Abstract
Osteoarthritis (OA), affecting > 500 million people worldwide, profoundly affects the quality of life and ability to work. The mitogen-activated protein kinase (MAPK) signaling pathway plays an essential role in OA. To address the lack of studies focused on synovial cells in OA, we evaluated the expression patterns and roles of the MAPK signaling pathway components in OA synovial tissues using bioinformatics. The JUN, MYC, and VEGFA expression levels were significantly higher in the synovial tissues of patients with OA than in control tissues. These loci were closely related to abnormal proliferation, inflammation, and angiogenesis in the synovial tissues of patients with OA. We speculate that Myc and VEGFA activate the p38-MAPK signaling pathway to further activate Jun, thereby promoting abnormal inflammation, proliferation, and angiogenesis in OA synovial tissue. The high MYC, JUN, and VEGFA expression was positively correlated with the patients’ K-L score, pain time, and synovial score. Furthermore, the high p38-MAPK and P-p38-MAPK expression confirmed that the abnormal expression and activation of the MAPK signaling pathway occurred in the synovial tissue of patients with OA. Our findings may provide a new direction for the clinical diagnosis and treatment of OA and insights into its pathogenesis.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-80551-7.
Keywords: Osteoarthritis, MAPK, MYC, VEGFA, JUN, Synovial tissue
Subject terms: Computational biology and bioinformatics, Data mining, Databases, Functional clustering, Genome informatics
Introduction
Osteoarthritis (OA) is the most common chronic degenerative joint disease. The main clinical symptoms include chronic pain, joint instability, stiffness, and radiographic joint space narrowing1. OA pathogenesis mainly includes different degrees of synovial inflammation, the degeneration of the knee ligaments and meniscuses, the degeneration and destruction of articular cartilage, the thickening of subchondral bone, secondary bone hyperplasia, joint capsule hypertrophy, and inflammation of the infrapatellar fat pad1–5. According to the latest Global Burden of Disease study, the global age-standardized prevalence of knee OA is 3.8%; among 291 diseases evaluated, knee OA was the 11th largest contributor to disability worldwide (when measured in years lived with disability) and ranked 38th in disability-adjusted life years. OA is the most common form of arthritis, affecting more than 500 million people worldwide (approximately 7% of the global population), with a particularly high incidence in older people (> 65 years)6–8. However, treatment methods and drugs for OA are very limited; except for joint replacement in patients with advanced disease, current therapies only alleviate and slow the progression of the disease, and there is currently no cure9–12.
The molecular mechanism underlying the occurrence and development of OA is still unclear13–16. Under the influence of multiple risk factors, including age, obesity, trauma, and mechanical load, OA can activate synovial inflammation by various mechanisms, such as mitochondrial dysfunction, injury-related molecular patterns, cytokines, metabolites, and crystals in the synovium17–25. Synovial inflammation leads to further changes in the biological function of the synovium and damages the cartilage and bone tissue, eventually leading to the irreversible development of OA26,27. Baseline synovitis detected by magnetic resonance imaging or ultrasound is associated with the radiographic progression of OA, as defined by worsening of the Kellgren and Lawrence (KL) Grade or joint space narrowing8,28–34. The mitogen-activated protein kinase (MAPK) signaling pathway is found in all eukaryotes MAPK is a member of the intracellular serine-threonine protein kinase superfamily and is the central node in various signal transduction pathways35,36. and it is plays an important role in a variety of inflammatory diseases. Jun belongs to the activator protein 1 (AP-1) family of proteins37. It plays a key role in regulating cell proliferation and apoptosis38 and contributes to the regulation of a variety of other cellular processes including survival, differentiation, migration, and transformation39. It is also involved in the occurrence and development of OA, where it may drive chondrocyte hypertrophy and the differentiation of osteoprogenitor cells into osteoblasts, mediate the synthesis of cartilage-specific extracellular matrix components, and alter angiogenesis in bones40. In addition to Jun, c-Myc and vascular endothelial growth factor A (VEGFA) may have key roles in OA. C-Myc, a member of the Myc family, is a helix-loop-helix leucine zipper pleiotropic transcription factor involved in a variety of cellular processes including cell growth, apoptosis, metabolism, differentiation, and DNA repair41. Myc overexpression is associated with OA as well as small-cell lung cancer, myeloid leukemia, breast cancer, and cervical cancer42. Silencing the c-Myc-encoding gene reduces the effects of IL-1β on cell cycle progression and apoptosis in the chondrocytes of rats with OA43. VEGFA is a member of the vascular endothelial growth factor family and has a wide range of functions including the stimulation of angiogenesis, monocyte chemotaxis, vascular permeability, and vasodilation44,45; it is also an important mediator of osteogenesis46. High VEGFA expression is associated with OA, tumor angiogenesis, rheumatoid arthritis, psoriasis, atherosclerosis, brain edema, age-related macular degeneration, diabetic retinopathy, retinopathy of prematurity, and sepsis44,47. VEGFs appear to be involved in specific pathological processes in OA, including cartilage degeneration, osteophyte formation, subchondral bone cysts and sclerosis, synovitis, and pain. In addition, extensive research has shown that the inhibition of VEGF signaling slows the progression of OA48.
Therefore, Jun, c-Myc, and VEGFA may play a key role in OA. Nevertheless, up until the present moment, the majority of investigations have predominantly centered around osteocytes, with comparatively fewer studies directed toward the synovium. Concurrently, in an endeavor to establish greater clinical relevance and facilitate the expeditious evolution of OA diagnostic and therapeutic methodologies, the present study procured specimens from 20 OA patients alongside 10 samples of normative human synovial tissue, obtained through knee arthroscopy, to undergo iterative validation processes. This undertaking entailed an assessment of the expression profiles and functional attributes pertaining to Jun, c-Myc, and VEGFA within human synovial tissues.
Materials and methods
Dataset acquisition
A search against the Gene Expression Omnibus (GEO) database, created by the National Center for Biotechnology Information, was performed with “osteoarthritis” as the keyword. The screening requirements were as follows: (1) the species was Homo sapiens; (2) a healthy control group and an OA group were included, each with sample sizes not less than three; (3) the OA group was not treated with drugs or other therapies; and (4) complete, original gene expression data files and complete chip annotation data were available.
Principal component analysis
R Studio was used to preprocess the data. After the conversion of probes and gene names, the average value for a gene corresponding to multiple probes was obtained. After normalizing the dataset using the “limma” package in R, a principal component analysis was performed using the “FactoMineR” and “factoextra” packages.
Differential expression analysis
R Studio was used to compare expression levels between OA synovial tissues and normal synovial tissues to obtain differentially expressed genes (DEGs). The original data set was preprocessed using the “limma” package in R. A fold change (FC) value ≥ 2, log2(FC) > 1, and P < 0.05 were used as screening criteria to filter out DEGs in OA synovial tissues.
Intersection analysis
An intersection analysis was performed to identify DEGs obtained from the three gene expression datasets (i.e., DEGs shared between the healthy control group and the OA group). The results were displayed in an UpSet graph.
PPI network construction and visualization
The common DEGs in the OA synovial tissues were imported into the STRING online database to study protein–protein interactions (PPIs). The species was set to Homo sapiens, with default conditions used for other parameters. Nodes with no connections to other proteins were removed. The results were imported into Cytoscape for visualization, and the number of proteins interacting with each protein was calculated and expressed as a degree value.
Biomarker analysis
Using clinical information for samples and the expression levels of indicators, the key targets obtained by the PPI network analysis were analyzed for biomarker identification. Receiver operating characteristic (ROC) curves for the key genes were drawn using the R package “pROC,” and the areas under the curve (AUC) were calculated. AUC > 0.50 indicated that genes had a diagnostic value for OA, with values closer to 1.0 indicating a higher diagnostic value.
Gene ontology functional enrichment analysis
Common DEGs in OA synovial tissues were imported into the DAVID database for a gene ontology (GO) enrichment analysis. The species was set as Homo sapiens, with P < 0.05 as the screening criterion. The results were divided into three parts: cellular component (CC), molecular function (MF), and biological process (BP).
Kyoto Encyclopedia of genes and genomes (KEGG) pathway enrichment analysis
The common DEGs in the OA synovial tissues were imported into the DAVID database for a KEGG enrichment analysis. The species was set as Homo sapiens, and P < 0.05 was used as the screening criterion to identify related pathways.
Specimen collection
We obtained synovial tissue from 20 patients (nine male and 11 female) with OA who underwent knee arthroplasty at the Joint and Microsurgical Surgery Department of the First Affiliated Hospital of Anhui Medical University. Of the patients, four were at KL Grade 2, six were at KL Grade 3, and 10 were at KL Grade 4; none of the cases had used drug intervention before surgery. Meanwhile, the synovial tissues of 10 healthy people (six men and four women) were obtained from knee arthroscopy of sprain patients in the Arthroscopy Department of the First Affiliated Hospital of Anhui Medical University. No drug intervention was used before arthroscopy. This work was approved by the Human Research Ethics Committee of Anhui Medical University (Ethics number: S20210085, date: 2021/03/31).
Quantitative real-time PCR analysis
The total RNA was extracted using TRIzol reagent (Thermo Fisher, Waltham, MA, USA) according to the manufacturer’s protocol. Total RNA (500 ng) was reverse-transcribed into cDNA using a PrimeScript RT Reagent Kit (Takara, Kusatsu, Japan). Quantitative real-time PCR (qRT-PCR) was performed using SYBR Green PCR Mix (Thermo Fisher). Primer sequences are listed in Table 1. ACTB served as an internal reference, and the relative expression levels of messenger RNAs (mRNAs) were quantified using the 2−ΔΔCt method. In order to better visualize the results, we normalized the results with the average mRNA expression of the normal group.
Table 1.
Primers used for RT-qPCR.
| Gene symbol | Primer | Sequence |
|---|---|---|
| JUN | Forward | TCCAAGTGCCGAAAAAGGAA |
| Reverse | CGAGTTCTGAGCTTTCAAGGT | |
| MYC | Forward | GTCAAGAGGCGAACACACAAC |
| Reverse | TTGGACGGACAGGATGTATGC | |
| VEGFA | Forward | AGGGCAGAATCATCACGAAGT |
| Reverse | AGGGTCTCGATTGGATGGCA | |
| ACTB | Forward | CATGTACGTTGCTATCCAGGC |
| Reverse | CTCCTTAATGTCACGCACGAT |
Western blot
Tissues were lysed on ice using NP40 buffer containing a protease and phosphatase inhibitor cocktail (Beyotime Biotechnology, China) for 30 min. The protein concentration was determined using the BCA protein assay kit (Beyotime Biotechnology). Total protein was further separated by SDS-PAGE before transferring to polyvinylidene difluoride membranes (Merck, Rahway, NJ, USA). The membranes were blocked with 5% non-fat milk at room temperature for 1 h, and then incubated with primary antibodies (C-MYC, VEGFA, JUN, p38-MAPK, β-actin; Cell Signaling Technology) overnight at 4 °C. After five washes, the membranes were incubated with HRP-conjugated secondary antibodies at room temperature for 1 h. Protein bands were visualized using an ECL solution (NCM Biotech, Suzhou, China) and detected with an automatic chemiluminescence imaging analysis system (Bio-Rad Laboratories, Hercules, CA, USA). The intensity of each band was quantified using ImageJ software (National Institute of Mental Health, Bethesda, MD, USA).
Hematoxylin-eosin staining and immunohistochemistry
Human OA synovium or normal synovium were fixed with 4% paraformaldehyde overnight and embedded in paraffin before sectioning into 5-µm slices. The sections were then deparaffinized with xylene before being rehydrated in water using an ethanol gradient. After staining with hematoxylin and eosin, they were incubated with primary antibodies for over 4 h at 4 °C. Finally, the sections were washed and incubated with appropriate secondary antibodies for 1 h. Antigen-positive cells were visualized using the DAB Substrate kit (Beyotime Biotechnology).
Immunofluorescence staining
Paraffin-embedded tissue sections were deparaffinized by heating in a 60 °C oven for 15 min and then hydrated through a series of xylene and graded alcohols. For antigen retrieval, heat-induced epitope retrieval was performed using a vegetable steamer. Slides were incubated in 10 mM citrate buffer at 60 °C for 40 min followed by washing in PBS. At room temperature, cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min. Subsequently, the appropriate primary antibodies were applied and incubated overnight. The following day, corresponding fluorescent secondary antibodies and DAPI were applied, and the samples were observed using laser confocal microscopy (Zeiss LSM 780). Details of the primary or secondary antibodies used are shown in Table 2.
Table 2.
Antibody details list.
| Antibodies | SOURCE | IDENTIFIER |
|---|---|---|
| VEGFA | Abcam | EP1176Y |
| C-MYC | CST | #18,583 |
| JUN | CST | #9165 |
| P38-MAPK | CST | #9212 |
| P-P38-MAPK | CST | #4511 |
| Goat anti-Rabbit IgG (H + L) Alexa Fluor 647 | CST | #4414 |
Kellgren’s score
The severity of knee osteoarthritis (KOA) is assessed and scored by a specialized orthopedic surgeon based on specific features on the radiograph. These features include joint space narrowing, osteophyte formation, subchondral sclerosis, and cystic degeneration. KOA is categorized into five grades, ranging from 0 (normal) to 4 (severe lesions). A score of 1 indicates the possibility of joint space narrowing and osteophyte formation; a score of 2 indicates definite osteophyte formation and possible joint space narrowing; a score of 3 indicates the formation of severe osteophytes, definite joint space narrowing, sclerosis, and possibly bone deformity; and a score of 4 indicates severe osteophyte formation, joint space narrowing (sometimes bone-on-bone contact), large osteophytes, severe sclerosis, and end-stage OA.
Synovitis score
In active OA patients, the synovial tissue of the knee joint was obtained by fine needle biopsy and HE staining. The degree of hyperplasia of the lining layer, the degree of infiltration of inflammatory cells in the underlining layer, and the degree of matrix activation were evaluated by 400× light semi-quantitative method (0 ~ 3 points), and the total synovitis score (0 ~ 9 points) was obtained. According to the synovitis score, the patients were divided into the no synovitis (0 points), low synovitis (1–4 points), moderate synovitis (4–6 points), and high synovitis groups (> 6 points).
Statistical methods
Comparisons between multiple groups were performed using one-way analysis of variance (ANOVA) in GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA). The t-test was used to compare the two sets of data. Data were presented as means ± SD. P-values < 0.05 (two-sided) were considered significant (*P < 0.05, **P < 0.01, ***P < 0.001).
Results
Raw data processing
The GEO database was searched using the keywords “osteoarthritis” and “synovium,” and a total of three datasets were eligible for inclusion in the analysis. Information about the datasets is shown in Table 3. In GSE12021, two duplicate samples were taken from each volunteer and tested by GPL96 and GPL97 microarrays, respectively. The other two datasets included in this paper are based on GPL96, and we choose only the data obtained from GPL96 detection in GSE12021.
Table 3.
Dataset information.
Principal component analysis and DEGs
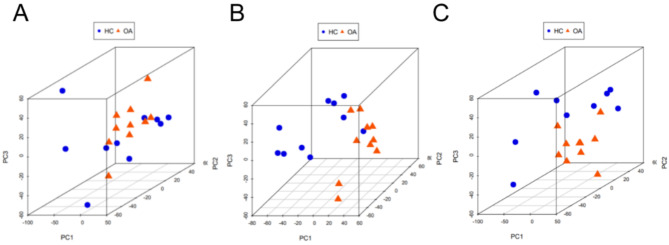
Illustrated in Fig. 1 is the application of principal component analysis to three distinct datasets, namely GSE55457 (depicted in Fig. 1A), GSE55235 (depicted in Fig. 1B), and GSE12021 (depicted in Fig. 1C). In each of these datasets, the assessment of intra-group distances pertaining to samples from the normal control and OA groups revealed minimal dispersion, accompanied by distinct demarcations between the said groups. This observation is suggestive of substantial dissimilarity between the normal control and OA cohorts. As such, these three datasets hold promise for subsequent investigative endeavors.
Fig. 1.
PCA analysis was performed to indicate the difference between OA and HC synovial tissues. GSE55457(A), GSE55235(B), and GSE12021(C). Note: blue nodes represent samples in the healthy control group, orange nodes represent samples in the osteoarthritis group, and the ellipse represents a 95% confidence interval.
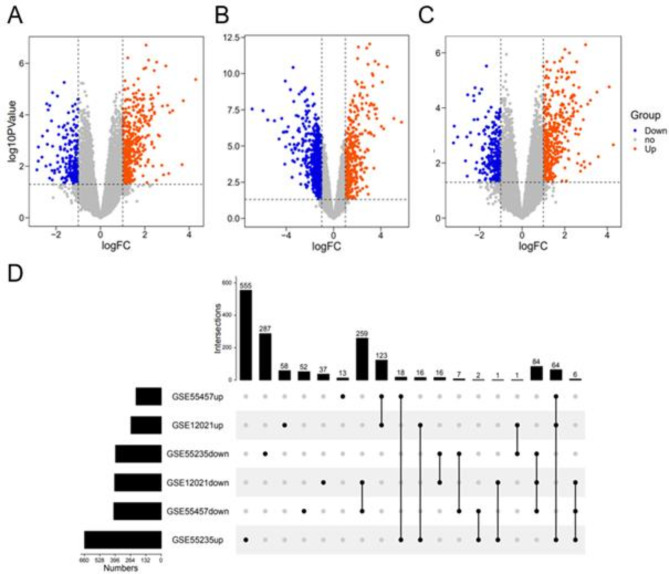
DEGs identified in the GSE55457 (Fig. 2A), GSE55235 (Fig. 2B), and GSE12021 (Fig. 2C) datasets are shown in volcano plots, where blue indicates genes that are downregulated compared with levels in the healthy control group (Log2FC < − 1, P < 0.05) and red indicates genes that are upregulated compared with levels in the healthy control group (Log2FC > 1, P < 0.05). There were 410 downregulated genes and 218 upregulated genes in GSE55457, 395 downregulated genes and 662 upregulated genes in GSE55235, and 403 downregulated genes and 262 upregulated genes in GSE12021. The results of an intersection analysis are shown in Fig. 2D. There were 64 upregulated genes, and 84 downregulated genes shared among all three datasets.
Fig. 2.
DEGs in OA synovial tissues were identified via differential expression analysis. Volcano plots show DEGs in GSE55457(A), GSE55235(B), and GSE12021(C). Upset diagram of the overlapping DEGs(D). Note: blue nodes represent up-regulated genes, orange nodes represent down-regulated genes, and grey nodes represent no significant genes.
PPI network analysis
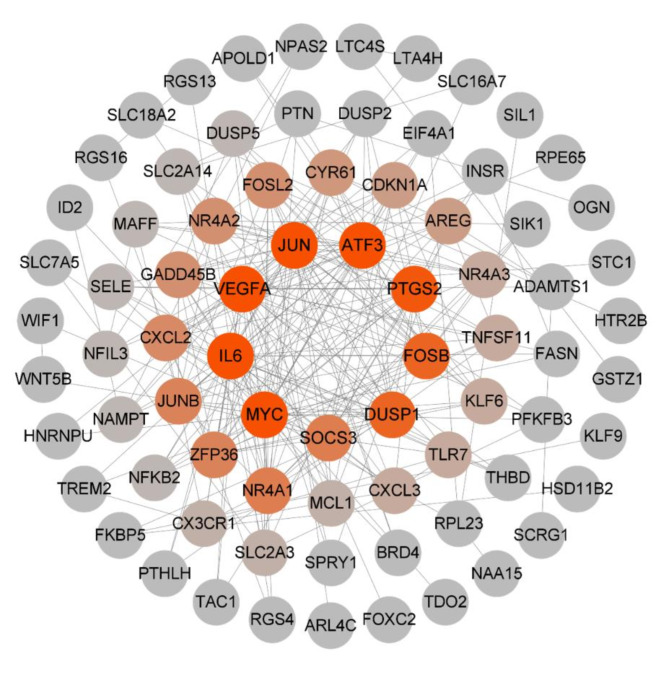
Exploration of the interdependent relationships among DEGs, which exhibit overlaps, was facilitated through queries within the STRING database. Subsequently, a protein–protein interaction (PPI) network was meticulously constructed and visually represented employing Cytoscape software. Within this network framework, the quantum of associations that individual proteins engendered with their counterparts was quantified as their degree values. Notably, these degree values were color-coded, transitioning from a gradient ranging from red to gray, symbolizing a descending order of magnitude. The chromatic gradient manifested red hues for higher degree values. The outcome of this network analysis prominently highlighted escalated degree values for pertinent proteins such as MYC, IL6, VEGFA, JUN, ATF3, and PTGS2 (Fig. 3). This augmented numerical magnitude strongly implies a heightened propensity for reciprocal interactions among these aforementioned proteins.
Fig. 3.
PPI network analysis of the overlapping DEGs between the OA and NC. Note: The darker the node, the greater the degree.
Core protein expression analysis and biomarker analysis
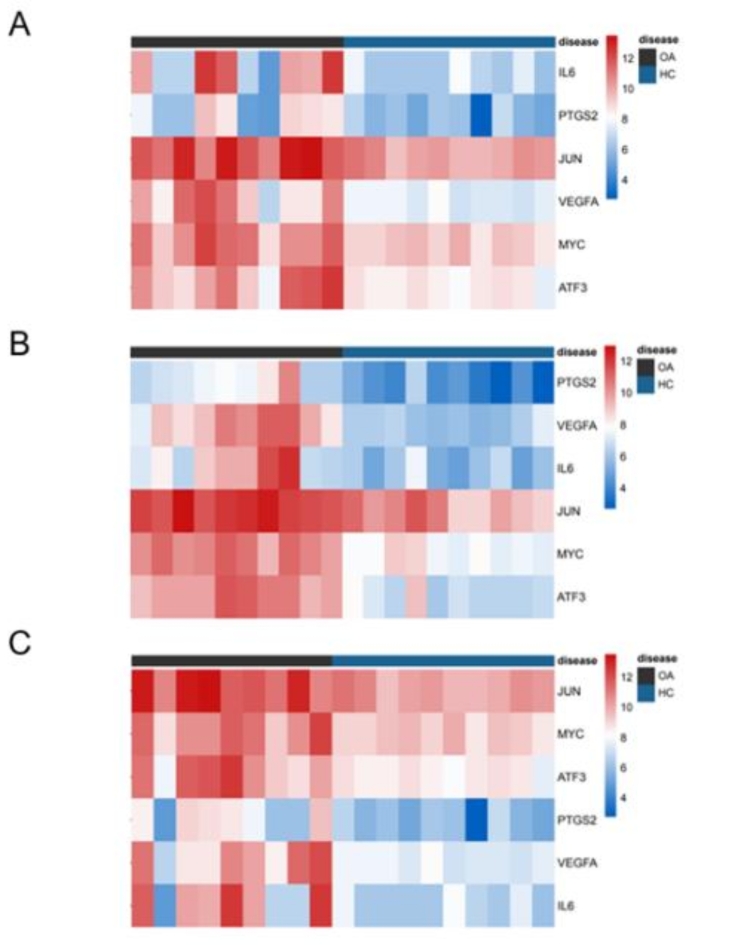
Heatmaps depicting the expression profiles of pivotal genes, namely MYC, IL6, VEGFA, JUN, ATF3, and PTGS2, within the datasets GSE55457 (depicted in Fig. 4A), GSE55235 (depicted in Fig. 4B), and GSE12021 (depicted in Fig. 4C), were generated. These heatmaps effectively portray the gene expression dynamics across distinct datasets, whereby the spectrum of expression intensities ranging from minimal to maximal is graphically represented through a color gradient spanning from blue to red. A discernible trend is evident from these visualizations, illustrating that the expression levels of IL6, PTGS2, and VEGFA in the context of OA surpass those within the normal control group. Conversely, for the genes, JUN, MYC, and ATF3, a marginally reduced expression magnitude is observed within the normal control group as compared to the OA cohort.
Fig. 4.
Expression of hub genes was presented by heatmap in microarray data. GSE55457(A), GSE55235(B), and GSE12021(C). (A - C) : use the online tool bioinformatics analysis, the link is: http://www.bioinformatics.com.cn/plot_basic_cluster_heatmap_plot_024.
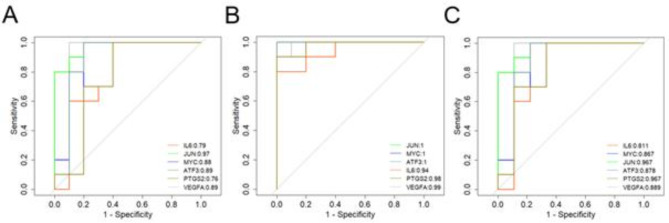
Indicators with AUC values > 0.5 for the three datasets were as follows: GSEA55457 (Fig. 5A), MYC (AUC = 0.88), IL6 (AUC = 0.79), VEGFA (AUC = 0.89), JUN (AUC = 0.97), ATF3 (AUC = 0.89), and PTGS2 (AUC = 0.76); GSEA55235 (Fig. 5B), MYC (AUC = 1), IL6 (AUC = 0.94), VEGFA (AUC = 0.99), JUN (AUC = 1), ATF3 (AUC = 1), and PTGS2 (AUC = 0.98); GSEA12021 (Fig. 5C), MYC (AUC = 0.867), IL6 (AUC = 0.811), VEGFA (AUC = 0.889), JUN (AUC = 0.967), ATF3 (AUC = 0.878), and PTGS2 (AUC = 0.967).
Fig. 5.
Diagnostic effectiveness of the biomarkers for OA. GSE55457(A), GSE55235(B), and GSE12021(C) were used to evaluate the diagnostic effectiveness of the biomarkers for OA by ROC analysis.
Functional enrichment analysis
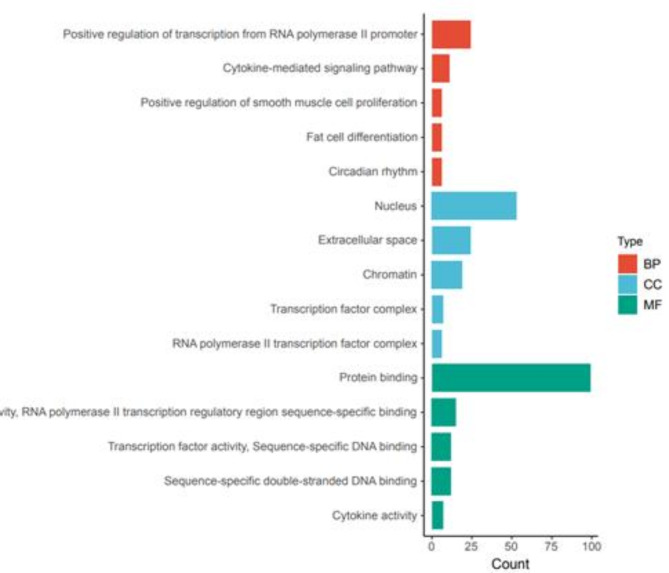
The results of the GO enrichment analysis are shown in Fig. 6. In the BP category, DEGs were enriched for various terms, including positive regulation of transcription from RNA polymerase II promoter, cytokine-mediated signaling pathway, positive regulation of smooth muscle cell proliferation, fat cell differentiation, and circadian rhythm. Enrichment in the CC category included chromatin, RNA polymerase II transcription factor complex, extracellular space, nucleus, and transcription factor complex. In the MF category, enriched terms included transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding, transcription factor activity, sequence-specific DNA binding, sequence-specific double-stranded DNA binding, cytokine activity, and protein binding.
Fig. 6.
GO functional enrichment analysis of the overlapping DEGs.
Pathway enrichment analysis
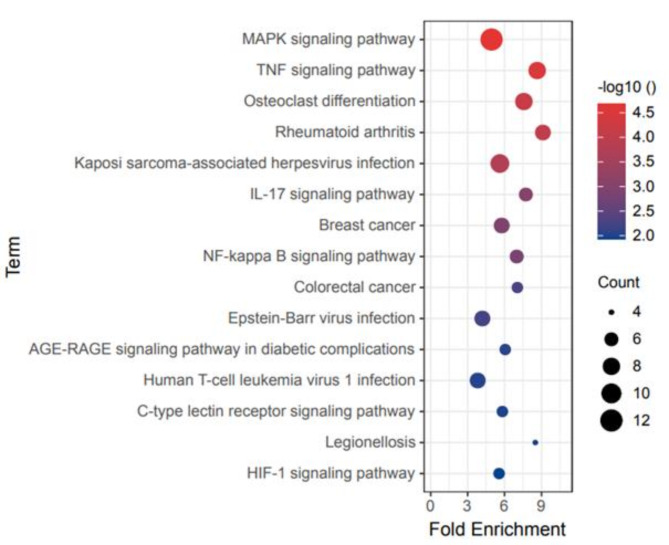
The results of the KEGG pathway enrichment analysis are shown in Fig. 7. The DEGs were involved in various pathways including the MAPK signaling pathway, tumor necrosis factor (TNF) signaling pathway, osteoclast differentiation, rheumatoid arthritis, Kaposi sarcoma-associated herpesvirus infection, IL-17 signaling pathway, breast cancer, NF-κB signaling pathway, colorectal cancer, Epstein-Barr virus infection, AGE-RAGE signaling pathway in diabetic complications, human T-cell leukemia virus 1 infection, C-type lectin receptor signaling pathway, legionellosis, and HIF-1 signaling pathway.
Fig. 7.
KEGG enrichment analysis of the overlapping DEGs.
Analysis of DEGs in pathways
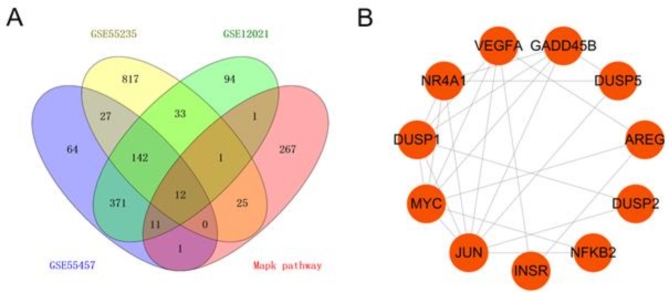
Gene information on the MAPK signaling pathway was collected from the GSEA database and KEGG database, and the intersection of genes in the MAPK pathway that had been identified in the three datasets was obtained (Fig. 8A). A total of 12 DEGs in the pathway were obtained and analyzed through a PPI network (Fig. 8B). We found three important targets: JUN, MYC, and VEGFA. These targets play important roles in the pathological process of OA by regulating the MAPK signaling pathway.
Fig. 8.
The overlapping DEGs in the MAPK pathway. Venn diagram of the overlapping DEGs in microarray data and MAPK pathway(A). PPI network analysis of the overlapping genes(B).
JUN, MYC, and VEGFA are abnormally elevated in the synovial tissue of OA patients and correlated with disease severity
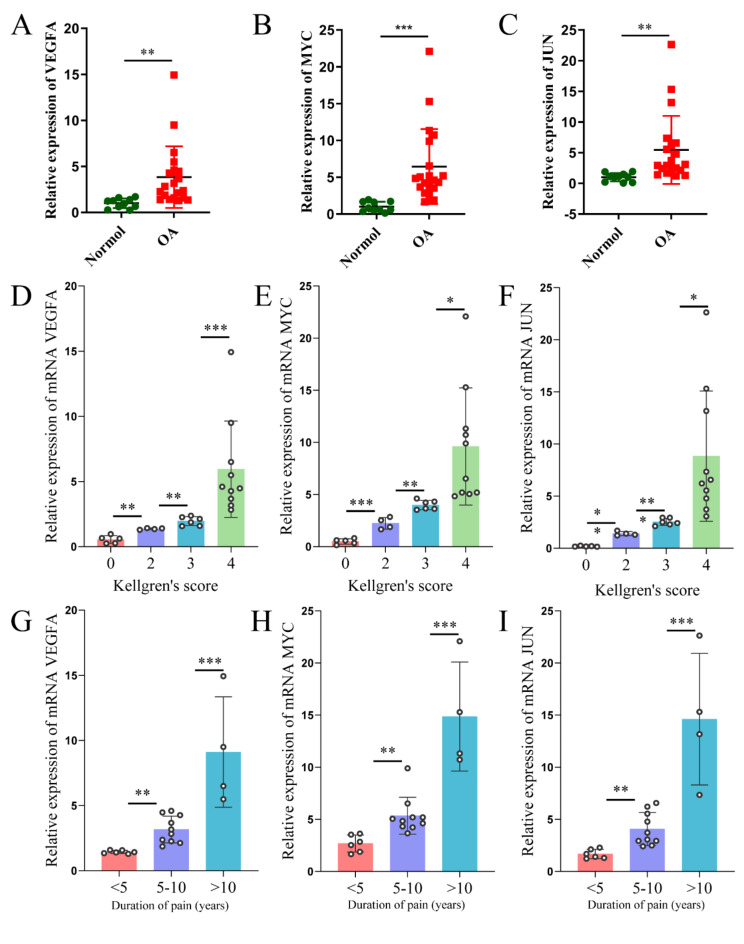
To investigate the differential mRNA expression of JUN, MYC, and VEGFA in OA, we examined synovial tissues from 20 OA patients and 10 healthy controls. Quantitative real-time PCR (qRT-PCR) results confirmed the bioinformatics findings, showing significantly elevated mRNA expression levels of JUN, MYC, and VEGFA in the synovial tissues of OA patients compared to controls (Fig. 9A-C). To assess the clinical relevance of these elevated levels, we analyzed their correlation with patients’ KL scores, age, and pain duration. The analysis revealed a significant positive correlation between JUN, MYC, and VEGFA mRNA expression levels and increasing K-L scores (Fig. 9D-F). However, no significant correlation was observed between age and the mRNA levels of these genes (S1A-C). Additionally, as the mRNA expression levels of JUN, MYC, and VEGFA increased, patients reported longer pain duration (Fig. 9G-I). In summary, JUN, MYC, and VEGFA are not only abnormally overexpressed in the synovial tissues of OA patients but also show a positive correlation with disease severity, as indicated by higher K-L scores and prolonged pain duration.
Fig. 9.
mRNA expression levels of VEGFA, MYC, and JUN in the synovial tissues of OA patients and healthy controls, and their correlation with OA severity. (A-C) Relative mRNA expression levels of (A) VEGFA, (B) MYC, and (C) JUN in synovial tissues from 20 OA patients and 10 healthy controls. (D–F) mRNA expression levels of VEGFA, MYC, and JUN in synovial tissues of patients with varying Kellgren-Lawrence (K–L) scores. (G–I) mRNA expression levels of VEGFA, MYC, and JUN in synovial tissues of patients with different durations of pain. Data are presented as mean ± SD, with statistical significance between normal and OA groups calculated using the Student’s t-test. Significance statistics for Kellgren-Lawrence scores and durations of pain using ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001.
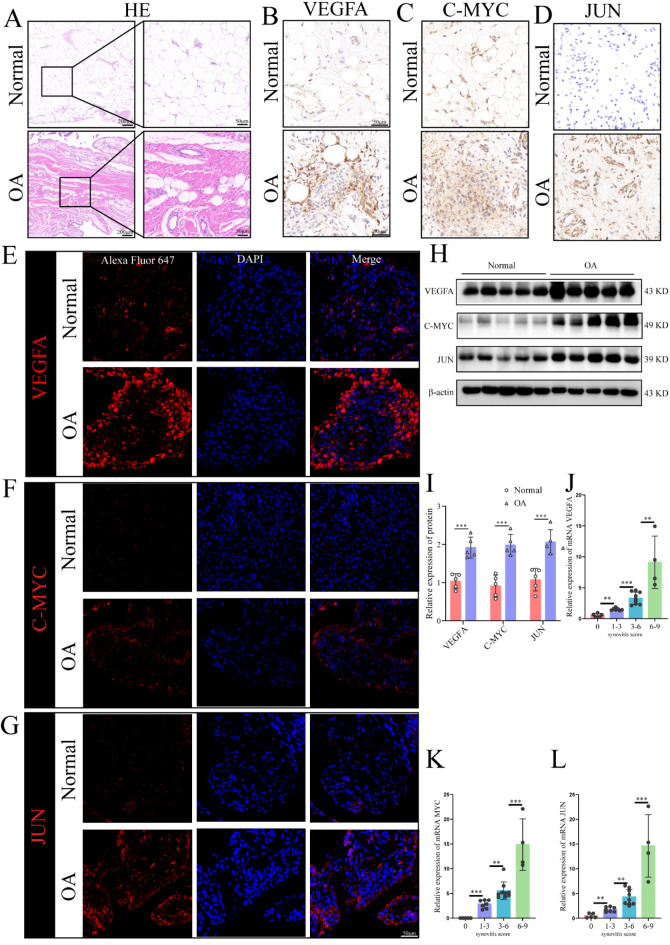
To further evaluate the differential expression of JUN, MYC, and VEGFA at the protein level, we employed immunohistochemistry, immunofluorescence, and western blot analyses on synovial tissues from five OA patients and five healthy controls. Hematoxylin and eosin (H&E) staining confirmed the presence of significant OA in the tissue, providing the basis for synovial scoring (Fig. 10A). Results from immunohistochemistry, immunofluorescence, and western blot analysis showed significantly higher protein expression levels of JUN, C-MYC, and VEGFA in the synovial tissues of OA patients compared to controls (Fig. 10B-I). Additionally, we examined the correlation between synovial scores and the mRNA expression levels of JUN, MYC, and VEGFA in corresponding patients. Our findings demonstrated that higher JUN, MYC, and VEGFA mRNA expression levels were associated with higher synovial scores (Fig. 10J-L).
Fig. 10.
Protein expression levels of C-MYC, VEGFA, and JUN in the synovial tissues of OA patients and healthy controls, and their correlation with synovitis severity. (A) Hematoxylin and eosin (H&E) staining of synovial tissues; (B–D) Immunohistochemical analysis of synovial tissues showing (B) VEGFA expression, (C) C-MYC expression, and (D) JUN expression; (E–G) Immunofluorescence staining of synovial tissues displaying (E) VEGFA expression, (F) C-MYC expression, and (G) JUN expression; (H,I) Western blot analysis of VEGFA, C-MYC, and JUN protein expression levels; (J-L) mRNA expression levels of VEGFA, C-MYC, and JUN in synovial tissues across different synovitis score ranges. *p < 0.05; **p < 0.01; ***p < 0.001. Data are presented as mean ± SD, with statistical significance determined by one-way ANOVA. Differential.
Expression and activation of p38-MAPK
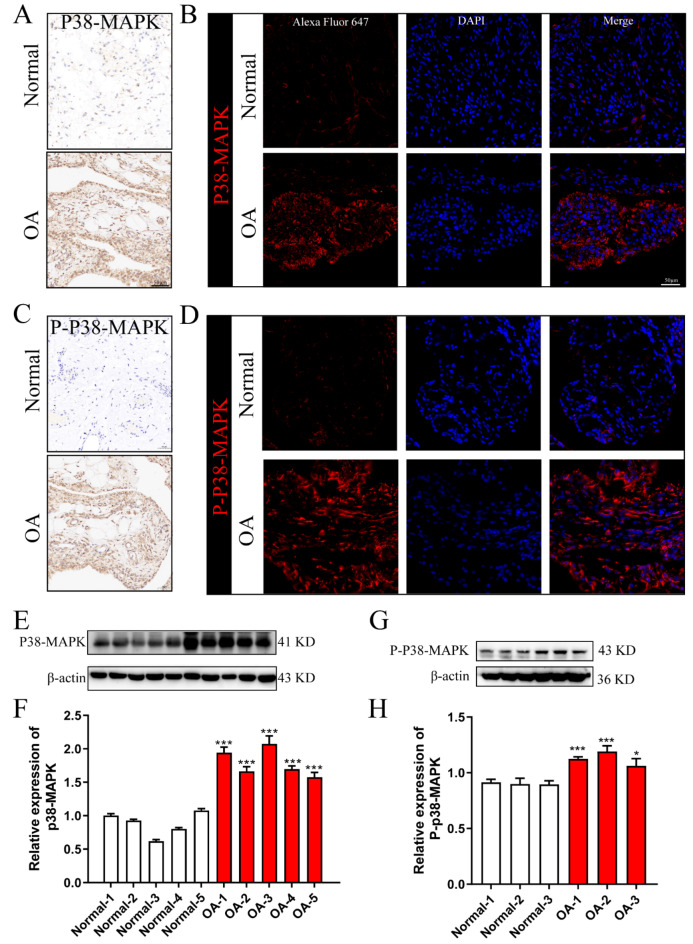
To determine whether p38-MAPK exhibits differential expression and activation in OA, we analyzed synovial tissue from OA patients and healthy controls. Immunohistochemistry, immunofluorescence, and western blotting were employed to assess the expression levels of both p38-MAPK and its activated form, phosphorylated p38-MAPK (P-p38-MAPK). The results revealed a significantly higher expression of p38-MAPK in the synovial tissues of OA patients compared to normal tissues (Fig. 11A,B,E,F). Similarly, the expression of P-p38-MAPK was markedly elevated in OA synovial tissues relative to controls (Fig. 11C,D,G,H). In summary, the expression and activation levels of P38-MAPK were markedly elevated in the synovial tissue of patients with OA, suggesting an intrinsic correlation between elevated p38 levels and the increased expression of myc, jun, and VEGF.
Fig. 11.
Protein expression and activation of p38-MAPK in synovial tissues from OA patients and healthy controls. (A) p38-MAPK protein expression detected by immunohistochemistry; (B) p38-MAPK protein expression detected by immunofluorescence; (C) Phosphorylated p38-MAPK (P-p38-MAPK) protein expression detected by immunohistochemistry; (D) P-p38-MAPK protein expression detected by immunofluorescence; (E,F) p38-MAPK protein expression levels assessed via Western blotting; (G,H) P-p38-MAPK protein expression levels assessed via Western blotting. *p < 0.05; **p < 0.01; ***p < 0.001. Data are presented as mean ± SD, and statistical significance was determined using one-way ANOVA.
Discussion
As a degenerative disease, OA is associated with a poor quality of life and can eventually lead to a loss of the ability to work49–51. However, its pathogenesis and risk factors are not clear. Most research has focused on chondrocytes, osteoblasts, and osteoclasts. Multiple studies have demonstrated that synovial tissue in individuals with osteoarthritis (OA) exhibits a heightened proliferative capacity compared to that of healthy individuals, accompanied by more pronounced inflammatory alterations and notable synovitis [16; 52; 53]. A particular study revealed that synovitis contributes to cartilage loss and exacerbates pain, where each 0.1 mm of cartilage lost over a 24-month period correlates with a Western Ontario and McMaster Universities Osteoarthritis Index (WOMAC) pain subscale score of 0.32 (95% CI: 0.21–0.44)54. Consequently, pathological modifications in synovial tissue and cells may hold significant implications for cartilage health and OA progression. Recent bioinformatics research has identified altered expression levels of several genes, including JUN, ATF3, VEGFA, FOSB, NR4A2, IL6, MYC, CDKN1A, IL1B, and EGR1, in the synovial tissues of OA patients, potentially linking them to the disease’s development52–56. These discoveries align with our findings and further emphasize the crucial roles of genes such as VEGFA, JUN, and MYC in OA. However, it is noteworthy that current research primarily relies on bioinformatics approaches, with limited laboratory studies reported to date.
We analyzed 30 patients with OA and 29 normal controls based on three datasets in GEO and identified 662 upregulated genes and 410 downregulated genes in OA. After obtaining the intersection of the three sets, we detected 64 upregulated genes and 84 downregulated genes in OA. The six proteins encoded by MYC, IL6, VEGFA, JUN, ATF3, and PTGS2 had more interactions based on a PPI network analysis and were closely associated with OA based on a ROC curve analysis. These are candidate biomarkers and may have important roles in OA. GO and KEGG pathway enrichment analyses revealed that the DEGs were involved in the MAPK signaling pathway. Based on the intersection between the MAPK signaling pathway and our DEG set, we obtained 12 indicators related to MAPK with significant expression differences in OA. Combined with the six key indicators of OA identified in the DEG analysis, three important targets were obtained, namely JUN, MYC, and VEGFA. Our study employs bioinformatics methods to ascertain the potential regulatory functions that Jun, c-Myc, and VEGFA may play in the context of OA. The alignment of our results with previous findings underscores the credibility of our discoveries56,57.
It is crucial to highlight that the majority of previous studies have predominantly focused on bioinformatics analyses, often lacking essential experimental validation. To enhance the clinical diagnosis and treatment strategies for OA, we meticulously collected 30 human synovial tissue samples for rigorous experimental validation. Employing a combination of multimodal techniques, including quantitative polymerase chain reaction (qPCR), western blotting, immunohistochemistry, and immunofluorescence, we thoroughly examined the expression profiles of Jun, c-Myc, and VEGFA at both the mRNA and protein levels. Our findings demonstrated a significant upregulation of Jun, c-Myc, and VEGFA in the synovial tissue of OA patients, suggesting that these genes may play a critical role in OA pathogenesis. To further investigate the clinical relevance of Jun, c-Myc, and VEGFA in OA diagnosis and treatment, we analyzed the correlation between their mRNA expression levels and key clinical parameters, including KL scores, patient age, and synovitis scores. Our results revealed no significant correlation between the mRNA expression levels of Jun, c-Myc, and VEGFA and patient age. However, a significant positive correlation was observed between the mRNA expression levels of these genes and both the KL scores and synovitis scores, indicating that their expression levels are associated with the severity of OA. This comprehensive experimental study addressed the current gap in empirical validation, particularly with respect to human samples. Moreover, our findings provide valuable insights into the molecular underpinnings of OA and establish a stronger foundation for improving the precision of clinical diagnosis and the development of targeted therapeutic interventions.
In pursuit of a more comprehensive comprehension of the intricate interplay among multiple DEGs, an exhaustive investigation was conducted. Through an exhaustive analysis of pertinent literature, it became evident that Jun, c-Myc, and VEGFA exhibit pronounced associations with the MAPK pathway3,16,39,49,56–62. Significantly, the MAPK signaling cascade is intrinsically implicated in the progression of OA. In its basal state, the MAPK pathway comprises three distinct signaling cascades, namely, c-Jun N-terminal kinase (JNK), p38, and extracellular signal-regulated kinase (ERK) pathways37. Activation of the MAPK pathway elicits the transcriptional upregulation of proinflammatory cytokines including TNF, interleukins (IL)-1, IL-6, alongside select chemokines. Furthermore, this activation prompts the expression of matrix metalloproteinases (MMPs) such as MMP-1, MMP-13, and cyclooxygenase-238–40. MAPK activation significantly contributes to cartilage collagen degradation, chondrocyte apoptosis, and inflammatory processes characteristic of OA41–43. Perturbation of the MAPK signaling pathway engenders aberrant synovial tissue proliferation, inflammation, and the development of heterogeneous pannus across the synovium. It is noteworthy that p38, an integral component of the MAPK pathway, is implicated in the pathogenesis of OA. Constituting a pivotal link in transmitting extracellular cues to cellular entities, the p38-MAPK pathway orchestrates a cascade involving diverse kinases56. Recent studies have underscored a mutual regulatory relationship between VEGFA, c-Myc, JUN, and the MAPK signaling pathways8–10,13,43,46,63,64. Based on these findings, we hypothesized that p38-MAPK exerts a substantial influence on the differential expression of Jun, c-Myc, and VEGFA in the OA context. To further explore this hypothesis, we conducted a comprehensive analysis of p38-MAPK protein expression in human synovial tissue. Western blot analysis, immunohistochemistry, and immunofluorescence revealed a significant increase—approximately 1.5-fold—in p38-MAPK expression in the synovial tissue of OA patients compared to healthy controls. Although this finding confirms the differential expression of p38-MAPK in OA, we postulate that the activation status of the p38-MAPK signaling pathway may provide more valuable insights into disease progression. Therefore, we assessed the levels of phosphorylated p38-MAPK (P-p38-MAPK) using immunohistochemistry, immunofluorescence, and western blotting. The results demonstrated a significant elevation in both P-p38-MAPK and total p38-MAPK expression. These findings indicate not only increased expression but also heightened activation of p38-MAPK in OA synovial tissue, supporting its role in the dysregulated expression of VEGFA, c-Myc, and JUN.
Conclusions
Our study found that the gene and protein expressions of MYC, VEGFA and JUN in the knee synovial tissue of OA group were significantly higher than those of control group. In addition, abnormal expression of JUN, Myc, and VEGFA may be associated with the p38-MAPK signaling pathway, ultimately leading to continued proliferation, inflammation, and angiogenesis of the OA synovium. Therefore, the combination therapy targeting the above three genes may bring certain progress for the clinical treatment of OA.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
SCL and RJJ conceived and designed the study. ZZY, ZWJ CMN drafted, edited, and revised the manuscript. All authors approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant number: 81472088).
Data availability
The data that support the findings of this study are available in ncbi at /www.ncbi.nlm.nih.gov/geo, reference number GSE55457, GSE55235, GSE12021. These data were derived from the following resources available in the public domain: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE55457, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE55235, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12021.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The study was conducted in accordance with the Declara-tion of Helsinki, and approved by the Ethics Committee of Anhui Medical University (S20210085 and date of approval 2021/03/31).Informed Consent Statement: Informed consent was obtained from all subjects involved in the study prior to participation.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Genxiang Rong, Zhenyu Zhang, and Wenjing Zhan contribute equally to this work.
Contributor Information
Jingjing Ruan, Email: ruan-jingjing@hotmail.com.
Cailiang Shen, Email: shencailiang@ahmu.edu.cn.
References
- 1.Loeser, R. F., Goldring, S. R., Scanzello, C. R. & Goldring, M. B. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum.64, 1697–1707 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glyn-Jones, S. et al. Osteoarthritis. Lancet386 376–387. (2015). [DOI] [PubMed] [Google Scholar]
- 3.Sharma, L. Osteoarthritis of the knee. N Engl. J. Med.384, 51–59 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Loeser, R. F. Aging and osteoarthritis: The role of chondrocyte senescence and aging changes in the cartilage matrix. Osteoarthr. Cartil.17, 971–979 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clockaerts, S. et al. The infrapatellar fat pad should be considered as an active osteoarthritic joint tissue: A narrative review. Osteoarthr. Cartil.18, 876–882 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Qin, J. et al. Lifetime risk of symptomatic Hand Osteoarthritis: The Johnston County Osteoarthritis Project. Arthritis Rheumatol.69, 1204–1212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter, D. J., March, L. & Chew, O. Lancet Commission on, Osteoarthritis in 2020 and beyond - authors’ reply. Lancet397, 1060 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Wang, Y. et al. Knee effusion volume assessed by magnetic resonance imaging and progression of knee osteoarthritis: Data from the Osteoarthritis Initiative. Rheumatology58, 246–253 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng, C. et al. Relative efficacy and safety of topical non-steroidal anti-inflammatory drugs for osteoarthritis: A systematic review and network meta-analysis of randomised controlled trials and observational studies. Br. J. Sports Med.52, 642–650 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang, W. et al. The efficacy and safety of Disease-modifying osteoarthritis drugs for knee and hip Osteoarthritis-a systematic review and network Meta-analysis. J. Gen. Intern. Med.36, 2085–2093 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mobasheri, A. & Batt, M. An update on the pathophysiology of osteoarthritis. Ann. Phys. Rehabil Med.59, 333–339 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Ebell, M. H. Osteoarthritis: Rapid Evid. Rev. Am. Fam Physician, 97 523–526. (2018). [PubMed] [Google Scholar]
- 13.Xia, B. et al. Osteoarthritis pathogenesis: A review of molecular mechanisms. Calcif Tissue Int.95, 495–505 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geyer, M. & Schonfeld, C. Novel insights into the pathogenesis of Osteoarthritis. Curr. Rheumatol. Rev.14, 98–107 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Bliddal, H. [Definition, pathology and pathogenesis of osteoarthritis]. Ugeskr Laeger 182 (2020). [PubMed]
- 16.Scanzello, C. R. & Goldring, S. R. The role of synovitis in osteoarthritis pathogenesis. Bone51, 249–257 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sacitharan, P. K. Ageing and osteoarthritis. Subcell. Biochem.91, 123–159 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Roos, E. M. Joint injury causes knee osteoarthritis in young adults. Curr. Opin. Rheumatol.17, 195–200 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Malfait, A. M. Osteoarthritis year in review 2015: Biology. Osteoarthr. Cartil.24, 21–26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, J. et al. Mitochondrial DNA damage is involved in apoptosis caused by pro-inflammatory cytokines in human OA chondrocytes. Osteoarthr. Cartil.18, 424–432 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Jordan, J. M. et al. Prevalence of knee symptoms and radiographic and symptomatic knee osteoarthritis in African americans and caucasians: The Johnston County Osteoarthritis Project. J. Rheumatol.34, 172–180 (2007). [PubMed] [Google Scholar]
- 22.Felson, D. T. et al. The prevalence of knee osteoarthritis in the elderly. The Framingham Osteoarthritis Study. Arthritis Rheum.30, 914–918 (1987). [DOI] [PubMed] [Google Scholar]
- 23.Felson, D. T. et al. Evidence for a mendelian gene in a segregation analysis of generalized radiographic osteoarthritis: Te Framingham Study. Arthritis Rheum.41, 1064–1071 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Dillon, C. F., Rasch, E. K., Gu, Q. & Hirsch, R. Prevalence of knee osteoarthritis in the United States: Arthritis data from the Third National Health and Nutrition Examination Survey 1991-94. J. Rheumatol.33, 2271–2279 (2006). [PubMed] [Google Scholar]
- 25.Anandacoomarasamy, A., Caterson, I., Sambrook, P., Fransen, M. & March, L. The impact of obesity on the musculoskeletal system. Int. J. Obes. (Lond). 32, 211–222 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Rim, Y. A. & Ju, J. H. The role of fibrosis in osteoarthritis progression. Life (Basel) 11 (2020). [DOI] [PMC free article] [PubMed]
- 27.Loeser, R. F., Collins, J. A. & Diekman, B. O. Ageing and the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol.12, 412–420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atukorala, I. et al. Synovitis in knee osteoarthritis: A precursor of disease? Ann. Rheum. Dis.75, 390–395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ayral, X., Pickering, E. H., Woodworth, T. G., Mackillop, N. & Dougados, M. Synovitis: a potential predictive factor of structural progression of medial tibiofemoral knee osteoarthritis -- results of a 1 year longitudinal arthroscopic study in 422 patients. Osteoarthr. Cartil.13, 361–367 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Collins, J. E. et al. Semiquantitative imaging biomarkers of knee osteoarthritis progression: Data from the Foundation for the National Institutes of Health Osteoarthritis Biomarkers Consortium. Arthritis Rheumatol.68, 2422–2431 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felson, D. T. et al. Synovitis and the risk of knee osteoarthritis: The MOST study. Osteoarthr. Cartil.24, 458–464 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacFarlane, L. A. et al. Association of Changes in Effusion-Synovitis with progression of cartilage damage over eighteen months in patients with osteoarthritis and Meniscal tear. Arthritis Rheumatol.71, 73–81 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roemer, F. W. et al. Presence of MRI-detected joint effusion and synovitis increases the risk of cartilage loss in knees without osteoarthritis at 30-month follow-up: The MOST study. Ann. Rheum. Dis.70, 1804–1809 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roemer, F. W. et al. Can structural joint damage measured with MR imaging be used to predict knee replacement in the following year? Radiology274, 810–820 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kciuk, M., Gielecinska, A., Budzinska, A., Mojzych, M. & Kontek, R. Metastasis and MAPK pathways. Int. J. Mol. Sci. 23 (2022). [DOI] [PMC free article] [PubMed]
- 36.Zeyen, L., Seternes, O. M. & Mikkola, I. Crosstalk between p38 MAPK and GR Signaling. Int. J. Mol. Sci.23 (2022). [DOI] [PMC free article] [PubMed]
- 37.Karin, M., Liu, Z. & Zandi, E. AP-1 function and regulation. Curr. Opin. Cell. Biol.9, 240–246 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Kappelmann, M., Bosserhoff, A. & Kuphal, S. AP-1/c-Jun transcription factors: Regulation and function in malignant melanoma. Eur. J. Cell. Biol.93, 76–81 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Shaulian, E. & Karin, M. AP-1 in cell proliferation and survival. Oncogene20, 2390–2400 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Blaney Davidson, E. N., van der Kraan, P. M. & van den Berg, W. B. TGF-beta and osteoarthritis. Osteoarthr. Cartil.15, 597–604 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Meyer, N. & Penn, L. Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer. 8, 976–990 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Pelengaris, S., Khan, M. & Evan, G. c-MYC: More than just a matter of life and death. Nat. Rev. Cancer2, 764–776 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Wu, Y. H. et al. Effects of microRNA-24 targeting C-myc on apoptosis, proliferation, and cytokine expressions in chondrocytes of rats with osteoarthritis via MAPK signaling pathway. J. Cell. Biochem.119, 7944–7958 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Ferrara, N. Vascular endothelial growth factor: Basic science and clinical progress. Endocr. Rev.25, 581–611 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Parikh, A. A. & Ellis, L. M. The vascular endothelial growth factor family and its receptors. Hematol. Oncol. Clin. North. Am.18, 951–971 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Zelzer, E. & Olsen, B. R. Multiple roles of vascular endothelial growth factor (VEGF) in skeletal development, growth, and repair. Curr. Top. Dev. Biol.65, 169–187 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Maharaj, A. S. & D’Amore, P. A. Roles for VEGF in the adult. Microvasc Res.74, 100–113 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma, K. et al. Targeting vascular endothelial growth factor receptors as a therapeutic strategy for Osteoarthritis and Associated Pain. Int. J. Biol. Sci.19, 675–690 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sulzbacher, I. Osteoarthritis: histology and pathogenesis. Wien Med. Wochenschr. 163, 212–219 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Miniaci, A. & Scarcella, M. J. Shoulder resurfacing for treatment of focal defects and diffuse osteoarthritis. Orthopade50, 112–118 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Cibulka, M. T. et al. Hip Pain and mobility deficits-hip osteoarthritis: Revision 2017. J. Orthop. Sports Phys. Ther.47, A1–A37 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Piaotao, C. et al. Exploration of effective biomarkers and infiltrating Immune cells in Osteoarthritis based on bioinformatics analysis. Artif. Cells Nanomed. Biotechnol.51 (2023). [DOI] [PubMed]
- 53.Jie, L. et al. Bioinformatics analysis to identify key genes and pathways influencing synovial inflammation in osteoarthritis. Mol. Med. Rep. 18 (2018). [DOI] [PMC free article] [PubMed]
- 54.Lin, X. & Ningji, G. Identification and verification of ferroptosis-related genes in the synovial tissue of osteoarthritis using bioinformatics analysis. Front. Mol. Biosci. 9 (2022). [DOI] [PMC free article] [PubMed]
- 55.JiangFei, Z. et al. Identification of aging-related biomarkers and immune infiltration characteristics in osteoarthritis based on bioinformatics analysis and machine learning. Front. Immunol. 14 (2023). [DOI] [PMC free article] [PubMed]
- 56.Lilei, X. et al. Identification of key hub genes in knee osteoarthritis through integrated bioinformatics analysis. Sci. Rep. 14 (2024). [DOI] [PMC free article] [PubMed]
- 57.Bacon, K., LaValley, M. P., Jafarzadeh, S. R. & Felson, D. Does cartilage loss cause pain in osteoarthritis and if so, how much? Ann. Rheum. Dis.79, 1105–1110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scott, L. J. Etanercept: A review of its use in autoimmune inflammatory diseases. Drugs74, 1379–1410 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Sluzalska, K. D. et al. Interleukin-1beta affects the phospholipid biosynthesis of fibroblast-like synoviocytes from human osteoarthritic knee joints. Osteoarthr. Cartil.25, 1890–1899 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Sur, S., Nakanishi, H., Steele, R. & Ray, R. B. Depletion of PCAT-1 in head and neck cancer cells inhibits tumor growth and induces apoptosis by modulating c-Myc-AKT1-p38 MAPK signalling pathways. BMC Cancer19, 354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wancket, L. M., Frazier, W. J. & Liu, Y. Mitogen-activated protein kinase phosphatase (MKP)-1 in immunology, physiology, and disease. Life Sci.90, 237–248 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, H., Shan, X. B. & Qiao, Y. J. PDK2 promotes chondrogenic differentiation of mesenchymal stem cells by upregulation of Sox6 and activation of JNK/MAPK/ERK pathway. Braz J. Med. Biol. Res.50, e5988 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Zeyen, L., Seternes, O. M. & Mikkola, I. Crosstalk between p38 MAPK and GR signaling. Int. J. Mol. Sci.23, 3322 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, Y., Xu, M., Zhang, X., Chu, F. & Zhou, T. MAPK/c-Jun signaling pathway contributes to the upregulation of the anti-apoptotic proteins Bcl-2 and Bcl-xL induced by Epstein-Barr virus-encoded BARF1 in gastric carcinoma cells. Oncol. Lett.15, 7537–7544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available in ncbi at /www.ncbi.nlm.nih.gov/geo, reference number GSE55457, GSE55235, GSE12021. These data were derived from the following resources available in the public domain: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE55457, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE55235, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12021.