Abstract
Activation of androgen receptor (AR) via androgen in muscle cells has been closely linked to their growth and differentiation. Here, we report the cloning and characterization of supervillin (SV), a 205-kDa actin-binding protein, as an AR coregulator from the skeletal muscle cDNA library. Mammalian two-hybrid and glutathione S-transferase pull-down assays indicate a domain within SV (amino acids 594-1268) can interact with AR N terminus and DNA-binding domain–ligand-binding domain in a ligand-enhanced manner. Subcellular colocalization studies with fluorescence staining indicate SV can colocalize with AR in the presence of 5α-dihydrotestosterone in COS-1 cells. The functional reporter assays showed full-length SV and the SV peptide (amino acids 831-1281) within the interaction domain can enhance AR transactivation. Furthermore, SV can enhance the endogenous AR target gene, p27KIP1, expression in prostate PC-3(AR2) cells. SV preferentially enhanced AR rather than other tested nuclear receptors and could be induced by natural androgens better than other steroids. SV can also cooperate with other AR coregulators, such as ARA55 or ARA70, to enhance AR transactivation further. Unlike SRC-1 that can enhance the interaction between AR N terminus and AR C terminus, SV shows a mild suppressive effect on N–C interactions, suggesting SV may go through a different mechanism to enhance AR transactivation. Together, our data demonstrate that SV is an AR coregulator that can enhance AR transactivation in muscle and other cells.
Androgen receptor (AR) belongs to the steroid receptor superfamily, which is activated by androgens, testosterone (T), or 5α-dihydrotestosterone (DHT), to regulate gene transcription. After AR binds to androgens, it dissociates from chaperone proteins with subsequent processes, including nuclear translocation, dimer formation, and DNA response element binding, that result in regulation of its target genes (1). Since 1995, yeast two-hybrid system and complex pull-down methods have discovered several coregulators of steroid receptors. Many of these coregulators have either intrinsic histone acetylation/deacetylation transferase activity, or are able to recruit proteins with histone acetylation/deacetylation transferase activities to regulate the transcriptional activity of the receptor (2). Other coregulators modulate receptor functions by posttranslational modifications, such as phosphorylation or sumoylation (3, 4). In addition, coregulators can also modulate receptors' activity by interfering with their nuclear translocation (5, 6). Overall, various coregulators can modulate steroid receptor activity through multiple mechanisms.
Because the expression of coregulators varies among different cell types, AR functions depend on the availability of expressed coregulators in the same cell. Although it is well documented that SRC-1 can enhance estrogen receptor (ER) transactivation in many reporter assays, immunohistochemistry studies demonstrated that SRC-1 and ER are not located in the same subset of epithelial cells within the adult mammary gland (7). This finding excludes any possibility for SRC-1 to bind to ER and modulate ER function in those cells. Moreover, FHL2 and ARIP3 are two AR coregulators reported to express mostly in myocardium and testes, respectively (8, 9). It is therefore important to identify which AR coregulators are present in various androgen target organs.
Skeletal muscle is an AR target organ (10, 11). To understand how T induces AR function in skeletal muscle, we applied yeast two-hybrid screening to identify T-responsive AR-interacting proteins from the skeletal muscle cDNA library. One of the clones identified from this screening encodes the partial sequence of supervillin (SV).
SV is an actin-binding protein identified from blood cells. In addition to blood cells, it also expresses in muscle-enriched tissues, especially skeletal muscles, and several cancer cell lines (12). The roles of SV in muscle and cancer are still under investigation. Although its C terminus shows high homology to gelsolin and villin (13), functional domain studies determined that the N terminus of SV represents the strong actin-binding activity (14). The nuclear localization signal located in the middle of this protein is functional and may contribute to its nuclear translocation (14). However, the functions of SV in the cytoskeleton network and the nucleus remain unclear. Early studies also found that SV is a T-down-regulated gene in dermal papilloma cells, which may contribute to male baldness syndrome (15). Recently, the use of systematic RNA-mediated interference in Caenorhabditis elegans has demonstrated that the SV homologue plays a role in sex determination (16). Here we present SV as an AR-interacting protein and demonstrate that SV can function as an AR coregulator by enhancing AR transactivation.
Materials and Methods
Expression Plasmids.
pCMX-VP16-hSVn and pCMX-VP16-hSVc were constructed by releasing fragments from pACTII-hSV(558–1788) by using restriction enzyme digestion and inserted into the pCMX-VP16 vector. pEGFP-bSV, pEGFP-bSV(831–1792), pEGFP-bSV(1010–1792), and pEGFP-bSV(831–1281) were kindly provided by E. J. Luna, Univ. of Massachusetts Medical School, Worcester. pSG5-bSV was constructed by inserting bSV cDNA, which was released from pEGFP-bSV, into the pSG5 vector. The p(ARE)4-Luc plasmid has been described (17). The pGL3-PSA6.0Luc plasmid was kindly provided by A. Mizokami (Kanazawa University, Kanazawa, Japan).
Yeast Two-Hybrid Screening.
A fusion protein (Gal4-AR) containing Gal4 DNA-binding domain [Gal4(DBD)] and C terminus of AR (amino acids 595–918) was used as bait to screen from 3 × 106 transformants of MATCHMAKER human skeletal muscle library (CLONTECH). Transformants were selected for growth on nutrition selection plates containing synthetic dropout media lacking histidine, leucine, and tryptophan (−3SD) with 25 mM 3-aminotriazole and 10 nM T. The yeast was cultured in a humidified 30°C chamber for 3 days. Colonies were also filter-assayed for β-galactosidase activity. Plasmids isolated from candidate clones were cotransformed into Y190 with bait, and the ligand-dependent interaction was then confirmed further by filter assay for β-galactosidase activity with EtOH or 10 nM T treatment. The plasmid pACTII or pACTII-SV(558–1788) was cotransformed into yeast with bait and plated on −2SD plates (lacking leucine and tryptophan). The yeast colonies that grew on −2SD plates were selected and plated on −3SD plates with or without 10 nM DHT to test for growth ability.
Cell Culture and Transfection.
Mouse myoblast cell line (C2C12), human prostate cancer cell lines (PC-3 and DU145), and monkey kidney fibroblast cell line (COS-1) were maintained in DMEM containing penicillin (25 units/ml), streptomycin (25 mg/ml), and 10% FBS. In mammalian two-hybrid assay, transfections were performed by the calcium phosphate precipitation method as described (15). In brief, 1.5–3 × 105 cells were plated on 35-mm dishes for 24 h, and the medium was changed to DMEM containing 10% charcoal-dextran-stripped FBS (CD-FBS) 2 h before transfection. Cells were transfected with 0.5-μg plasmids expressing Gal4(DBD) and VP-16 fusion proteins as indicated. The Gal4 response element-controlled Firefly luciferase expression plasmid, pG5-Luc, was used as reporter gene. A Renilla luciferase expression plasmid pRL-SV40 was used as an internal control for transfection efficiency. The total amount of DNA was adjusted to 5 μg with pCMX-VP16 vectors. After 16 h transfection, cells were treated with ligands as described for another 24 h.
In AR transactivation activity assays, transfections were performed by using SuperFect (Qiagen, Chatsworth, CA) following protocols described in the manual provided by Qiagen. Cells were plated on 35-mm dishes and after 24 h were transfected by using the SuperFect kit. The total DNA amount was adjusted to 2 μg with pSG5 or pEGFP vectors. The medium was changed to DMEM with 10% CD-FBS 2 h after transfection. After 24 h, the DMEM with 10% CD-FBS was changed again, and the cells were treated with various steroids. Cells were harvested after 24 h for dual-luciferase assay as described in the protocol provided by Promega. At least three independent experiments were performed in each case.
Glutathione S-Transferase (GST) Pull-Down Assay.
GST-ARN, GST-AR-DBD-ligand-binding domain (LBD) (AR-DL) fusion proteins, and GST control protein were purified as instructed by the manufacturer (Amersham Pharmacia). Plasmids containing GST-fusion protein-expressing cDNA were transformed into the BL21(DE3)pLysS bacteria strain and selected for ampicillin- and chloramphenicol-resistant colonies. Selected colonies were grown in LB medium (bacteria expressing GST-AR-DL were cultured under 1 μM DHT treatment) at 30°C until OD600 reached 0.6 to 1. Then 0.4 mM isopropyl β-D-thiogalactopyranoside was added into medium for 3 h. Bacteria were lysed by three cycles of freezing-thawing in NETN buffer (20 mM Tris, pH 8.0/100 mM NaCl/6 mM MgCl2/1 mM EDTA/0.5 mM Nonidet P-40/1 mM DTT/8% glycerol/1 mM PMSF). Lysed bacteria were spun down, and the supernatants were collected. The GST fusion proteins were pulled down by glutathione beads in 4°C for 1 h and then washed three times with NETN buffer. The purified GST fusion proteins and beads were suspended in 100 μl NETN buffer. Resuspended GST proteins and beads were incubated with 5 μl in vitro-translated 35S-methionine-labeled VP16-hSVn or VP16-hSVc expressed from pCMX-VP16-hSVn or pCMX-VP16-hSVc by TNT-coupled reticulocyte lysate system (Promega). After incubating for 1 h at 4°C in the presence or absence of 1 μM DHT, glutathione beads were washed with NETN buffer four times and then the protein complexes were loaded in SDS/PAGE and visualized by using a PhosphorImager (Molecular Dynamics).
Immunocytofluorescence and Confocal Microscopy.
COS-1 cells were seeded on 2-well Lab Tek Chamber slides (Nalge) in DMEM with 10% CD-FBS for 18 h before being transfected with 2 μg DNA/105 cells by the FuGENE6 transfection reagent (Boehringer Mannheim). Transfected cells were treated with 10 nM DHT or vehicle for 16 h, then fixed in fixation solution (3% formaldehyde and 10% sucrose in PBS) for 15 min on ice and permeabilized by methanol. Immunostaining was performed by incubating slides with blocking solution (2% BSA in PBS) for 15 min at room temperature, stained with 1:200 dilution of anti-AR polyclonal antibody (NH27) for 45 min, followed by Texas red-conjugated goat anti-rabbit antibody (ICN) for 45 min at room temperature. Stained slides were washed and mounted (Vectashield; Vector Laboratories). The slides were photographed under 40-fold magnification with a Leica TCS SP Spectral Confocal Microscope (Leica, Deerfield, IL).
Western Blotting.
Protein samples extracted from the cell were separated on SDS/15% polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were incubated 1 h with 5% nonfat milk in TBST buffer (0.9% NaCl/20 mM Tris, pH 7.4/0.5% Tween 20) at room temperature, followed by the antibodies against p27(KIP1) (Santa Cruz Biotechnology), followed by AP-conjugated goat-anti-mouse antibody. Blots were developed with the AP-developing reagent from Bio-Rad. Band intensity was quantitated by COLLAGE image analysis software (Fotodyne, New Berlin, WI).
Results
SV Is an AR-Associated Protein.
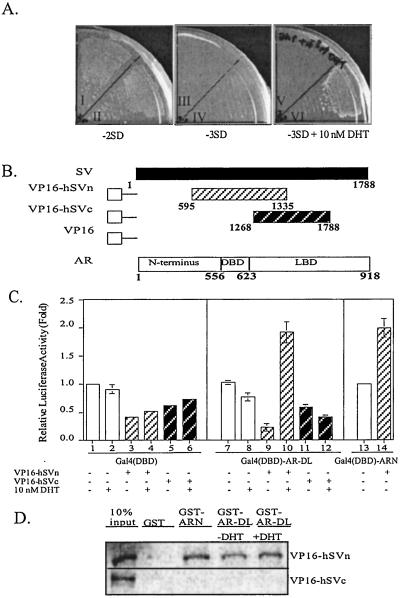
The human AR LBD was used as a bait to screen AR interaction proteins in a human skeletal muscle cDNA library in the presence of 10 nM T. Several positive clones were selected by nutrition deprivation and confirmed by the β-galactosidase assay. Further analysis indicated that five clones containing cDNA inserts match well with various segments of SV cDNA. As shown in Fig. 1A, one of these clones, encoding amino acids 558-1788 of SV, interacted well with AR-peptide bait in the presence of 10 nM DHT. This SV cDNA was then truncated and fused with VP16 as indicated in Fig. 1B. Mammalian two-hybrid indicated that hSVn peptide (amino acids 594-1335), but not hSVc peptide (amino acids 1268–1788), could interact with the AR-DBD-LBD (AR-DL) in a DHT-dependent manner (Fig. 1C). The hSVn can also interact with the AR N-terminal domain (ARN) (Fig. 1C, lane 14). GST pull-down assay further confirmed that VP16-hSVn but not VP16-hSVc can be pulled down by GST-AR-DL and GST-ARN (Fig. 1D). Together, data from yeast two-hybrid, mammalian two-hybrid, and GST pull-down assays all suggest that hSV peptide (amino acids 594-1268) can interact with the ARN as well as the AR-DL in a DHT-enhanced manner.
Figure 1.
SV fragments interact with AR in yeast two-hybrid, mammalian two-hybrid, and GST pull-down assays. (A) Yeast two-hybrid assay demonstrated the interaction between AR and SV. Yeast strain Y190 was cotransformed with pAS-AR and pACTII or pACTII-SV(595–1788). After transformation, yeast was plated on −2SD nutrition selection plates and cultured in 30°C incubator for 3 days. Colonies were selected and plated on −2SD, −3SD, and −3SD + 10 nM DHT nutrition selection plates. I, III, V are the yeast transformed with pAS-AR and pACTII; II, IV, and VI are the yeast transformed with pAS-AR-DL and pACTII-SV(595–1788). The growth of yeast was observed after 3 days culture in 30°C incubator. (B) Diagram of VP16-hSV constructs and AR functional domains. (C) Plasmids expressing Gal4(DBD), Gal4(DBD)-AR-DL, or Gal4(DBD)-ARN were cotransfected with VP16-hSVn or VP16-hSVc expression plasmids into COS-1 cells. Gal4 response element-controlled luciferase reporter gene, pG5-Luc, was used to detect the interaction and pRL-SV40 was used for internal control. After 16 h transfection, 10 nM DHT or EtOH were added for another 16 h. Cells were harvested and assayed for luciferase activity. The activities relative to VP16 alone without ligand were calculated. Results are the mean ± SD of three independent experiments. (D) GST protein and two GST fusion proteins containing AR N terminus (GST-ARN) and AR DBD plus LBD (GST-AR-DL) were expressed in bacteria and purified by glutathione beads. SV fragments were expressed by in vitro translation and labeled by [35S]methionine. After incubation of SV fragment and GST-AR with EtOH or 1 μM DHT, pulled-down proteins were loaded on gel and detected by PhosphorImager.
Nuclear Localization and Enhancement of AR Transactivation by SV Domain (Amino Acids 831-1281).
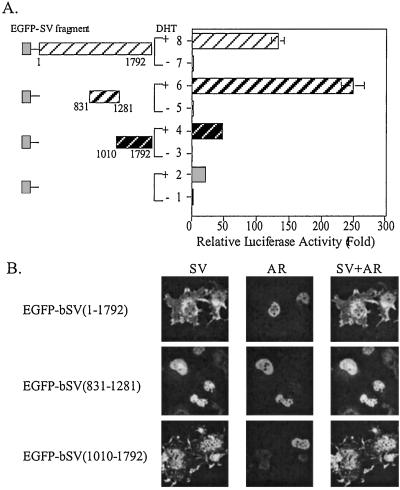
Results from Fig. 1 demonstrate that the SV peptide, amino acids 594-1268, can interact with AR. To test further whether this interaction also influences AR transactivation, we cotransfected plasmids encoding various domains of bSV along with AR-expressing plasmid and mouse mammary tumor virus-luciferase (MMTV-Luc) reporter in COS-1 cells. The bSV contains 1,792 amino acids sharing 92.7% homology with human SV (12). Fragments of bSV were conjugated with enhanced green fluorescent protein (EGFP), which emits fluorescence under light elicitation. As shown in Fig. 2A, addition of 10 nM DHT induced AR transactivation 25-fold (lane 1 vs. 2) when AR was coexpressed with EGFP. The full-length bSV (amino acids 1–1792) further enhanced AR transactivation to 132-fold (lane 2 vs. 8). A peptide containing amino acids 831-1281 of bSV, which is within the interaction domain, can further enhance AR transactivation to 248-fold (lane 6 vs. 8). In contrast, the other domain within SV (amino acids 1010–1792) had only a marginal effect on the AR transactivation (lane 2 vs. 4). These data strongly suggest that bSV(831–1281) in the interaction domain is sufficient to enhance AR transactivation function. As shown in Fig. 2B, subcellular colocalization studies with a confocal microscope further demonstrated that bSV(831–1281) is located exclusively in the nucleus and colocalizes with DHT-bound AR in the nucleus. In contrast, bSV(1010–1792) is located mainly in the cytosol. Together, the results in Fig. 2 demonstrated that full-length bSV and the domain (amino acids 831-1281) could enhance AR transactivation and colocalize with AR in the nucleus.
Figure 2.
Functional domain and cellular localization of SV fragment with AR. (A) Plasmids (1.5 μg) expressing EGFP only or EGFP-bSV fragments were coexpressed with 30 ng pCMV-AR, 0.5 μg MMTV-Luc, and 1 ng pRL-SV40 into COS-1 cells. Cells were treated with EtOH or 10 nM DHT, as indicated, for 20 h. The Firefly luciferase activity from AR reporter gene, MMTV-Luc, was normalized by Renilla luciferase activity. After measuring the luciferase activity, values relative to lane 1 were calculated. Results are the mean ± SD of three independent experiments. (B) EGFP-bSV fragments were coexpressed in the COS-1 cell line with AR. After transfection and treatment with 10 nM DHT for 16 h, cells were stained with AR antibody (NH27), followed by Texas red-conjugated secondary antibody, and analyzed under confocal microscope. Signals of the single focal plane are scanned and computerized to images. Merged images are shown as indicated in labels.
SV Enhances AR Transactivation.
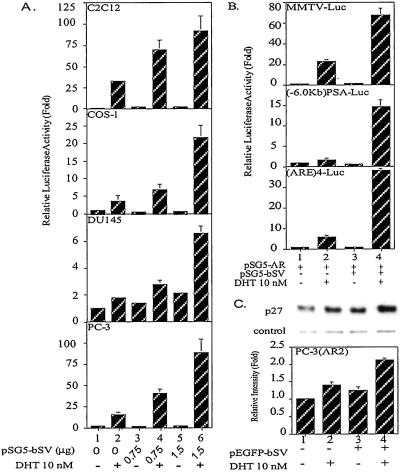
Cotransfection of the full-length bSV and AR expression plasmids at 25:1 and 50:1 ratios enhanced AR transactivation 3- to 8-fold in C2C12 muscle cells in the presence of DHT. Similar results were also observed when we replaced C2C12 cells with COS-1, DU145, and PC-3 (Fig. 3A). In addition to MMTV-Luc, two other AR reporter genes, prostate-specific antigen-Luc and androgen response element-Luc, were applied to demonstrate the coactivation function of SV. All results demonstrate that regardless of different androgen response element-containing promoters, SV can enhance AR transactivation function in PC-3 cells (Fig. 3B). To further rule out the possible artifact effect by using reporter gene assays, we analyzed the effect of SV on AR endogenous target genes expression, such as p27KIP (18), in the PC-3 cells stably transfected with AR expression plasmid, PC-3(AR2) cells (19). As shown in Fig. 3C, 10 nM DHT induced p27(KIP1) protein expression (lane 1 vs. 2). Addition of bSV further enhanced p27(KIP1) protein expression (lane 2 vs. 4). The less effect compared with reporter gene assays may be caused by the interference from multiple factors involved in the regulation of endogenous gene. Feedback regulation may also happen in the controlling of the homeostasis in cells. Therefore, the simplified reporter gene usually has a more obvious effect. Overall, these data clearly demonstrate that SV can function as an AR coregulator to enhance AR transactivation.
Figure 3.
SV enhanced AR transcriptional activity. (A) C2C12, COS-1, DU145, and PC-3 cell lines were cotransfected with 30 ng pSG5-AR, 0.5 μg MMTV-Luc, 1 ng pRL-SV40, various amounts of pSG5-bSV as indicated, and adjusted to total amount of 2 μg DNA with pSG5. The assay method was the same as Fig. 2. After measuring the luciferase activity, values relative to lane 1 were calculated. Results are the mean ± SD of three independent experiments. (B) PC-3 was cotransfected with 30 ng pSG5-AR, 1.5 μg pSG5-bSV, 1 ng pRL-SV40, and 0.5 μg reporter gene as indicated by using SuperFect. After 20 h, cells were treated with EtOH or 10 nM DHT for another 24 h and then lysed for luciferase activity assay. (C) PC-3(AR2) cell line was transfected with EGFP or EGFP-bSV expressing vector by using SuperFect. After 20 h, cells were treated with EtOH or 10 nM DHT for another 30 h. Proteins extracted from cells were loaded on SDS/15% polyacrylamide gels and analyzed by Western blotting. The intensity of each p27 band was quantified and normalized with control protein which is a nonspecific band pick up by the antibody in the same blot. The relative intensities to lane 1 were calculated. PSA-Luc, prostate-specific antigen-Luc; (ARE)4-Luc, androgen response element-Luc.
The Specificity of SV Coregulator Activity.
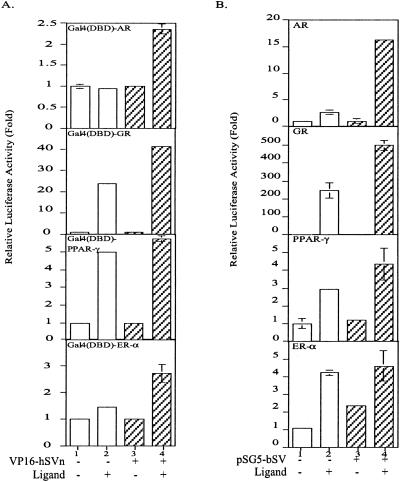
By using mammalian two-hybrid assay, our data indicated that SV could also interact with other steroid receptors such as glucocorticoid receptor (GR), estrogen receptor-α (ER-α), and peroxisome-proliferator activated receptor-γ (PPAR-γ). The interaction of SV with these receptors was similar (ER-α) or relatively weaker (GR and PPAR-γ) as compared with the interaction with AR (Fig. 4A), which could be caused by the different coregulator context in the cell. The activation function-2 domain of GR and PPAR-γ might be able to recruit more coactivators or have stronger affinity to certain coactivators that results in the less coactivation activity of SV with these two receptors. SV-modulated transcriptional activities of nuclear receptors were then assayed by using AR and GR reporter gene (MMTV-Luc) and PPAR-γ and ER-α reporter genes. The results show that SV has less enhancement effect on the transactivation of GR than with AR, and has little effect on PPAR-γ and ER-α (Fig. 4B).
Figure 4.
SV interacted with other steroid receptors and enhanced their function. (A) The interaction of SV with AR, GR, PPAR-γ, and ER-α is tested in mammalian two-hybrid assay. One-microgram plasmids expressing Gal4(DBD)-AR, GR, PPAR-γ, or ER-α were cotransfected with 4-μg plasmids expressing VP16 or VP16-SVn to COS-1 cells. 10 nM DHT, 10 nM dexamethasone, 1 μM 15-deoxy-Δ12,14-prostaglandin J2, and 10 nM 17β-estradiol were applied to AR, GR, PPAR-γ, and ER-α, respectively. The assay method was the same as described in Fig. 1. Relative activities of ligand treatment to EtOH treatment are shown. (B) The coactivation function of SV in different receptors was assayed by using the reporter gene study. MMTV-Luc and PPAR-γ and ER-α reporter genes are the reporter genes for AR, GR, PPAR-γ, and ER-α, respectively. Lane 1 is regarded as 1-fold.
Comparison of Cooperative Effect and Ligand Enhancement Effect Between SV and Other ARAs.
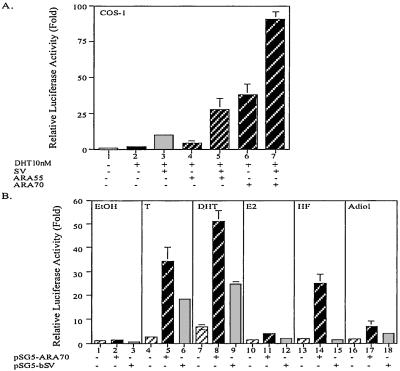
To compare the coregulator function of SV and other known AR coregulators, we tested the cooperative effect between SV and two other AR coregulators, ARA55 and ARA70N (amino acids 1–401). The combination of SV and ARA55 or ARA70N shows the additive effect better than the enhancement of SV, ARA55, or ARA70N alone (Fig. 5A), which indicates that these coactivators may modulate AR activity through multiple yet cooperative mechanisms to potentiate AR function.
Figure 5.
SV cooperates with other ARAs and affects various steroid-induced AR transactivations. (A) COS-1 cells were cotransfected with 0.5 μg MMTV-Luc, 1 ng pRL-SV40, pSG5-AR (30 ng), and combination of 1.4 μg pSG5-bSV, 0.1 μg ARA55, or 0.1 μg ARA70N as described in the figure. The total amount of DNA was adjusted to 2 μg with pSG5. The assay was carried out as in Fig. 2. (B) COS-1 cells were transfected with 0.5 μg MMTV-Luc, 1 ng pRL-SV40, 30 ng pSG5-AR with 1.5 μg pSG5, 0.1 μg pSG5-ARA70N, or 1.5 μg pSG5-bSV. The total amount of DNA was adjusted to 2 μg with pSG5. After 16 h transfection, cells were treated with vehicle (EtOH) or steroids [10 nM T, DHT, 17β-estradiol (E2), hydroxyflutamide (HF), or androst-5-ene-3β,17β-diol (Adiol)] for 20 h as indicated. The assay was conducted as described in Fig. 2.
Coregulators can enhance AR transactivation under various steroid treatments. For example, ARA70N could enhance AR transactivation in the presence of T and DHT, as well as 17β-estradiol, hydroxyflutamide, and androst-5-ene-3β,17β-diol (20–22). Here we compare the effect of SV with ARA70N in the induction of AR function under these steroids. The results show that SV significantly enhances T- and DHT-induced AR transactivation, slightly enhances androst-5-ene-3β,17β-diol-induced AR transactivation, but has a marginal effect on 17β-estradiol- or hydroxyflutamide-induced AR transactivation. These data therefore again demonstrated that only selective AR coregulators were able to enhance AR transactivation induced by various steroids.
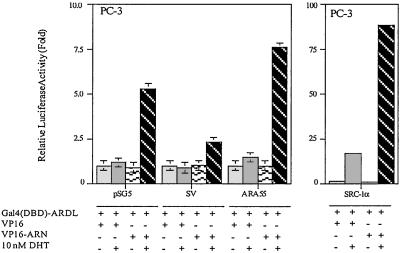
The Interaction Between AR N Terminus and C Terminus Is Suppressed by SV.
Early reports suggested that interaction between ARN and C terminus (ARC) may help to stabilize the dimer complexes of AR (23). Because SV can interact with both ARN and AR-DL (Fig. 1 C and D), it is possible that SV may stabilize the dimer complexes by holding the ARN and ARC together. By using mammalian two-hybrid assays, we demonstrated AR N–C interaction in a DHT-dependent manner (Fig. 6). Selective AR coregulators, such as SRC-1, could further enhance this N–C interaction. Addition of SV showed a mild suppressive effect on this N–C interaction. The contrasting effects between SV and SRC-1 strongly suggest that different AR coregulators may go through different mechanisms to enhance AR transactivation.
Figure 6.
AR N–C interaction is reduced by bSV. PC-3 cells were transfected with 30 ng plasmids expressing Gal4(DBD)-AR-DL, VP16, or VP16-ARN combined with 1.5 μg pSG5, pSG5-bSV, pSG5-ARA55, or pSG5-SRC-1α as indicated. The reporter plasmid pG5-Luc (0.5 μg) and control plasmid pRL-Luc (1 ng) were transfected to every sample. The assay was performed as described in Fig. 1C.
Discussion
We have identified SV as an AR coregulator from skeletal muscle. SV binds to actin and increases the amount of F-actin and vinculin when overexpressed (14), suggesting it functions in cell adhesion and motility. On the other hand, actin itself was proposed to be the key regulator of serum response factor that could modulate gene expression by functioning as a suppressor to sequester the coregulators of serum response factor (24). Because we were able to show that SV, an actin-binding protein, can function as an AR coregulator to enhance AR transactivation, it will be interesting to determine in future studies whether actin can play any roles to modulate SV-enhanced AR transactivation.
Among identified AR coregulators, ARA24 and ARA160 interact with ARN (25, 26); ubc-9 and SNURF interact with AR DBD (27, 28); and ARA54, ARA55, and ARA70 interact with AR LBD (29–31). SV and some nuclear receptor coregulator members, such as NCoA, can interact with both N-terminal activation function-1 and C-terminal activation function-2 of AR (32, 33). The LXXLL motif of several coregulators plays an essential role for the interaction and coactivation function with most receptors except AR (34, 35). We found that the SV peptide (amino acids 594-1335), which does not contain the LXXLL motif, can still interact with ARN and ARC, further indicating that the interaction mechanism between coregulators and AR is distinct from that of coregulators and other steroid receptors. Recently, the motifs important for AR N–C interaction have been reported (36). Those motifs, including FXXLF and WXXLF, that play important roles for the interaction with AR C terminus, are located in ARN. It is possible that AR N–C interactions may stabilize the dimer of AR and promote its activity. Because SV interacts with both the N and C termini of AR, it is reasonable to hypothesize that SV may play some roles in the AR dimerization. However, the results in Fig. 6 indicate SV can suppress AR N–C interaction. It is possible that SV may replace the ARN effect in ARC to stabilize ligand binding.
Our data show that SV(amino acids 831-1281) has a better enhancing effect on AR transactivation than full-length SV and SV(amino acids 1010–1792). Immunostaining shows this peptide is mainly in the nucleus and colocalizes with DHT-bound AR in contrast to SV(amino acids 1010–1792) which remains in the cytosol. It is possible that by remaining within the nucleus, SV may increase the interaction frequency with AR, hence resulting in changing the AR conformation to an activated form to facilitate the binding of the androgen response element located in the target genes. Another possibility is that a suppressive domain exists in either the N or C terminus of SV that diminishes the full-length function. The consequence of these events may then result in the increase of AR transactivation. The fact that SV(amino acids 831-1281) peptide colocalizes with AR in the nucleus better than full-length SV could be due to the existence of multiple factors to modulate full-length SV translocation, which could then make this event transient and less detectable. Whether the SV fragment exists as an endogenous product is not presently clear. However, many studies have shown that caspases regulate protein activity by digesting their substrates. It is possible that degraded SV peptide is available in cells.
We further ask whether the AR nuclear translocation rate could be altered by coexpression with SV. By fixing cells every 5 min followed by immunocytostaining, we observed the amount of AR in the nucleus and cytosol. Results indicate that AR translocated into the nucleus within 30 min in the presence of DHT. The importing rate of DHT-AR was not altered by SV coexpression (data not shown). Whether the exporting rate of AR from the nucleus could be altered with SV overexpression, however, remains unclear. We also used a pulse–chase labeling assay to measure the AR degradation rate. The half-life of AR is 3.5 h (23). After cotransfection of SV and AR, the half-life of AR showed no significant difference within 6 h after DHT treatment (data not shown). The mechanism, therefore, is still not clear and needs further study.
Because of the differences of transcription-translation efficiency of transfected genes, we may need to adjust the amount of transfected plasmids expressing coregulators and steroid receptors to an optimal ratio to show maximum coactivator activity. For example, SRC-1 needs a ratio of expression plasmids up to 100:1 as compared with steroid receptors to show the significant coactivator activity (37, 38). In contrast, other coregulators, such as ARA55 or ARA70N, may require lower ratios of expression plasmids (coregulator/AR up to 3–5:1) for their maximal coactivator activities (30, 31). Because different cells have various amounts of endogenous coregulators that may affect the impact of exogenously transfected SV, we expect that the amount of transfected SV plasmids for maximum AR activity varies between cells. Similarly, we may not expect to see that SV always functions as a coregulator to enhance AR transactivation preferentially as compared with other steroid receptors. Considering that any given cell may have multiple coregulators interacting with multiple steroid receptors, we expect to see squelching effects occur in some cells resulting in less coregulator effect for any particular receptor. Furthermore, in varying physiological environments and clinical situations, cells are exposed to multiple steroid hormones. Compared with ARA70N, SV is generally much weaker in promoting non-androgen steroid-mediated AR transactivation. SV, however, is able to coordinate with other AR coregulators, such as ARA70N and ARA55, to enhance AR transactivation. These results again suggest that the final AR activity may be the balance and coordination of multiple coregulators in any given cell. It is well documented that different concentrations of DHT and various amounts of AR within one cell may change the androgen-AR function to either promote cell proliferation or stimulate cell apoptosis. For example, although 0.1 nM DHT can stimulate LNCaP cell proliferation, 10 nM DHT promotes LNCaP cell apoptosis (39, 40). Similarly, 10 nM DHT can also arrest PC-3(AR2) cell growth and promote cells into apoptosis (19, 41). Although details of the mechanisms remain unclear, it is possible that, because of AR interacting with various amounts of coregulators, the combination of such interactions may then trigger cells into either proliferation or apoptosis. As an earlier report demonstrated, androgen can down-regulate the SV gene expression (15), SV may provide a nice feedback mechanism for cells to determine how AR and SV perform their physiological function in muscle and other cells.
In conclusion, we demonstrate that SV can function as an AR coregulator to enhance AR transactivation. Because androgen-AR has been suggested to play important roles for muscle development and function, further studies may be able to reveal how SV and AR perform in muscle development and function, benefiting those patients who need to rebuild their muscle strength, such as hypogonadal patients and AIDS patients with wasting syndrome.
Acknowledgments
We thank Drs. E. J. Luna and T. J. Brown for their reagents and discussion. This work was supported by National Institutes of Health Grant CA71570 and a George Whipple Professorship endorsement.
Abbreviations
- AR
androgen receptor
- T
testosterone
- ER
estrogen receptor
- GR
glucocorticoid receptor
- DHT
5α-dihydrotestosterone
- SV
supervillin
- bSV
bovine supervillin
- EGFP
enhanced green fluorescent protein
- GST
glutathione S-transferase
- MMTV-Luc
mouse mammary tumor virus-luciferase
- Gal4(DBD)
Gal4 DNA-binding domain
- LBD
ligand-binding domain
- −3SD
synthetic dropout media lacking histidine, leucine, and tryptophan
- −2SD
synthetic dropout media lacking leucine and tryptophan
- PPAR-γ
peroxisome-proliferator activated receptor-γ
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Chang C, Saltzman A, Yeh S, Young W, Keller E, Lee H J, Wang C, Mizokami A. Crit Rev Eukaryotic Gene Expression. 1995;5:97–125. doi: 10.1615/critreveukargeneexpr.v5.i2.10. [DOI] [PubMed] [Google Scholar]
- 2.Glass C K, Rosenfeld M G. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 3.Yeh S, Lin H K, Kang H Y, Thin T H, Lin M F, Chang C. Proc Natl Acad Sci USA. 1999;96:5458–5463. doi: 10.1073/pnas.96.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poukka H, Karvonen U, Janne O A, Palvimo J J. Proc Natl Acad Sci USA. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozanne D M, Brady M E, Cook S, Gaughan L, Neal D E, Robson C N. Mol Endocrinol. 2000;14:1618–1626. doi: 10.1210/mend.14.10.0541. [DOI] [PubMed] [Google Scholar]
- 6.Sengupta S, Vonesch J L, Waltzinger C, Zheng H, Wasylyk B. EMBO J. 2000;19:6051–6064. doi: 10.1093/emboj/19.22.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shim W S, DiRenzo J, DeCaprio J A, Santen R J, Brown M, Jeng M H. Proc Natl Acad Sci USA. 1999;96:208–213. doi: 10.1073/pnas.96.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller J M, Isele U, Metzger E, Rempel A, Moser M, Pscherer A, Breyer T, Holubarsch C, Buettner R, Schule R. EMBO J. 2000;19:359–369. doi: 10.1093/emboj/19.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotaja N, Aittomaki S, Silvennoinen O, Palvimo J J, Janne O A. Mol Endocrinol. 2000;14:1986–2000. doi: 10.1210/mend.14.12.0569. [DOI] [PubMed] [Google Scholar]
- 10.Mooradian A D, Morley J E, Korenman S G. Endocr Rev. 1987;8:1–28. doi: 10.1210/edrv-8-1-1. [DOI] [PubMed] [Google Scholar]
- 11.Doumit M E, Cook D R, Merkel R A. Endocrinology. 1996;137:1385–1394. doi: 10.1210/endo.137.4.8625915. [DOI] [PubMed] [Google Scholar]
- 12.Pope R K, Pestonjamasp K N, Smith K P, Wulfkuhle J D, Strassel C P, Lawrence J B, Luna E J. Genomics. 1998;52:342–351. doi: 10.1006/geno.1998.5466. [DOI] [PubMed] [Google Scholar]
- 13.Pestonjamasp K N, Pope R K, Wulfkuhle J D, Luna E J. J Cell Biol. 1997;139:1255–1269. doi: 10.1083/jcb.139.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wulfkuhle J D, Donina I E, Stark N H, Pope R K, Pestonjamasp K N, Niswonger M L, Luna E J. J Cell Sci. 1999;112:2125–2136. doi: 10.1242/jcs.112.13.2125. [DOI] [PubMed] [Google Scholar]
- 15.Pan H J, Uno H, Inui S, Fulmer N O, Chang C. Endocrine. 1999;11:321–327. doi: 10.1385/ENDO:11:3:321. [DOI] [PubMed] [Google Scholar]
- 16.Fraser A G, Kamath R S, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Nature (London) 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 17.Lu M L, Schneider M C, Zheng Y, Zhang X, Richie J P. J Biol Chem. 2001;276:13442–13451. doi: 10.1074/jbc.M006598200. [DOI] [PubMed] [Google Scholar]
- 18.Ling M T, Chan K W, Choo C K. J Endocrinol. 2001;170:287–296. doi: 10.1677/joe.0.1700287. [DOI] [PubMed] [Google Scholar]
- 19.Yuan S, Trachtenberg J, Mills G B, Brown T J, Xu F, Keating A. Cancer Res. 1993;53:1304–1311. [PubMed] [Google Scholar]
- 20.Yeh S, Miyamoto H, Shima H, Chang C. Proc Natl Acad Sci USA. 1998;95:5527–5532. doi: 10.1073/pnas.95.10.5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeh S, Miyamoto H, Chang C. Lancet. 1997;349:852–853. doi: 10.1016/S0140-6736(05)61756-4. [DOI] [PubMed] [Google Scholar]
- 22.Miyamoto H, Yeh S, Lardy H, Messing E, Chang C. Proc Natl Acad Sci USA. 1998;95:11083–11088. doi: 10.1073/pnas.95.19.11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Z X, Lane M V, Kemppainen J A, French F S, Wilson E M. Mol Endocrinol. 1995;9:208–218. doi: 10.1210/mend.9.2.7776971. [DOI] [PubMed] [Google Scholar]
- 24.Sotiropoulos A, Gineitis D, Copeland J, Treisman R. Cell. 1999;98:159–169. doi: 10.1016/s0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- 25.Hsiao P W, Chang C. J Biol Chem. 1999;274:22373–22379. doi: 10.1074/jbc.274.32.22373. [DOI] [PubMed] [Google Scholar]
- 26.Hsiao P W, Lin D L, Nakao R, Chang C. J Biol Chem. 1999;274:20229–20234. doi: 10.1074/jbc.274.29.20229. [DOI] [PubMed] [Google Scholar]
- 27.Poukka H, Aarnisalo P, Karvonen U, Palvimo J J, Janne O A. J Biol Chem. 1999;274:19441–19446. doi: 10.1074/jbc.274.27.19441. [DOI] [PubMed] [Google Scholar]
- 28.Poukka H, Karvonen U, Yoshikawa N, Tanaka H, Palvimo J J, Janne O A. J Cell Sci. 2000;113:2991–3001. doi: 10.1242/jcs.113.17.2991. [DOI] [PubMed] [Google Scholar]
- 29.Fujimoto N, Yeh S, Kang H Y, Inui S, Chang H C, Mizokami A, Chang C. J Biol Chem. 1999;274:8316–8321. doi: 10.1074/jbc.274.12.8316. [DOI] [PubMed] [Google Scholar]
- 30.Kang H Y, Yeh S, Fujimoto N, Chang C. J Biol Chem. 1999;274:8570–8576. doi: 10.1074/jbc.274.13.8570. [DOI] [PubMed] [Google Scholar]
- 31.Yeh S, Chang C. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bevan C L, Hoare S, Claessens F, Heery D M, Parker M G. Mol Cell Biol. 1999;19:8383–8392. doi: 10.1128/mcb.19.12.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alen P, Claessens F, Verhoeven G, Rombauts W, Peeters B. Mol Cell Biol. 1999;19:6085–6097. doi: 10.1128/mcb.19.9.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 35.Leo C, Chen J D. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- 36.He B, Kemppainen J A, Wilson E M. J Biol Chem. 2000;275:22986–22994. doi: 10.1074/jbc.M002807200. [DOI] [PubMed] [Google Scholar]
- 37.McInerney E M, Tsai M J, O'Malley B W, Katzenellenbogen B S. Proc Natl Acad Sci USA. 1996;93:10069–10073. doi: 10.1073/pnas.93.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeshita A, Cardona G R, Koibuchi N, Suen C S, Chin W W. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 39.Langeler E G, van Uffelen C J, Blankenstein M A, van Steenbrugge G J, Mulder E. Prostate. 1993;23:213–223. doi: 10.1002/pros.2990230304. [DOI] [PubMed] [Google Scholar]
- 40.Sonnenschein C, Olea N, Pasanen M E, Soto A M. Cancer Res. 1989;49:3474–3481. [PubMed] [Google Scholar]
- 41.Heisler L E, Evangelou A, Lew A M, Trachtenberg J, Elsholtz H P, Brown T J. Mol Cell Endocrinol. 1997;126:59–73. doi: 10.1016/s0303-7207(96)03970-6. [DOI] [PubMed] [Google Scholar]