle R. Genome sequence of a chimpanzee herpesvirus and its relation to other primate alphaherpesviruses. Arch. Virol. 2013;158:1825–1828. doi: 10.1007/s00705-013-1666-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wertheim J.O., Hostager R., Ryu D., Merkel K., Angedakin S., Arandjelovic M., et al. Discovery of Novel herpes simplexviruses in wild gorillas, bonobos, and chimpanzees supports zoonotic origin of HSV-2. Mol. Biol. Evol. 2021;38:2818–2830. doi: 10.1093/molbev/msab072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rawls W.E., Laurel D., Melnick J.L., Glicksman J.M., Kaufman R.H. A search for viruses in smegma, premalignant and early malignant cervical tissues. The isolation of herpesviruses with distinct antigenic properties. Am. J. Epidemiol. 1968;87:647–655. doi: 10.1093/oxfordjournals.aje.a120855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Further information and requests for resources, data, and reagents should be directed to and will be fulfilled by the Lead Contact, Barry Slobedman (barry.slobedman@sydney.edu.au). Cells, viral mutants, and constructs are available upon request, subject to our institutional and Material Transfer Agreements, and those with the host institution at which these reagents were generated.