Abstract
tRNAs, the adapter molecules in protein synthesis, also serve as metabolic cofactors and as primers for viral RNA-directed DNA synthesis. The genomic and subgenomic RNAs of some plant viruses have a 3′-terminal tRNA-like structure (TLS) that can accept a specific amino acid and serve as a site for initiation of replication and as a simple telomere. We report a previously undescribed role for the TLS of brome mosaic virus (BMV), and potentially for cellular tRNA, in mediating the assembly of its icosahedral virions. BMV genomic RNAs and subgenomic RNA lacking the TLS failed to assemble into virions when incubated with purified BMV coat protein. Assembly was restored by addition of a 201-nt RNA containing the BMV TLS. TLSs from two other plant viruses as well as tRNAs from wheat germ and yeast were similarly active in the BMV virion assembly reaction, but ribosomal RNA and polyadenylate did not facilitate assembly. Surprisingly, virions assembled from TLS-less BMV RNA in the presence of tRNAs or TLS-containing short RNA did not incorporate the latter molecules. Consistent with a critical role for the BMV TLS in virion assembly, mutations in the BMV genomic RNAs that were designed to disrupt the folding of the TLS also abolished virion assembly. We discuss the likely roles of the TLS in early stages of virion assembly.
Keywords: tRNA-like structure‖bromoviruses‖virus assembly‖RNA packaging
Transfer RNAs (tRNAs) are multifunctional. Their primary role is to be an adapter molecule that translates the codon sequences in mRNA into the amino acid sequence of a protein. tRNAs also participate in specialized functions in cellular metabolism such as biosynthesis of the bacterial cell wall (1), chlorophyll, and heme (2). tRNAs and tRNA-related activities are associated with a variety of RNA viruses, including several plant viruses and the retroviruses (3, 4). Host tRNAs found in the virions of retroviruses function as primers for RNA-directed DNA synthesis (3). The tRNA-like structures (TLSs) found at the 3′ end of the genomes of some plant viruses serve as efficient origins of replication and as primitive telomeres to ensure that the 3′-terminal CCA nucleotides are not lost during replication (4, 5). It has been suggested that these viral TLSs are molecular fossils that may relate to a primordial role for tRNA in RNA replication in the ancient RNA world (6).
Brome mosaic virus (BMV) is an example of an RNA virus that has an ≈170-nt-long TLS covalently bound to the 3′ end of its genomic and subgenomic RNAs. BMV is a member of the plant virus family Bromoviridae (7) and the alphavirus-like superfamily of human-, animal-, and plant-infecting positive-strand RNA viruses (8). Mature BMV virions encapsidate three genomic RNAs (B1–B3) and a single subgenomic RNA, B4 (7). Physical and biochemical data suggest that B1–B4 are packaged into three morphologically indistinguishable virus particles: B1 (3.2 kb) and B2 (2.9 kb) are packaged individually into separate particles, whereas the genomic B3 (2.1 kb) and the subgenomic B4 (0.9 kb) are copackaged into a single particle (9). All four BMV RNAs contain a strongly conserved TLS at their 3′ end that mimics the biochemical functions of tRNATyr (4), which is known to be aminoacylated during an infection in cells (10). The role of the aminoacylation of BMV RNA remains unknown, but the TLS serves as a 3′ telomere by recruiting the tRNA-specific host CCA nucleotidyltransferase to maintain intact 3′ CCA termini (11, 12). The TLS also contains signals necessary for minus-strand initiation and synthesis by the BMV replicase (5, 13).
BMV coat protein (CP) is composed of 189 amino acids and assembles into mature icosahedral virions with T = 3 quasisymmetry (9, 14). Purified RNA and CP subunits can be reassembled in vitro to produce infectious particles indistinguishable from those assembled in vivo (9, 15). Although empty capsids can assemble in vitro at low pH (≈5), they do not form in vitro under physiological conditions of low salt and neutral pH and are not observed in vivo (9, 16, 17). At neutral pH, various polyanions can act as nucleating agents to stimulate CP polymerization (17), and non-BMV RNAs can be assembled into capsids in vitro, but when BMV RNA is present in RNA mixtures, it is selectively encapsidated (18). Further, differential encapsidation has been discerned among the BMV RNAs in vitro, with RNA4, followed by RNA3, showing the greatest affinity for assembly (19).
These observations suggest that the encapsidation of BMV RNAs involves interaction with selective packaging signals in the viral RNAs. There is support for the existence of such signals within the coding region of RNA1 (20), but the basis for the selective encapsidation of BMV RNAs remains unknown. We describe in this report a crucial role for the 3′ TLS that can be satisfied in cis or in trans in the encapsidation of the BMV RNAs, and a possible role for cellular tRNAs in stimulating encapsidation.
Materials and Methods
Plasmid Construction and in Vitro Transcription.
The construction and characteristic features of full-length cDNA clones corresponding to the three genomic RNAs of BMV, pT7B1, pT7B2, and pT7B3, from which wild-type (wt) infectious RNAs 1 (B1), 2 (B2), and 3 (B3), respectively, can be transcribed in vitro have been described (12). Plasmid pT7B4 contains a cDNA copy of BMV RNA4 (B4) from which full-length transcripts of B4 identical to those found in wt BMV virions can be transcribed in vitro (21). Plasmid pT7B1H (22) is a variant clone of pT7B1 with an engineered HindIII site 201 nt upstream of the 3′ terminus, analogous to those naturally occurring in pT7B2 and pT7B3 (11, 12). Four variant clones, pT7B1/ΔTLS, pT7B2/ΔTLS, pT7B3/ΔTLS, and pT7B4/ΔTLS, were constructed by precisely deleting the 3′ 201-nt sequence as HindIII and BamHI fragments (11). Two mutations referred to as 5′Psk and 5′+3′Psk present within the TLS of pT7B3 (21) were transferred to pT7B2 and pT7B1H as HindIII and BamHI fragments. Because in vitro assembly of BMV RNA does not require a 5′ cap structure (15), noncapped transcripts (sometimes 32P-labeled) from wt and variant clones were synthesized in vitro by using T7 RNA polymerase (24). Plasmids pT73TR, pT73CT, and pT73TT, respectively, were used to synthesize transcripts encompassing the BMV TLS, cucumber mosaic virus (CMV), and tobacco mosaic virus (TMV; ref. 23). tRNAs from wheat germ and yeast (Sigma) used for trans complementation experiments were 5′-end-labeled with polynucleotide kinase (24).
CP Preparation, in Vitro Virion Assembly Assays, and Electron Microscopy (EM).
BMV virions were purified from symptomatic barley leaves as described (25). Preparation of CP subunits and in vitro assembly assays, performed in a buffer containing 50 mM Tris⋅HCl (pH 7.2), 50 mM NaCl, 10 mM KCl, 5 mM MgCl2, and 1 mM DTT, were done as described (21). Each assembly reaction (100 μl) contained 14 pmols of each genomic RNA and 3 μg of purified BMV CP subunits. For trans complementation experiments, 14 pmols of the viral TLS or nonviral RNA was also present, unless otherwise stated. Each in vitro assembled virion preparation was negatively stained with 1% uranyl acetate and examined with a Hitachi transmission electron microscope. Every experiment was repeated at least three times with independently prepared CP subunits and RNA substrates.
Quantification of in Vitro Assembly Efficiencies.
After assembly, virions were examined by EM and visually quantitated by counting within an area of 7.5 μm2 on each EM negative taken at a magnification of ×30,000. The average number of virions (from at least eight individual grid areas) assembled with purified CP and either virion RNA or wt transcripts was considered as the reference for 100% assembly efficiency. Virions assembled from purified CP subunits and BMV RNA variant sequences were subjected to a similar quantification procedure.
Virion RNA Analysis.
RNA was recovered from in vitro assembled virions as described (21). When 32P-labeled RNA transcripts were used in assembly reactions, virion RNA was denatured with glyoxal, electrophoresed on 1% agarose gels, and subjected to autoradiography (25). When nonradioactive transcripts were used, virion RNA was subjected to Northern blot hybridization analysis after glyoxal denaturation (25). The blot was subjected to hybridization with riboprobes for appropriate strand and species specificity (25). Reverse transcription (RT)-PCR was performed as described (21, 25).
Results
The TLS-Containing 3′ End Is Obligatory for the Assembly of BMV RNAs into Icosahedral Virions.
The observations that in vitro reassembled virions retain wt levels of infectivity and are physically indistinguishable from virions recovered from natural wt infections (9, 16) suggest that in vitro and in vivo assembly are fundamentally similar. In vitro assembly studies using wt CP subunits and variants of each BMV RNA should permit precise identification of sequences that are required for efficient assembly of virions in the context of an infection in cells. Throughout these studies, in vitro assembly assays were performed by using low salt and near-neutral pH (50 mM NaCl and pH 7.2) conditions that not only yield exclusively RNA-containing virions but also reflect a natural in vivo situation (21). Under these conditions, BMV CP selectively packages BMV RNAs from RNA mixtures (18).
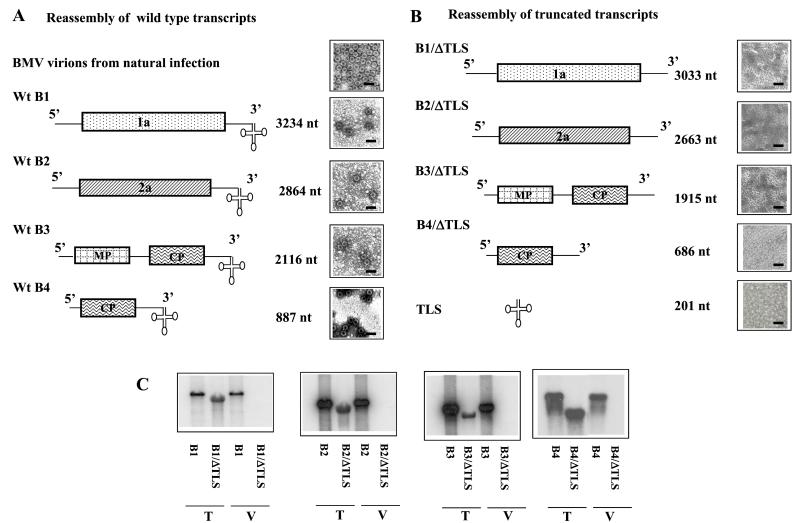
Despite the fact that the highly conserved TLS of the four BMV RNAs would be a likely candidate for harboring sequence elements involved in encapsidation, no experimental evidence exists to support this possibility (20). To examine the extent to which the TLS is involved in the assembly of viral RNAs, we began by assessing the in vitro packaging of each of the four BMV RNAs lacking the 3′ 201 nucleotides (ΔTLS). Noncapped, 32P-labeled transcripts of B1/ΔTLS, B2/ΔTLS, B3/ΔTLS, and B4/ΔTLS (Fig. 1B) were allowed to assemble in vitro with wt CP subunits. Control assembly assays were performed with the respective noncapped wt full-length transcripts. As shown in Fig. 1A, the interaction between wt CP subunits and each of the four wt full-length BMV RNAs resulted in the formation of icosahedral virions morphologically identical to native BMV virions (Fig. 1A). Efficient assembly of these wt transcripts into virions was confirmed by Northern blot assays (Fig. 1C). In contrast, each of the four BMV RNA transcripts lacking its TLS was unable to assemble into virions (Fig. 1 B and C and Fig. 2B). No virion assembly was observed when transcripts representing the 3′ 201-nt sequence was present as the only RNA (Fig. 1B). These observations were consistently reproduced in at least four independent assays performed with different transcript and dissociated CP subunit preparations.
Figure 1.
BMV RNAs lacking the 3′ 201 nt (ΔTLS) fail to assemble into virions. Schematic representations of plasmid clones of wt BMV genomic RNAs 1 (B1), 2 (B2), 3 (B3), and subgenomic RNA4 (B4) are shown in A, and their respective variants lacking the 3′ 201 nt (ΔTLS) are shown in B (B1/ΔTLS, B2/ΔTLS, B3/ΔTLS, and B4/ΔTLS). The cloverleaf structure at the 3′ end represents the highly conserved TLS. The noncoding sequences are represented as solid lines, with coding regions shown as rectangular boxes. B1 and B2 respectively encode 1a and 2a replication proteins. B3 encodes the 3a movement protein (MP) and coat protein (CP) expressed from subgenomic RNA B4. The lengths of wt BMV RNAs and their respective variant transcripts are shown. In each panel, electron micrographs display the negatively stained preparations of virions assembled in vitro from purified CP and individual 32P-labeled transcripts of either wt or a variant sequence (ΔTLS) corresponding to BMV RNAs B1–B4. (Scale bar = 50 nm.) Before EM analysis, the in vitro assembled virion preparations shown in A were diluted 500-fold, whereas those in B were undiluted. Note that wt BMV virions as well as RNA-containing virions assembled in vitro exhibit small dark centers because of the penetration of uranyl acetate (A). (C) Analysis of BMV RNAs assembled into virions. Autoradiograph of an agarose gel showing 32P-labeled transcripts (T) used in assembly assays and RNA recovered from in vitro assembled virions (V). RNA was denatured with glyoxal and subjected to electrophoresis in 1% agarose gels before autoradiography.
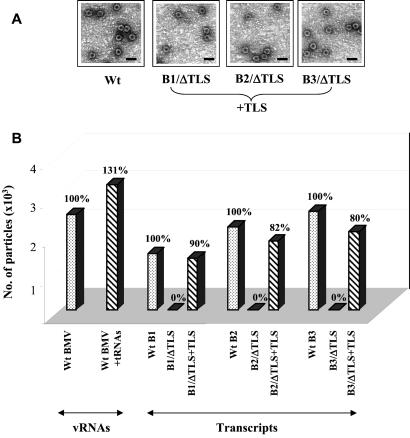
Figure 2.
The 3′ 201-nt (TLS RNA) of BMV RNA support encapsidation when present in trans. (A) Virions assembled in vitro with TLS RNA present in trans are morphologically normal. Electron micrographic images showing the negatively stained preparations of particles assembled in vitro from purified CP and transcripts of wt BMV RNA, or from CP in the presence of each 3′-truncated (ΔTLS) BMV genomic RNA as well as the B3 TLS RNA. (Scale bar = 50 nm.) For EM analysis, virions were diluted 500-fold before analysis. (B) Efficiency of cis vs. trans TLS-based assembly. Purified wt BMV CP subunits were allowed to assemble in vitro with either BMV virion RNA or individual transcripts. Stippled bars represent the number of assembled virions formed in the presence of the various genomic RNAs indicated; in each case the efficiency of assembly was assigned as 100%. Hatched bars represent the number of virions assembled for the indicated ΔTLS variants of the genomic RNAs in the presence or absence of trans-complementing wheat germ tRNAs or 3′ 201-nt-long BMV RNA (indicated as TLS); the percent efficiencies of assembly relative to the respective control are indicated.
Defective Assembly of Truncated BMV RNAs Is Rescued by Trans Complementation with Short TLS-Containing RNAs.
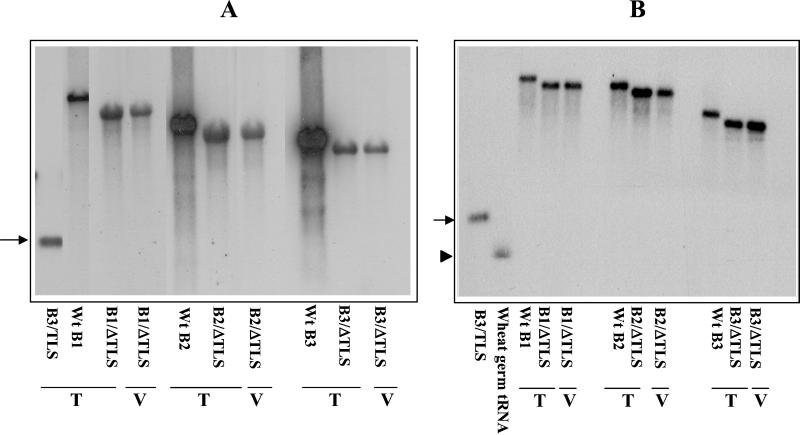
The above experiments demonstrated that the TLS is obligatory for assembly of each of the four BMV RNAs into virions. To further verify whether this TLS-dependent assembly is cis-regulated, 32P-labeled transcripts of the 3′ 201-nt sequence encompassing the TLS of B3 were synthesized from pT73TR (11) and added in equimolar ratio to an assembly reaction mixture containing transcripts of each truncated BMV RNA B1–B3 and wt CP subunits. Surprisingly, assembly in each case was restored, yielding virions indistinguishable from native BMV virions under the electron microscope (Fig. 2A). Quantitative analysis of reassembled virions demonstrated that the assembly efficiency of each truncated transcript mediated by the trans-complementing RNA was 80–90% relative to the control assays performed with full-length transcripts (Fig. 2B). The presence of the truncated RNA transcripts in these assembled virions was confirmed by agarose gel analysis (Fig. 3A). Stoichiometric analysis of trans-complementation by the B3 3′ 201-nt sequence (B3ΔTLS RNA) revealed that assembly of each truncated BMV RNA (present at 140 nM) could be achieved with subequimolar amounts of the 3′ RNA (assembly efficiency was 50% relative to control assays when B3ΔTLS RNA was present at 1% the concentration of genomic RNA; data not shown). These observations indicate that the defective assembly of truncated BMV RNAs shown in Fig. 1B was not caused by the reduction in the size of the transcripts but solely by the absence of the TLS-containing 3′ domain either in cis or in trans. To explore the essential properties provided in trans by the 3′ 201-nt fragment of BMV RNA, assembly experiments were performed with heterologous viral 3′ fragments. CMV is a tripartite icosahedral virus with genome organization similar to that of BMV (26), and belongs to a different genus within the same family. The TLS found on all three genomic and single subgenomic RNAs of CMV exhibits extensive similarities in primary and secondary structures to the BMV TLS (27), and likewise is specifically aminoacylated with tyrosine (4, 26). TMV is a rod-shaped virus with a TLS found at the 3′ end of its single genomic RNA that is distinct in primary and secondary structure from the BMV or CMV TLS; the TMV TLS is specifically aminoacylated with histidine (4). We wanted to examine whether 3′ fragments encompassing the TLS from these two heterologous viral genomes could functionally substitute for the BMV 3′ 201-nt RNA in mediating virion assembly. Two transcriptional plasmids, pT73CT (28) and pT7TT (23), carrying a cDNA copy corresponding to the TLS-containing 3′ end of CMV RNA3 and TMV genomic RNA, respectively, were used.
Figure 3.
Effect of trans complementation of TLS RNA (A) and wheat germ tRNAs (B) on the assembly of truncated BMV/ΔTLS RNAs. The indicated 32P-labeled in vitro transcribed RNAs were separated by agarose gel electrophoresis. RNAs run in lanes marked T show the transcripts of genomic RNAs used in assembly assays. RNAs run in lanes marked V show the genomic RNAs recovered from virions assembled in the presence of the trans-complementing B3 TLS RNA (A) or wheat germ tRNA (B). (A) The migration position of the 3′ 201-nt TLS RNA is indicated by an arrow. (B) The position of 32P-end-labeled wheat germ tRNA is indicated by an arrowhead. wt transcripts were loaded as size markers.
In vitro transcripts representing 3′ fragments of CMV RNA (197 nt) and TMV RNA (250 nt) were added to the independent assembly mix containing each truncated BMV RNA transcript and wt CP subunits. Both the CMV and TMV TLS-containing RNAs were able to complement in trans the assembly of truncated BMV RNAs. Furthermore, the efficiency with which the assembly occurred in the presence of these heterologous RNAs was indistinguishable from that of the homologous BMV 3′ 201-nt RNA, and the resultant virions were morphologically identical (data not shown). Given the fact that the 3′ ends of BMV and CMV RNAs display extensive similarities in their primary and secondary structures, the assembly of truncated BMV RNAs by the CMV-derived TLS RNA is not surprising. However, there is only minimal sequence and structural homology between the BMV and TMV RNA 3′ termini, indicating that the observed trans-complementation of assembly is not dependent on extensive defined RNA sequences. Because the obvious shared characteristic between the BMV, CMV, and TMV RNA 3′ ends is the TLS, our results suggested that the TLSs rather than other parts of the 3′ RNAs are responsible for the promotion of virion assembly. It is interesting to note that the observed role of the TMV 3′ RNA in in vitro assembly corroborates our previous in vivo observations that three subgenomic TMV RNAs terminating in the 3′ TMV TLS are packaged in plants by BMV CP expressed from a TMV-based vector (23). Apparently, some features shared between the BMV and TMV TLSs are recognized by the BMV CP subunits.
tRNAs from Nonviral Sources Are Equally Competent in Promoting Virion Assembly.
Although the 3′ end of BMV, CMV, and TMV genomic RNAs exhibit tRNA-like secondary structures and mimic several tRNA-associated activities, they are structurally distinguished from cellular tRNAs by the presence of a pseudoknotted acceptor stem (4). To verify whether the defective assembly of truncated BMV RNAs could be rescued by canonical tRNAs of nonviral origin, in vitro assembly assays similar to those described above were performed with tRNAs from plant (wheat germ) or nonplant (yeast) origin. Control assembly assays were performed with rRNA and transcripts of poly(A)50. Consistent with the requirement for a generic TLS as suggested by the experiments in the previous section, efficient virion assembly of truncated BMV RNAs by wt CP subunits was observed in the presence of wheat germ and yeast tRNAs, but not with either rRNA or poly (A)50. Interestingly, addition of wheat germ and yeast tRNAs to an assembly mix containing BMV virion RNAs stimulated the assembly and resulted in a 30% increase in the yield of virions (Fig. 2B). A similar synergistic effect on virion assembly was also observed with the BMV TLS-containing 3′ 201-nt RNA (data not shown).
Trans-Complementing TLS or tRNAs Are Not Packaged into Virions.
Three independent approaches were used to determine whether TLSs supplied in trans are copackaged with truncated BMV RNA transcripts. In the first approach, after assembly of truncated BMV RNAs complemented in trans by 32P-labeled viral TLS or tRNAs, virion RNAs were isolated, electrophoresed on 1% agarose gels and subjected to autoradiography. In the second approach, glyoxal-denatured encapsidated RNA was subjected to Northern hybridization using riboprobes specific for the TLS of BMV, CMV, or TMV. In the third approach, encapsidated RNA was subjected to RT-PCR using primers specific for each TLS (23, 28, 29). Agarose gel electrophoretic analysis revealed the presence of each truncated genomic RNA in assembled virions, but no indication of 32P-labeled TLS or wheat germ tRNAs supplied in trans (Fig. 3B). Likewise, no evidence was obtained by Northern blot or RT-PCR assays to indicate copackaging of TLS-containing 3′ RNA into virions. We conclude that, although TLSs and tRNAs efficiently promote virion assembly in trans, they are not copackaged to detectable levels into assembled virions.
TLS-Mediated Virion Assembly Depends on an Intact tRNA-Like Structure.
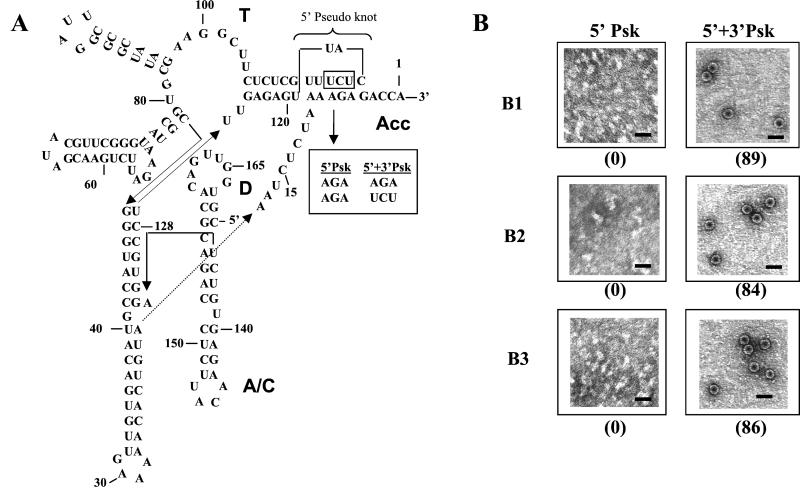
For BMV RNA, a tRNA-like conformation is essential to maintain near wt levels of minus-strand initiation by viral replicase as well as other tRNA-associated enzymatic activities (5, 30). Structural investigations performed with the BMV TLS revealed that a pseudoknot involving long-distance base pairing is obligatory in forming the aminoacyl acceptor stem and consequently to the overall tRNA-like conformation (Fig. 4A; see refs. 4 and 30). In a previously well-characterized mutant referred to as 5′Psk (Fig. 4A; ref. 30), three of the six base pairs that stabilize the acceptor stem pseudoknot were mutated. In another mutant, referred to as 5′+3′Psk, the pseudoknot-like conformation was restored by three compensating substitutions in the 5′Psk mutant (Fig. 4A; ref. 30). To test whether TLS-mediated virion assembly of BMV RNA is RNA structure- or sequence-dependent, 3′ 201-nt B3 sequences encompassing either the 5′Psk or 5′+3′Psk mutations were amplified in a PCR and subcloned into full-length B1, B2, and B3 cDNA clones. In vitro-synthesized transcripts of each of these mutant genomic clones were allowed to assemble with wt BMV CP subunits. Efficient assembly of virions was observed only with transcripts bearing the 5′+3′Psk mutation, but no virions were observed with those transcripts bearing the 5′Psk mutation (Fig. 4B). The defective packaging of each BMV RNA bearing the 5′Psk mutation was rescued by supplying transcripts of wt TLS in trans, in a fashion similar to that with truncated BMV RNAs (Fig. 4B). Collectively, these observations suggest that a tRNA-like conformation in the BMV RNAs plays a critical role in mediating the assembly of CP subunits into virions.
Figure 4.
An intact tRNA-like fold is obligatory for packaging. (A) Schematic representation of the secondary structure of the 3′ 155-nt TLS of BMV RNA3 (45). The location of the pseudoknot in the amino acid acceptor arm is shown by a bracket. The three nucleotides altered in mutant 5′Psk are enclosed in a stippled box; the mutations disrupt the pseudoknot by breaking 3 base pairs as indicated in the Inset. Mutation 5′+3′Psk is designed to reestablish these base-pairs and the pseudoknot. The domains analogous to those of canonical tRNAs are indicated. Acc, aminoacyl acceptor stem; T, TΨ arm; D, D-arm; A/C, anticodon arm. (B) In vitro assembly assays. Electron micrographic images showing the negatively stained preparations of particles assembled in vitro from purified CP and transcripts of each BMV genomic RNA bearing either the 5′Psk or 5′+3′Psk mutation. (Scale bar = 50 nm.) Virions assembled in the presence of 5′+3′Psk mutant RNA were diluted 500-fold before analysis, whereas those assembled in the presence of 5′Psk mutant RNA were undiluted. The number shown in parenthesis below each micrograph represents the percentage of assembly efficiency for each variant RNA sequence with respect to wt control transcripts.
Discussion
A Functional Role for the TLS in Virion Assembly.
Data presented in this paper show that BMV RNAs are packaged into icosahedral virions of T = 3 symmetry only if sequences encompassing the TLS are present either in cis or in trans. Our finding represents a previously undescribed role for the BMV TLS in virion assembly, in addition to functioning as an initiation site for replication and a simple telomere (4, 5, 11). Apart from the bromoviruses, the genomes of several genera of plant viruses and of an insect tetravirus have also been shown to contain TLSs (4, 5, 31, 32). Studies with these viruses have led to several other proposed functions for RNA viral TLSs (4, 5), but their involvement in capsid formation had not yet been experimentally demonstrated.
The contrasting abilities of the 5′Psk and 5′+3′Psk mutants to support encapsidation of BMV RNAs (Fig. 4B) indicate the requirement for an intact tRNA-like fold. There is no evidence for a BMV sequence-specific requirement concerning the TLS, however, because no specific interaction between BMV CP and the TLS is discernable in vitro (20) and BMV CP is able to encapsidate RNAs with a TMV 3′ TLS in vivo (23). Furthermore, the ability of 3′ RNAs bearing the CMV and TMV TLSs—and even of cellular tRNAs—to support in vitro encapsidation as effectively as homologous BMV sequences when present in trans, supports the recognition during encapsidation of a generic tRNA-like RNA domain rather than BMV RNA-specific features. It is interesting to consider whether the TLSs of other viruses are involved in capsid formation. Significantly, short subgenomic RNAs about 300 nt long from the 3′ ends of CMV and tomato aspermy virus RNAs are very efficiently copackaged into virions with genomic RNAs (33, 34), perhaps reflecting a TLS involvement in packaging in this sister genus to the bromoviruses.
Cellular tRNAs have in some instances been reported in the icosahedral capsids of RNA viruses. Two to three molecules of an RNA with the properties of tRNALys, with smaller amounts of other tRNAs, were reported in the top component (low buoyant density) particles of eggplant mosaic virus (35). Virion preparations from other tymoviruses also contain RNAs (presumably tRNAs) capable of accepting a range of amino acids other than the valine bound by the viral RNA (36). However, it is not known in these cases whether the tRNAs are accidentally copackaged or whether their presence reflects active selection by the viral CP. In any case, the situation differs from that which we have described here for BMV. Unlike BMV, tymoviruses are characterized by the occurrence of top component capsids lacking the genomic RNA, and there is no evidence for the inclusion of cellular tRNAs in the infectious capsids (35). Furthermore, the remarkable discovery that the TLS or tRNA can be provided in trans (Fig. 2) enabled us to demonstrate that the crucial involvement of short tRNA-like molecules is transient, because they are not present in the assembled capsids (Fig. 3).
Possible Mechanistic Role Played by the TLS and Perhaps Host tRNAs in Virion Assembly.
The transient yet critical involvement of the TLS in BMV assembly suggests a role in nucleating a higher-order arrangement of CP dimers that serves as an intermediate on the encapsidation pathway. Unlike capsids such as those of the tymoviruses, which are stabilized by strong protein–protein interactions, the assembly of BMV and other bromovirus virions is characterized at neutral pH by a strong involvement of polyanionic nucleating agents. RNA is presumably the physiological nucleating agent, but this role may be served even by the polymers poly(vinyl sulfate) or dextran sulfate in vitro (37).
If the assembly of BMV capsids at neutral pH proceeds by means of the pentameric complexes of dimers that have been implicated in the RNA-free assembly of cowpea chlorotic mottle virus capsids at low pH in vitro (38), then the TLS may serve to stabilize these pentamers or productive complexes of pentamers in favor of nonproductive aggregates that fail to associate with a genomic RNA and progress to full virions. In so helping to avoid kinetic traps that are a hallmark of the so-called EQ model of assembly used by the bromoviruses (38), the TLS may be viewed to act in the role of a chaperone. When this role is provided in trans, i.e., when a discrete TLS RNA or tRNA is present but is not encapsidated, this role as chaperone is clearly transient and possibly catalytic, because substoichiometric levels of TLS remain effective in supporting assembly. The transience of the interaction with the TLS is consistent with the absence of a tight binding site as assessed in vitro (20).
Specific RNA features that interact with CP and are involved in the assembly of icosahedral virions have been described for Sindbis virus (39) and turnip crinkle virus (40, 41), both monopartite viruses. Our studies with the tripartite BMV extend the observations of Duggal and Hall (20), who described a selective CP interaction domain in the coding region of BMV RNA1 and showed that this domain and others like it postulated to exist on RNAs 2 to 4 are not sufficient for encapsidation. We propose that the selective recognition of these domains occurs after or in concert with a TLS/CP interaction. The involvement in encapsidation of a common feature such as the TLS, which is present on each BMV RNA, would assist the selection of viral RNAs for encapsidation, even though the interaction itself is not highly specific (20). This specificity filter may be particularly important for BMV, a virus whose encapsidation pathway is not geared in cis to package newly replicated RNAs (42) as is the case for poliovirus (43).
The ability of tRNA to support the encapsidation of BMV RNA in trans raises the possibility that these host RNAs play a role in assembly in the cell. Significantly, we (Fig. 2B) and others (18) have observed stimulation of virion assembly in vitro when tRNAs are added to an assembly mix containing wt BMV CP subunits and RNAs (Fig. 2B). Perhaps tRNAs could stimulate encapsidation in vivo at early times when the concentrations of viral components are comparatively low, or serve to chaperone productive capsid formation late in the infection when high concentrations of CP could lead to nonproductive aggregates. Because BMV RNAs devoid of the TLS do not replicate in vivo (44), we have been unable at present to test this suggested involvement of tRNA in vivo. However, host tRNAs may have been instrumental in permitting a BMV CP mRNA lacking its TLS to encapsidate into icosahedra in yeast cells (42). Further studies addressing the role of tRNA-like domains in BMV assembly are needed to provide valuable insight into the mechanism of RNA packaging in viruses with icosahedral symmetry and the intriguing possible involvement of cellular tRNAs in this process.
Acknowledgments
We thank George Grantham for excellent technical help, Dan Gallie for providing a cDNA clone of poly(A)50 sequence, Cheng Kao for helpful discussions, and Allan Dodds, Shou-Wei Ding, and Mark Young for assistance with the manuscript. This research was supported by a grant from United States Department of Agriculture National Research Initiative Competitive Grants Program (9935303).
Abbreviations
- TLS
tRNA-like structure
- BMV
brome mosaic virus
- CP
coat protein
- CMV
cucumber mosaic virus
- TMV
tobacco mosaic virus
- RT
reverse transcription
- wt
wild type
- EM
electron microscopy
References
- 1.Strominger J L. Harvey Lect. 1970;64:179–213. [PubMed] [Google Scholar]
- 2.Jahn D, Verkamp E, Söll D. Trends Biochem Sci. 1992;17:215–218. doi: 10.1016/0968-0004(92)90380-r. [DOI] [PubMed] [Google Scholar]
- 3.Marquet R, Isel C, Ehresmann C, Ehresmann B. Biochimie. 1995;77:113–124. doi: 10.1016/0300-9084(96)88114-4. [DOI] [PubMed] [Google Scholar]
- 4.Florentz C, Giegé R. In: tRNA: Structure, Biosynthesis and Function. Söll D, RajBhandary U L, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 141–163. [Google Scholar]
- 5.Dreher T W. Annu Rev Phytopathol. 1999;37:151–174. doi: 10.1146/annurev.phyto.37.1.151. [DOI] [PubMed] [Google Scholar]
- 6.Weiner A M, Maizels N. Proc Natl Acad Sci USA. 1987;84:7383–7387. doi: 10.1073/pnas.84.21.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao A L N. In: Encyclopedia of Plant Pathology. Maloy O C, Murray T D, editors. Toronto: Wiley; 2001. pp. 155–158. [Google Scholar]
- 8.Ahlquist P. In: Encyclopedia of Virology. Webster R G, Granoff A, editors. San Diego: Academic; 1994. pp. 181–185. [Google Scholar]
- 9.Lane L C. Adv Virus Res. 1974;19:151–220. doi: 10.1016/s0065-3527(08)60660-0. [DOI] [PubMed] [Google Scholar]
- 10.Loesch-Fries L S, Hall T C. Nature (London) 1982;298:771–773. [Google Scholar]
- 11.Rao A L N, Dreher T W, Marsh L E, Hall T C. Proc Natl Acad Sci USA. 1989;86:5335–5339. doi: 10.1073/pnas.86.14.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreher T W, Rao A L N, Hall T C. J Mol Biol. 1989;206:425–438. doi: 10.1016/0022-2836(89)90491-9. [DOI] [PubMed] [Google Scholar]
- 13.Chapman M, Kao C C. J Mol Biol. 1999;286:709–720. doi: 10.1006/jmbi.1998.2503. [DOI] [PubMed] [Google Scholar]
- 14.Speir J A, Munshi S, Wang S, Baker T S, Johnson J E. Structure (London) 1995;3:63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao X, Fox J M, Olson N H, Baker T S, Young M J. Virology. 1995;207:486–494. doi: 10.1006/viro.1995.1108. [DOI] [PubMed] [Google Scholar]
- 16.Fox J M, Johnson J E, Young M J. Semin Virol. 1994;5:51–60. [Google Scholar]
- 17.Bancroft J B. Adv Virus Res. 1970;16:99–134. doi: 10.1016/s0065-3527(08)60022-6. [DOI] [PubMed] [Google Scholar]
- 18.Cuillel M, Herzog M, Hirth L. Virology. 1979;95:146–153. doi: 10.1016/0042-6822(79)90409-4. [DOI] [PubMed] [Google Scholar]
- 19.Herzog M, Hirth L. Virology. 1983;86:48–56. doi: 10.1016/0042-6822(78)90006-5. [DOI] [PubMed] [Google Scholar]
- 20.Duggal R, Hall T C. J Virol. 1993;67:6406–6412. doi: 10.1128/jvi.67.11.6406-6412.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi Y G, Rao A L N. Virology. 2000;275:207–217. doi: 10.1006/viro.2000.0513. [DOI] [PubMed] [Google Scholar]
- 22.Duggal R, Rao A L N, Hall T C. Virology. 1992;187:261–270. doi: 10.1016/0042-6822(92)90314-f. [DOI] [PubMed] [Google Scholar]
- 23.Choi Y G, Rao A L N. Virology. 2000;275:249–257. doi: 10.1006/viro.2000.0532. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Rao A L N, Duggal R, Lahser F C, Hall T C. In: Methods in Molecular Genetics: Molecular Virology Techniques. Adolph K W, editor. Vol. 4. Orlando, FL: Academic; 1994. pp. 216–236. [Google Scholar]
- 26.Palukaitis P, Roossinck M J, Dietzgen R G, Francki R I B. Adv Virus Res. 1992;41:281–348. doi: 10.1016/s0065-3527(08)60039-1. [DOI] [PubMed] [Google Scholar]
- 27.Ahlquist P, Dasgupta R, Kaesberg P. Cell. 1981;23:183–189. doi: 10.1016/0092-8674(81)90283-x. [DOI] [PubMed] [Google Scholar]
- 28.Schmitz I, Rao A L N. Virology. 1998;248:323–331. doi: 10.1006/viro.1998.9257. [DOI] [PubMed] [Google Scholar]
- 29.Rao A L N, Grantham G L. Virology. 1996;226:294–305. doi: 10.1006/viro.1996.0657. [DOI] [PubMed] [Google Scholar]
- 30.Dreher T W, Hall T C. J Mol Biol. 1988;201:41–56. doi: 10.1016/0022-2836(88)90437-8. [DOI] [PubMed] [Google Scholar]
- 31.Goodwin J B, Dreher T W. Virology. 1998;246:170–178. doi: 10.1006/viro.1998.9193. [DOI] [PubMed] [Google Scholar]
- 32.Gordon K H J, William M R, Hendry D A, Hanzlik T N. Virology. 1995;258:42–53. doi: 10.1006/viro.1999.9677. [DOI] [PubMed] [Google Scholar]
- 33.Blanchard C L, Boyce P M, Anderson B J. Virology. 1996;217:598–601. doi: 10.1006/viro.1996.0155. [DOI] [PubMed] [Google Scholar]
- 34.Shi B-J, Ding S-W, Symons R H. J Gen Virol. 1997;78:505–510. doi: 10.1099/0022-1317-78-3-505. [DOI] [PubMed] [Google Scholar]
- 35.Bouley J, Briand J, Genevaux M M, Pink M, Witz J. Virology. 1976;69:775–781. doi: 10.1016/0042-6822(76)90505-5. [DOI] [PubMed] [Google Scholar]
- 36.van Belkum A, Bingkun J, Rietveld K, Pleij C W A, Bosch L. Biochemistry. 1987;26:1144–1151. [Google Scholar]
- 37.Bancroft J B, Hiebert E, Bracker C E. Virology. 1969;39:924–930. doi: 10.1016/0042-6822(69)90029-4. [DOI] [PubMed] [Google Scholar]
- 38.Zlotnick A, Aldrich R, Johnson J M, Ceres P, Young M J. Virology. 2000;277:450–456. doi: 10.1006/viro.2000.0619. [DOI] [PubMed] [Google Scholar]
- 39.Weiss B, Geigenmuller-Gnirke U, Schlesinger S. Nucleic Acids Res. 1994;22:780–786. doi: 10.1093/nar/22.5.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei N, Morris T J. J Mol Biol. 1991;222:437–443. doi: 10.1016/0022-2836(91)90483-m. [DOI] [PubMed] [Google Scholar]
- 41.Wei N, Heaton L A, Morris T J, Harrison S C. J Mol Biol. 1991;214:85–95. doi: 10.1016/0022-2836(90)90148-F. [DOI] [PubMed] [Google Scholar]
- 42.Krol M A, Olson N H, Tate J, Johnson J E, Baker T S, Ahlquist P. Proc Natl Acad Sci USA. 1999;96:13650–13655. doi: 10.1073/pnas.96.24.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nugent C I, Johnson K L, Sarnow P, Kirkegaard K. J Virol. 1999;73:427–435. doi: 10.1128/jvi.73.1.427-435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao A L N, Hall T C. J Virol. 1990;64:2437–2441. doi: 10.1128/jvi.64.5.2437-2441.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Felden B, Florentz C, Westholf E, Geige R. Biochem Biolphys Res Commun. 1998;243:426–434. doi: 10.1006/bbrc.1997.7753. [DOI] [PubMed] [Google Scholar]