Highlights
-
•
LncRNA plays an important role in many life activities, such as epigenetic regulation, cell cycle regulation and cell differentiation regulation.
-
•
LncRNA WAC-AS1 was identified significantly upregulated in gastric cancer and affected the progression of gastric cancer.
-
•
LncRNA WAC-AS1 through miR-204-5p/HOXC8 axis significantly promoted the proliferation, migration and invasion of gastric cancer.
Keywords: lncRNA WAC-AS1, miR-204-5p, HOXC8, Gastric cancer, Mechanism
Abstract
LncRNAs affect tumorigenesis, and although the genesis, regulation and physiological mechanism of lncRNAs in gastric cancer (GC) have been reported, the research of lncRNAs still have a lot of value. Through comprehensive bioinformatics analysis, we screened the candidate lncRNA WAC-AS1(WAC-AS1). We analyzed WAC-AS1 expression in GC related tissues and cells using qRT-PCR. WAC-AS1’s impact on GC growth and metastasis was investigated. LncRNA WC-AS-miR-204-5p-HOXC8 interaction was established through dual-luciferase reporter, FISH, RIP and RNA pull-down assay. We observed substantial upregulation in WAC-AS1 expression in cells and tissues of GC. WAC-AS1 through miR-204-5p/HOXC8 axis promoted GC proliferation, invasion, and migration. WAC-AS1 plays a cancer-promoting role for promoting the progression of GC.
Introduction
According to the latest data from the statistical analysis of cancer epidemiology in China, the incidence of gastric cancer ranks fifth, and the mortality rate ranks third, posing a serious threat to the lives and health of the people [1]. The traditional treatment methods of GC are chemotherapy, surgery, and radiotherapy [2,3]. With precision medicine and targeted immunotherapy development [4], treatment and diagnosis of GC made rapid progress. However, the reappearance mortality rate of advanced GC is still high [3,5].
One of the challenges facing modern oncology is to improve our understanding of the role of transcription factors and epigenetic mechanisms, particularly non-coding Rnas (ncRNAs), in carcinogenesis, cancer progression, and recurrence. This maybe lead to the development of new pharmacological drugs or potentially provide new insights into genetic interventions. NcRNAs vary widely in size, shape, and function, and many types of ncRNAs have been found to be dysregulated in various cancers [[6], [7], [8], [9]]. In the ncRNAs family, lncRNA acts in cell cycle, cell differentiation and epigenetic regulation [[10], [11], [12]]. lncRNAs can affect the expression and/or function of DNAs, RNAs, and proteins by interacting with them, which participates in cancer development [13,14]. LncRNAs regulate the signaling pathways associated with autophagy, apoptosis, and stem cells [15]. Lnc RNA WAC-antisense RNA1 (lncRNA WAC-AS1, WAC-AS1) is a newly discovered lncRNA associated with the prognosis and development of several tumors [16]. For example, the expression of WAC-AS1 and seven other genes is significantly correlated with the survival rate of ovarian cancer patients [17]. WAC-AS1 has been identified as prognostic ferroptosis-related lncRNAs in glioma [18]. WAC-AS1 is highly expressed in liver cancer and associated with poor prognosis [19]. However, limited data suggests the specific role of WAC-AS1 in gastric cancer. This article aims to address this research question.
Mechanistically, lncRNAs typically exert their functional effects by regulating downstream microRNAs (miRNAs). Some miRNAs have been discovered acting on WAC-AS1. For instance, lncRNA WAC-AS1 regulates HBV replication by reinforcing the autophagy induced by miR-192–5p/ATG7 [20]. In present research, we discovered that miR-204–5p was the downstream miRNA of WAC-AS1. MiR-204–5p is related to clinicopathological features and cancer patients’ prognosis. miR-204–5p hinders tumor autophagy, proliferation, metastasis, and chemotherapy resistance in various cancer kinds [21,22]. It is found that miRNAs, together with RNA targets can form a regulatory network (ceRNA network) [23]. Moreover, miR-204–5p can promote apoptosis and inhibit GC cell migration [24,25]. The regulation of miR-204–5p is assumed to be the potential functional mechanism of WAC-AS1 in gastric cancer.
In this study, lncRNA WAC-AS1(WAC-AS1) was significantly upregulated from differentially expressed GC microarray datasets. WAC-AS1 interacts with miR-204–5p, promotes proliferation as well as hinders GC cell apoptosis. Moreover, miR-204–5p functions as a ceRNA network to inhibit HOXC8 expression. Inhibition of the WAC-AS1-miR-204–5p-HOXC8 axis can effectively inhibit GC cell occurrence and development.
Methods
Patient tissue samples
From GC patients, we acquired GC and non-tumor tissues (n = 25) at China-Japan Union Hospital, Jilin University. We have done tests following the Ethics Committee of China-Japan Union Hospital, Jilin University.
Culture of cells
The Cobioer Biosciences Co., LTD (Nanjing, China) and Cell Bank of Chinese Academy of Medical Sciences (Beijing, China) offered BGC-823, GES1, SGC-7901, MKN45, HGC-27, AGS, and cells. We cultivated AGS, MKN45, SGC-7901, BGC-823, and HGC-27 cells with RPMI1640 (Cat.No.31800, Solarbio, Beijing, China) mixed with 10% fetal bovine serum (FBS; Cat.No.11011–8611, TIANHANG, China). We cultured GES1 cells in dulbecco's modified eagle medium (DMEM) (Cat.No.11995, Solarbio, Beijing, China). We incubated cells at 5% CO2 environment and 37 °C.
Cell transfection
Lentivirus vectors overexpressing WAC-AS1 (LV- WAC-AS1) or knocking down WAC-AS1 (LV-shRNA- WAC-AS1), as well as lentivirus vectors overexpressing HOXC8 (LV-HOXC8) or silencing HOXC8 were constructed (OBiO, Shanghai, China), respectively. We bought miR-204-5p mimics and inhibitor from RiboBio (Guangzhou, China). Transfection and infection of cells were performed using Lipofectamine 2000 (Invitrogen, USA).
qRT-PCR
We isolated total RNA from the GC tissues and cell lines (including mouse tumor tissues and the patient's tissues). We reverse-transcribed total RNA into cDNA using the SweScript RT I First Strand cDNA Synthesis Kit (Cat.No. G3330, Servicebio, Wuhan, China). Then, we used the qRT-PCR three-step method. The 2-ΔΔCt method was used for calculating WAC-AS1, HOXC8, and miR-204–5p relative expression levels. We provided primer nucleotide sequences in Supplementary Table 1.
CCK-8 assay
We inoculated stably transfected GC cells into 96-well (5000cells/well) plates and cultured at 24 h, 48 h, 72 h, and 96 h. We detected the absorbance at 450 nm via CCK-8 assay (Cat. No. C0038, Beyotime, China).
Colony formation assay
We seeded suspended cells (1000 cells/well) into the 6-well plates and then cultivated for 2–3 wks. We changed the medium every 3 d after being fixed with PFA (4 %) (duration:15 min) and, after that, stained by means of crystal violet (1 %) for 1/2 h. The staining reagent was washed away. The plates were air-dried for taking photos. The colony number in each group was counted. Repeated 3 times.
EdU assay
We inoculated stably transfected 1 × 105 cells into 6-well plates, and used labeling/detection kit (Cat. No. G1602, Servicebio, Wuhan, China) was used for staining the GC cells. We used DAPI for staining cell nuclei and detected EdU positive cells. The findings were examined by EdU positive cell number to DAPI stained cell number ratio. Repeated 3 times.
Wound healing assay
We have seeded 5 × 105 cells into a 6-well plate. After that we cultivated them to 90 % confluence. A pipette tip (10 µL) was utilized for rubbing on cell monolayers. Photomicrographs were obtained with a biological microscope (OLYMPUS Japan) at 100x. Wound widths were measured by Image J software. Repeated 3 times.
Transwell assay
We seeded GC cells(5 × 105 cells/well) in the Matrigel-coated upper Transwell chamber. We fixed transwell chambers using 4 % PFA after 48 h culture and then stained by means of 1 % crystal violet [26]. We observed the cells under an Olympus microscope at a magnification of × 100 and analyzed the photos. Repeated 3 times.
Apoptosis assay
1.0 × 106 cells/6 wells digestion into a single-cell state, stained with propyl iodide (Cat.No. 40302ES20, PI, 10 μg /ml, YEASAN) and Annexin V-FITC (Cat.No. 40302ES20, 10 μl/ml, BD YEASAN). We analyzed the cells through flow cytometry after 10 min. Repeated 3 times.
Luciferase reporter assay
We mixed the WAC-AS1/HOXC8 3′UTR plasmid (Mut) using miR-204–5p mimic/ NC and lipofectamine 6000 (Cat.No. C0526, Beyotime, Shanghai, China) transfection reagent and added into 1.0 × 105 HGC-27/SGC-7901 cell/24 wells with RPMI1640 medium cultured at 37 °C along with 5 % CO2. Two days after transfection, we assessed relative luciferase activity using the Dual-Luciferase Reporter Assay Kit (Cat.No. 11402ES60, Yeasen, Shanghai, China).
RIP assay
According to the manufacturer's instructions, an RIP RNA-Binding protein immunoprecipitation kit (Cat. No. P2197M, Beyotime, Shanghai, China) was applied to perform the RIP experiments [27]. Briefly, 2 × 105 cells/6 wells were lysed in RIP lysis buffer before the addition of magnetic beads containing control IgG or AGO2 Monoclonal antibodies (PROTEINTECH, 67934-1-Ig). Enriched WAC-AS1 or HOXC8 was purified and analyzed. The primes were shown in the supplement Table 1.
Table 1.
The correlation between LncRNA WAC-AS1 and clinicopathological features in 25 GC patients.
| Characteristics | Case | LncRNA WAC-AS1 |
p value | |
|---|---|---|---|---|
| Low | High | |||
| Age at surgery(year) | 8 | 17 | 0.6533 | |
| <60 | 11 | 3 | 8 | |
| ≥60 | 14 | 5 | 9 | |
| Gender | 0.1516 | |||
| Male | 17 | 7 | 10 | |
| Female | 8 | 1 | 7 | |
| Tumor size (cm) | 0.9146 | |||
| ≥5 | 9 | 3 | 6 | |
| <5 | 16 | 5 | 11 | |
| T grade | 0.7433 | |||
| T1+T2 | 4 | 1 | 3 | |
| T3+T4 | 21 | 7 | 14 | |
| Lymph node invasion | 0.9360 | |||
| Negative (NO) | 6 | 2 | 4 | |
| Positive (N1-N3) | 19 | 6 | 13 | |
| TNM stage | 0.9579 | |||
| I-II | 3 | 1 | 2 | |
| III-IV | 22 | 7 | 15 | |
| Histological grade | 0.5201 | |||
| Low | 5 | 1 | 4 | |
| Middle-high | 20 | 7 | 13 | |
RNA pull-down assay
RNA 3′End Desthiobiotinylation Kit (Cat. No. 20,163, Thermo, USA) and RNA-Binding protein immunoprecipitation kit (Cat. No. P2197M, Beyotime, Shanghai, China) was used to perform RNA pull-down assay [28]. Briefly, 1.0 × 107 GC cells were harvested, lysed, and sonicated. The biotin-labelled probes was incubated with the supernatant at room temperature (RT) for 30 min to form an RNA‐protein complex. The cell lysates were incubated with the probe-coated beads mixture at 4 °C for 2 h. After washing with the wash buffer, the RNA complexes bound to the beads were eluted and purified with Trizol Reagent(Takara,Japan) for further analysis.
After purification, enriched RNAs were detected using qPCR. The probes and primes were shown in the supplement Table 2.
Table 2.
The correlation between miR-204-5p and clinicopathological features in 25 GC patients.
| Characteristics | Case | hsa-miR-204-5p |
p value | |
|---|---|---|---|---|
| Low | High | |||
| Age at surgery(year) | 18 | 7 | 0.9428 | |
| <60 | 11 | 8 | 3 | |
| ≥60 | 14 | 10 | 4 | |
| Gender | 0.0003 | |||
| Male | 17 | 16 | 1 | |
| Female | 8 | 2 | 6 | |
| Tumor size (cm) | 0.6294 | |||
| ≥5 | 9 | 7 | 2 | |
| <5 | 16 | 11 | 5 | |
| T grade | 0.285 | |||
| T1+T2 | 4 | 2 | 2 | |
| T3+T4 | 21 | 16 | 5 | |
| Lymph node invasion | 0.7386 | |||
| Negative (NO) | 6 | 4 | 2 | |
| Positive (N1-N3) | 19 | 14 | 5 | |
| TNM stage | 0.1118 | |||
| I-II | 3 | 1 | 2 | |
| III-IV | 22 | 17 | 5 | |
| Histological grade | 0.5040 | |||
| Low | 5 | 3 | 2 | |
| Middle-high | 20 | 15 | 5 | |
FISH
The WAC-AS1 subcellular localization was detected by FISH. We purchased the WAC-AS1 RNA probe labeled with Cyanine 5 from Servicebio (Cat.No. GDP1072, Wuhan, China). We spread the cells on overslips in a 12-well plate. On the next day, we washed coverslips using PBS and fixed them with 4 % PFA at 25 °C. We dehydrated cells with 80, 90, and 100 % ethanol for 2 min at each concentration and then incubated with the WAC-AS1 probe at 37 °C for 24 h in humid and dark chambers. We washed the coverslips 3 times in formamide (50 %) and 2 × SSC buffer (37 °C, 10 min). We incubated each slice with Rhodamine anti-digoxin antibody or FITC vitellin for 20 min at 25 °C and incubated at 25 °C for 20 min with a dropping anti-vitellin antibody after removing PBS. We counterstained cells with DAPI (Cat.No. G1012–100ML, Servicebio, Wuhan, China) and then mounted to be observed.
Western blotting
The protein lysates of cultured GC cells and mouse tumor tissues were extracted from the lysate buffer and the protein concentration was detected by a BCA Protein Assay Kit (Cat.No. PC0020, Solarbio, Beijing, China). We separated denatured protein lysates (10 ul) by means of SDS-PAGE electrophoresis, transported to PVDF membranes as well as blocked in BSA (bovine serum albumin, 5 %) for 2–3 h at 25 °C. We incubated membranes together with primary antibodies for 24 h at 4 °C. We incubated membranes using horseradish peroxidase-conjugated secondary antibody. Bands were noticed by Chemi Doc XRS System (Bio-Rad). Immunoblots were visualized by ECL (Cat.No. PE0010, Solarbio, Beijing, China) and analyzed using ImageJ software. The antibody against GAPDH (T004, Affinity Biosciences, USA) was utilized as a loading control. The experiment repeated 3 times.
Animal models
To construct the subcutaneous gastric tumor model, HGC-27 cells (1 × 108 cells per mouse) transfected with NC or WAC-AS1 knockdown lentivirus vector were injected through the BALB/c nude mice right armpit (n = 5, each group). Then, we examined lung tissues for metastasis. After 28 d, we sacrificed the mice and collected xenografted tumors for WB, PCR, HE staining, and IHC analysis. We calculated tumor volume and weight based on a previous study [29].
H&E staining
We fixed xenografted mouse tumor tissues with paraformaldehyde (4 %) and dehydrated paraffin-embedded. Continuous slices are cut to 4 um each. Dewaxing was done with xylene first, and then ethanol was used (100 %, 95 %, 75 %). We stained sections with Harris hematoxylin eosin. We sealed the slides with neutral glue. Sections are viewed under a microscope.
IHC
We performed IHC as mentioned previously [30]. We cut paraffin-embedded tissue blocks into slides (4-μm) cultured with primary antibodies against CD3, E Cadherin, ErbB2 / HER2, Ki67, N Cadherin, Slug, Snail or Vimentin at 4 °C for 24 h. We incubated sections and anti-mouse or rabbit horseradish peroxidase-conjugated secondary antibodies at 25 °C for 50 min duration after washing in PBS. We stained paraffin sections with DAB and then hematoxylin. We covered sections with coverslips for microscopic study. Antibody information in supplement Table 3.
Table 3.
The correlation between HOXC8 and clinicopathological features in 25 GC fresh-frozen tissues.
| Characteristics | Case | HOXC8 |
p value | |
|---|---|---|---|---|
| Low | High | |||
| Age at surgery(year) | 2 | 23 | 0.1912 | |
| <60 | 11 | 0 | 11 | |
| ≥60 | 14 | 2 | 12 | |
| Gender | 0.3118 | |||
| Male | 17 | 2 | 15 | |
| Female | 8 | 0 | 8 | |
| Tumor size(cm) | 0.2688 | |||
| ≥5 | 9 | 0 | 9 | |
| <5 | 16 | 2 | 14 | |
| T grade | 0.1715 | |||
| T1+T2 | 4 | 1 | 3 | |
| T3+T4 | 21 | 1 | 20 | |
| Lymph node invasion | 0.3694 | |||
| Negative(NO) | 6 | 1 | 5 | |
| Positive(N1-N3) | 19 | 1 | 18 | |
| TNM stage | 0.0847 | |||
| I-II | 3 | 1 | 2 | |
| III-IV | 22 | 1 | 21 | |
| Histological grade | 0.5199 | |||
| Low | 5 | 0 | 4 | |
| Middle-high | 20 | 2 | 19 | |
Bioinformatic analysis
We did differential analysis using GSE192468 and TCGA-STAD using R package “DESeq2”, with |log2 (fold change) |> 1 and P< 0.05 criteria, showing the lncRNA. ceRNA network was built through complex bioinformatic analysis. Log-rank tests having a P < 0.05 were considered significant. Detailed information of the database used in bioinformatic analysis is displayed in supplement Table 4.
Data analysis
Data were collected with GraphPad Prism 9. Each value was indicated as means ± SD. We applied the student's test and ANOVA. The association between WAC-AS1/HOXC8 or miR-204–5p with STAD prognosis was performed by Kaplan–Meier analysis. We used Pearson Correlation Analysis to analyze the association of WAC-AS1/HOXC8 with miR-204–5p. P < 0.05 was considered statistically significant. (*, P < 0.05. **, P ≤ 0.01. ***, P ≤ 0.001. ****, P ≤ 0.0001).
Results
WAC-AS1 upregulation in GC cells and tissues
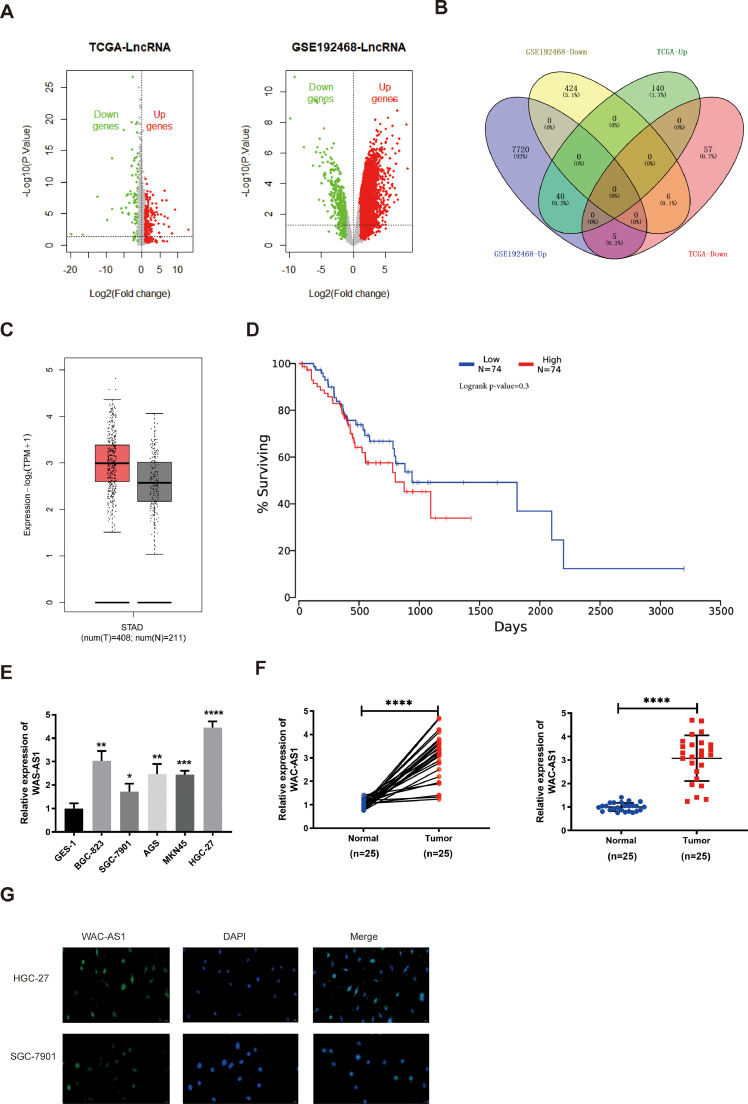
Based on the inclusion criteria, 247 and 8195 DEGs were extracted from the TCGA-STAD and GSE192468 lncRNA datasets using the R “Limma” package and visualized using volcano plots, respectively (Fig. 1A). Then, we analyzed 40 up-regulated and 6 down-regulated DEGs that were co-expressed in TCGA-STAD and GSE192468 based on the cutoff criteria (Fig. 1B). In our study, we focused on the WAC-AS1 upregulation in GC tissues and poor prognosis through literature research as well as database analysis (GEPIA 2 and Oncolnc software) (Fig. 1C-D).
Fig. 1.
The expression profile of WAC-AS1(WAC-AS1) in GC tissues and cell lines A. The LncRNAs volcano map of GC with TCGA and GSE192468 data B. The DEGs among the two databases by Venn diagram C. The expression level of WAC-AS1 in GC tissues from GEPIA 2 database D. The overall survival (OS) of WAC-AS1 in GC patients from Oncolnc online software E-F. The expression of WAC-AS1 in GC cells and tissues G. The prognosis of high or low expression of WAC-AS1 was analyzed by K-M Plotter, the cut-off point came from the median value. H. The subcellular localization of WAC-AS1 in HGC-27 and SGC-7901 cells was determined by FISH. Original magnification 400×, median WAC-AS1 value was used as cutoff. Data were showed as mean ± SD; *P < 0.05, **P < 0.01, ***P & ****P < 0.001.
Further qRT-PCR experimental confirmed that WAC-AS1 was expressed in 5 GC cells while comparing with normal GC (Fig. 1E). In addition, we investigated WAC-AS1 expression in 25 pairs of GC as well as normal gastric tissues, and WAC-AS1 was overexpressed in GC tissues linking with neighboring normal tissues (Fig. 1F). Furthermore, the relationship between lncRNA WAC-AS1 expression level and clinicopathological characteristics in GC patients is summarized in Table 1. WAC-AS1 was situated in the cytoplasm (Fig. 1G).
WAC-AS1 promotes the GC development and progression
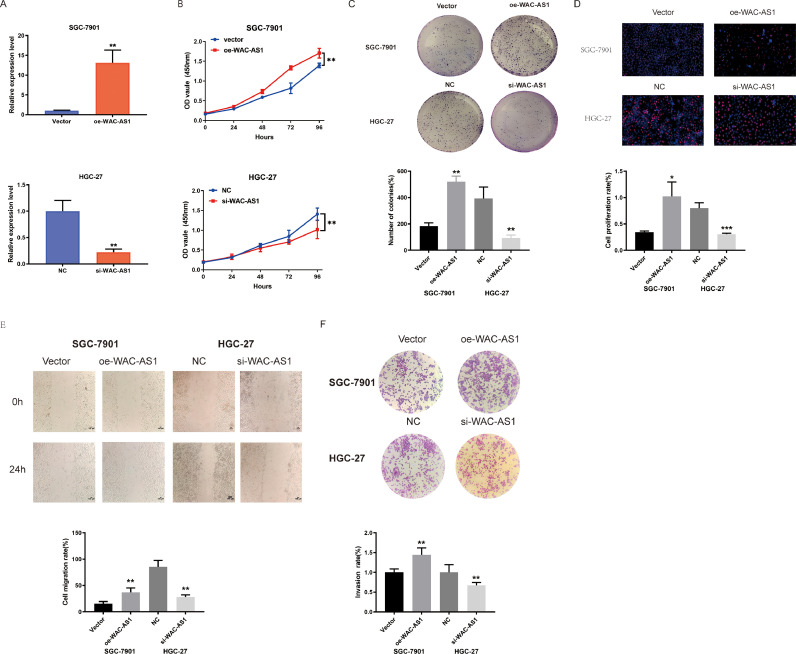
We knocked down and overexpressed WAC-AS1 in GC cells. The expression was confirmed through qRT-PCR (Fig. 2A). After WAC-AS1 upregulation, the proliferation, colony-forming ability, and EdU-positive cells of GC cells were significantly raised, but after WAC-AS1 knockdown, the effect was the opposite (Fig. 2B-D), which suggests that WAC-AS1 promoted GC cells proliferation.
Fig. 2.
WAC-AS1 promoted GC cells growth in vitro. A. The expression of si-WAC-AS1 or oe-WAC-AS1 in HGC-27 and SGC-7901 cells after stable transfection was detected by qRT-PCR assay B-D. The ability of proliferation was performed in SGC-7901 cells transfected with oe-lnc or vector, and HGC-27 cells transfected with si-lnc or NC by CCK-8 assays, colony formation assays and EdU assays, Scale bar, 50 μm. E-F. Cell migration and proliferation capacity were assessed by wound healing assays and transwell assays in SGC-7901 cells transfected with oe-lnc or vector, and HGC-27 cells transfected with si-lnc or NC. The black scale bar indicated 100 μm. Data were showed as mean ± SD. *P < 0.05, **P < 0.01, *** & ****P < 0.001, qRT-PCR, quantitative real-time polymerase chain reaction.
Furthermore, we determined GC cell invasion and migration. WAC-AS1 overexpression increased GC cell migration and invasion, while WAC-AS1 knockdown exerted opposite impacts (Fig. 2E-F).
Moreover, WAC-AS1 overexpression obviously decreased GC cell total apoptosis rate, while knockdown WAC-AS1 raised the total apoptosis rate of GC cells by flow cytometry assay (Supplement Fig. 1A). Finally, we used WB for detecting EMT marker expression. Knockdown of WAC-AS1 suppressed Vimentin, N-cadherin, Slug, and Snail1 protein expressions in HGC-27 cells and enhanced the E-cadherin protein expression, whereas upregulated WAC-AS1 enhanced Vimentin, N-cadherin, Slug, and Snail1 protein expressions in SGC-7901 cells, and suppressed E-cadherin protein expression. When WAC-AS1 was decreased, GC cells could not acquire an EMT phenotype (Supplement Fig. 1B).
MiR-204-5p downregulation in GC and progression and poor prognosis
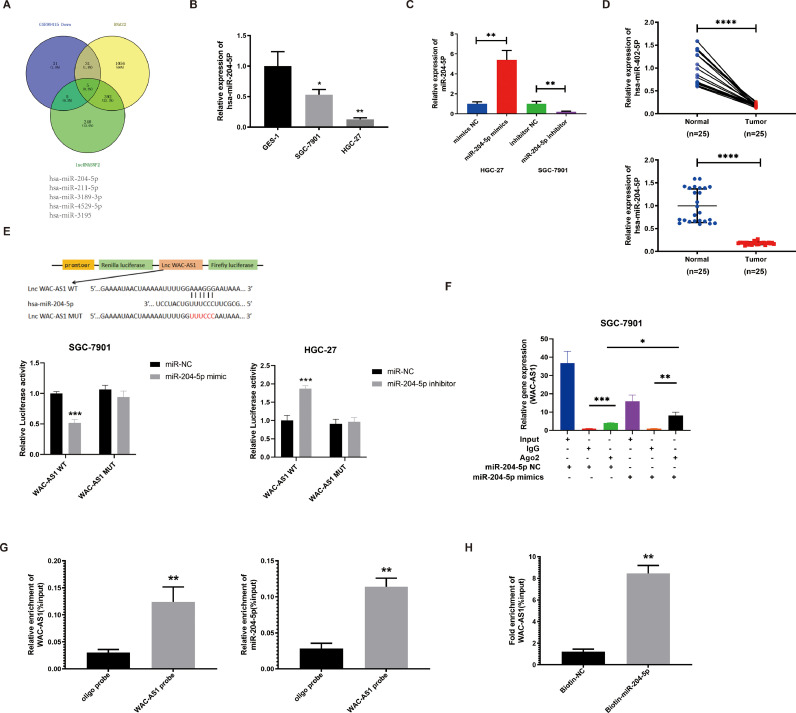
According to the Competing Endogenous RNA (ceRNA) theory, lncRNAs regulate miRNA expression [31]. Since WAC-AS1 is distributed in the cytoplasm, WAC-AS1 prevents miRNAs from binding to its target mRNAs. Then, GSE99415, RNA22 and lncRNASNP2 were used for predicting the WAC-AS1 targets. miR-204–5p was detected as a candidate target miRNA, which was expressed lower in the GC tissues (Supplement Fig. 2A & Fig. 3A). From the starBase and K-M plotter software, we found miR-204–5p downregulation in GC and showed a trend for poor prognosis (Supplement Fig. 2B&C). Then, it was also downregulated in GC cells and tissues than the control by PCR analysis (Fig. 3B-D). Additionally, the association between clinicopathological characteristics and miR-204–5p expression in GC patients is summarized in Table 2, it was found that the expression level of miR-204–5p significantly with gender (P < 0.0003).
Fig. 3.
WAC-AS1 interacts with miR-204-5p. A. Venn diagram exhibiting overlapping of the target miRNAs of WAC-AS1 predicted by GSE99415, RNA22 and lncRNASNP2. B-D. The relative expression of miR-204-5p in GC cells and tissues was detected by qRT-PCR. E. The direct interaction between WAC-AS1 and miR-204-5p was investigated by the luciferase assay. F. The interaction between WAC-AS1 and miR-204-5p was demonstrated by RIP assays in SGC-7901 cells. G-H. The enrichment of lWAC-AS1 and miR-204–5p was detected by RNA pull-down assay in SGC-7901 cells. Data were showed as mean ± SD. *P < 0.05, **P < 0.01, ***P & ****P < 0.001.
A highly negative correlation between miR-204–5p as well as WAC-AS1 expressions was observed (Supplement Fig. 3A). We compared WAC-AS1 in HGC-27 cell levels transfected with miR-204–5p mimic and control vector and SGC-7901 cells transfected with miR-204–5p inhibitor construct and NC. WAC-AS1 was significantly impaired in the miR-204–5p mimc group, whereas it enhanced in the miR-204–5p inhibitor group (Supplement Fig. 3B).
To confirm the relationship between miR-204–5p and WAC-AS1, dual-luciferase reporter assay was performed in GC cells. The results showed that the luciferase activity of WAC-AS1-WT group was significantly reduced in miR-204–5p mimic group compared with NC group, whereas the luciferase activity of miR-204–5p inhibitor group was increased while comparing with the NC. However, no difference in luciferase activity between WAC-AS1-MUT and miR-204–5p mimics, inhibitor, and NC groups was observed, suggesting that WAC-AS1 may interact with miR-204-5p (Fig. 3E). It is commonly believed that miRNAs regulate the expression of target genes by binding to Argonaute 2 (Ago2), an important component of the RNA-induced silencing complex (RISC). Subsequently, RNA immunoprecipitation (RIP) assays were performed in SGC-7901 cells to pull down the Ago2-bound RNA transcripts. qRT-PCR results showed that anti-Ago2 reduced the expression of WAC-AS1 while comparing with the input control. In addition, WAC-AS1 was highly enriched in cells transfected with miR-204-5p mimics compared to the NC (Fig. 3F). To further confirm the direct interaction between WAC-AS1 and miR-204-5p, RNA pull-down assay was performed using a biotin-labeled WAC-AS1 probe and miR-204-5p probe. Interestingly, WAC-AS1 and miR-204-5p were significantly enriched in the WAC-AS1 probe group compared with the control through qRT-PCR. Moreover, biotin-labeled miR-204-5p probes captured more WAC-AS1 than the control probes group (Fig. 3G-H). In summary, these data suggest that WAC-AS1 interacts with miR-204-5p to jointly regulate the disease progression of GC.
MiR-204-5p endorses GC cell migration, invasion, and proliferation by HOXC8 target
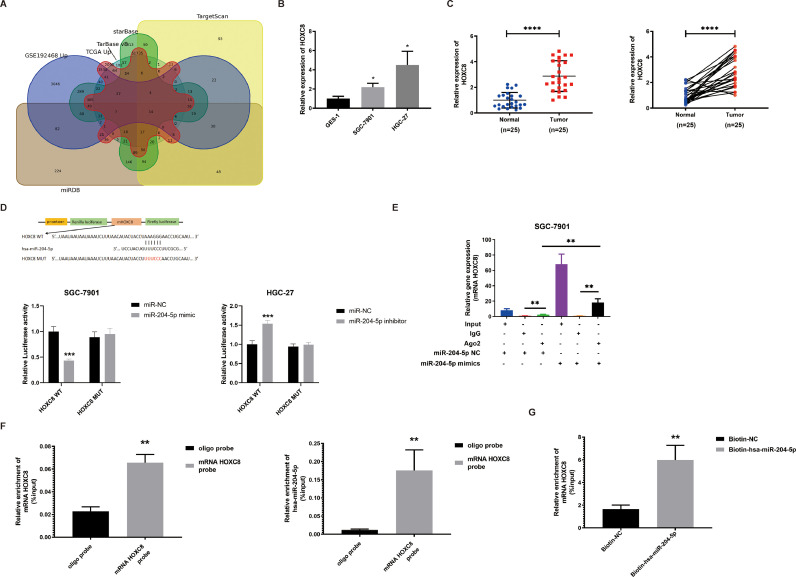
Some papers have reported that miR-204-5p played a role in the progression of GC, but its mechanism has not been fully studied. Our paper will deeply understand the mechanism and biological process of miR-204-5p in GC through the following studies. Then, we identified that HOXC8 was a candidate target of miR-204-5p through starBase, TarBase v8, miRDB, TargetScan, TCGA-STAD, and GSE192468 (Fig. 4A). The volcano map of TCGA-STAD and GSE192468 is shown in Supplement Fig. 4A. An association between HOXC8 expression level and GC patients’ clinical characteristics was observed (Table 3).
Fig. 4.
The interaction between miR-204-5p and HOXC8 A. HOXC8 was identified as a target of miR-204-5p by Venn diagram analysis. B-C. The expression of HOXC8 in GC cells and tissues by qRT-PCR. D. The direct interaction between WAC-AS1 and miR-204-5p was investigated by the luciferase assay. E. The interaction between WAC-AS1 and miR-204-5p was demonstrated by RIP assays in SGC-7901 cells. F-G. The enrichment of lWAC-AS1 and miR-204-5p was detected by RNA pull-down assay in SGC-7901 cells. Data were showed as mean ± SD. *P < 0.05, **P < 0.01, ***P&****P < 0.001.
In addition, using GEPIA 2, UALCAN, and TIMER databases, we found that high HOXC8 expression level was elevated in GC tissues compared with normal tissues (Supplement Fig. 4B), and high HOXC8 expression level was associated with a poor prognosis in GC patients (Supplement Fig. 4C). The result is verified by qRT-PCR, Kaplan-Meier method and survival information for the patients registered previously was gathered (n = 25) (Fig. 4B-C & Supplement Fig. 5A). Furthermore, we found that the HOXC8 and miR-204–5p expression level was negatively correlated in GC tissues (Supplement Fig. 5B). Therefore, HOXC8 was detected as the target gene of miR-204–5p for an in-depth study. The qRT-PCR and WB findings verified that HOXC8 protein and mRNA expression levels were decreased when over-expressed miR-204–5p, while the mRNA and protein expression of HOXC8 was increased when knockdown miR-204–5p (Supplement Fig. 5C-D). Luciferase assay showed that miR-204–5p overexpression reduced the luciferase activity of wild-type HOXC8 reporter gene but had no effect on mutant reporter gene in SGC-7901 cells, and miR-204-5p knockdown increased the luciferase activity of wild-type HOXC8 reporter gene but had no effect on mutant reporter gene in HGC-27 cells (Fig. 4D), indicating that HOXC8 was the direct target of miR-204-5p. Additionally, RIP assays &RNA Pull down assays could verify the direct miR-204–5p and HOXC8 network (Fig. 4E-G). Therefore, we can confirm that HOXC8 is the targeted gene for miR-204–5p.
Fig. 5.
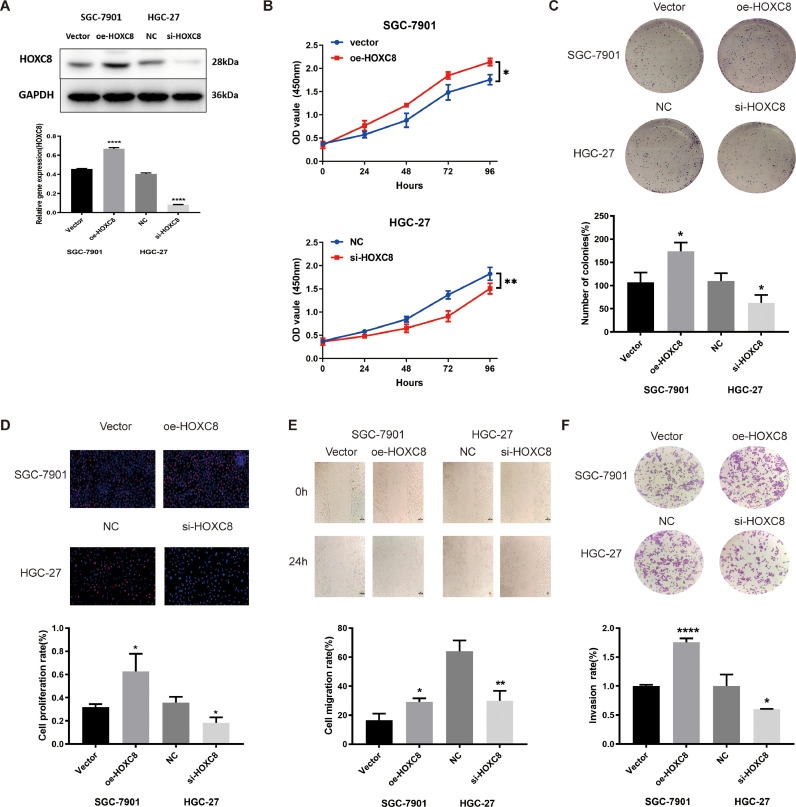
HOXC8 promoted GC cells growth in vitro. A. The expression of oe-HOXC8 or si-HOXC8 in SGC-7901 and HGC-27 cells after stable transfection was detected by WB assay B-D. The ability of proliferation was performed in SGC-7901 cells transfected with oe-HOXC8 or vector, and HGC-27 cells transfected with si-HOXC8 or NC by CCK-8 assays, colony formation assays and EdU assays, Scale bar, 50 μm. E-F. Cell migration and proliferation capacity were assessed by wound healing assays and transwell assays in SGC-7901 cells transfected with oe-HOXC8 or vector, and HGC-27 cells transfected with si-HOXC8 or NC. The black scale bar indicated 100 μm. G. Cell apoptosis were detected by Flow cytometry assay in SGC-7901 cells transfected with oe-HOXC8 or vector, and HGC-27 cells transfected with si-HOXC8 or NC. H. The protein expression levels of N-cadherin, E-cadherin, Vimentin, Slug, and Snail were detected in the overexpression and silencing HOXC8 by WB. Data were showed as mean ± SD. *P < 0.05, **P < 0.01, *** & ****P < 0.001.
We further examined HOXC8 biological function in GC cells. HOXC8 protein level was increased when SGC-7901 cells were transfected with oe-HOXC8, and HOXC8 levels were decreased when HGC-27 cells were transfected with si-HOXC8(Fig. 5A). CCK-8 assay, colony formation assay, and EdU assay showed that increased HOXC8 expression significantly promoted cell proliferation, while decreased HOXC8 expression had the opposite effect (Fig. 5B-D). Moreover, the impacts of HOXC8 on migration and invasion of GC cells were examined by wound healing and transwell assays. The results demonstrated that overexpression of HOXC8 promoted the migration and invasion of SGC-7901 cells, whereas knockdown of HOXC8 inhibited the migration and invasion of HGC-27 cells (Fig. 5E-F). Additionally, HOXC8 overexpression obviously decreased the total apoptosis rate of GC cells, while knockdown HOXC8 increased the total apoptosis rate of GC cells by flow cytometry assay (Supplement Fig. 6A). Finally, HOXC8 was transfected into HGC-27 and SGC-7901 cell lines for investigating the impact of HOXC8 on EMT factors in GC. N-cadherin, Vimentin, Slug, and Snail1 expression levels significantly decreased in the si-HOXC8 group in HGC-27 cells, and E-cadherin expression raised. The expression levels of Vimentin, N-cadherin, Slug, and Snail1 was significantly raised in the oe-HOXC8 group in SGC-7901 cells, and E-cadherin expression was significantly decreased (Supplement Fig. 6B). When HOXC8 was decreased, GC cells could not acquire an EMT phenotype (Supplement Fig. 6B). In summary, HOXC8 promotes the proliferation, migration and invasion of GC.
Fig. 6.
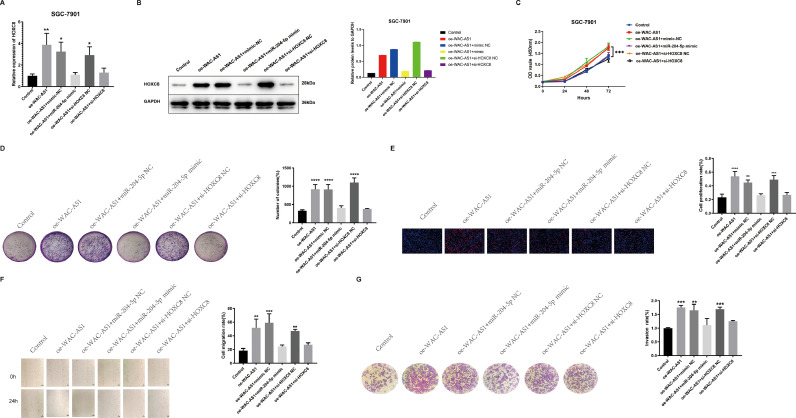
WAC-AS1 promote GC progression and development through WAC-AS1/miR-204-5p/HOXC8 axis. A-B. Relative mRNA and protein expression of HOXC8 in SGC-7901 cells transfected with miR-204-5p mimic, NC, or si-HOXC8 by qRT-PCR. C-E. The ability of proliferation in SGC-7901 cells transfected with miR-204-5p mimic, NC, or si-HOXC8 were performed by CCK-8 assays, colony formation assays and EdU assays, respectively. Scale bar, 50 μm. F-G. The migration and invasion in SGC-7901 cells transfected with miR-204-5p mimic, NC, or si-HOXC8 were determined by wound healing and transwell assays, respectively. The black scale bar indicated 100 μm. Data were showed as mean ± SD. *P < 0.05, **P < 0.01, *** & ****P < 0.001,.
WAC-AS1 serves as a miRNA sponge of miR-204-5p to regulate HOXC8 expression
Many articles have reported that lncRNAs act as competing endogenous RNAs (ceRNAs) that bind to miRNAs and act as miRNA sponges in cells [[32], [33]]. To study the relationship between WAC-AS1, miR-204–5p, and HOXC8, we conducted a series of rescue experiments in SGC-7901 cells, including oe-WAC-AS1, miR-204–5p mimic, and si-HOXC8. HOXC8 expression level in SGC-7901 cells was improved when upregulated WAC-AS1, while the effects of activating WAC-AS1 were reversed by miR-204–5p mimic or si-HOXC8 (Fig. 6A-B).
In addition, we attempted to explore whether the biological function of WAC-AS1 in GC cells could also be reversed by miR-204–5p mimics or si-HOXC8. The results of CCK-8 assay, colony formation assay, EdU assay, wound healing assay, transwell assay and flow cytometry apoptosis assay showed that miR-204–5p mimic and si-HOXC8 reversed the promotion of proliferation, migration and invasion and inhibition of apoptosis-induced by overexpression of lncRNAWAC-AS1 in SGC-7901 cells (Fig. 6C-G & Supplement Fig. 7A). Overexpression of WAC-AS1 increased the transcription of HOXC8 and mesenchymal markers N-cadherin, Vimentin, Slug, and Snail1. However, transcription of these markers was restored when co-transfected with miR-204–5p mimics or si-HOXC8. The E-cadherin transcription pattern was opposite to that of the other proteins (Supplement Fig. 7B).
Fig. 7.
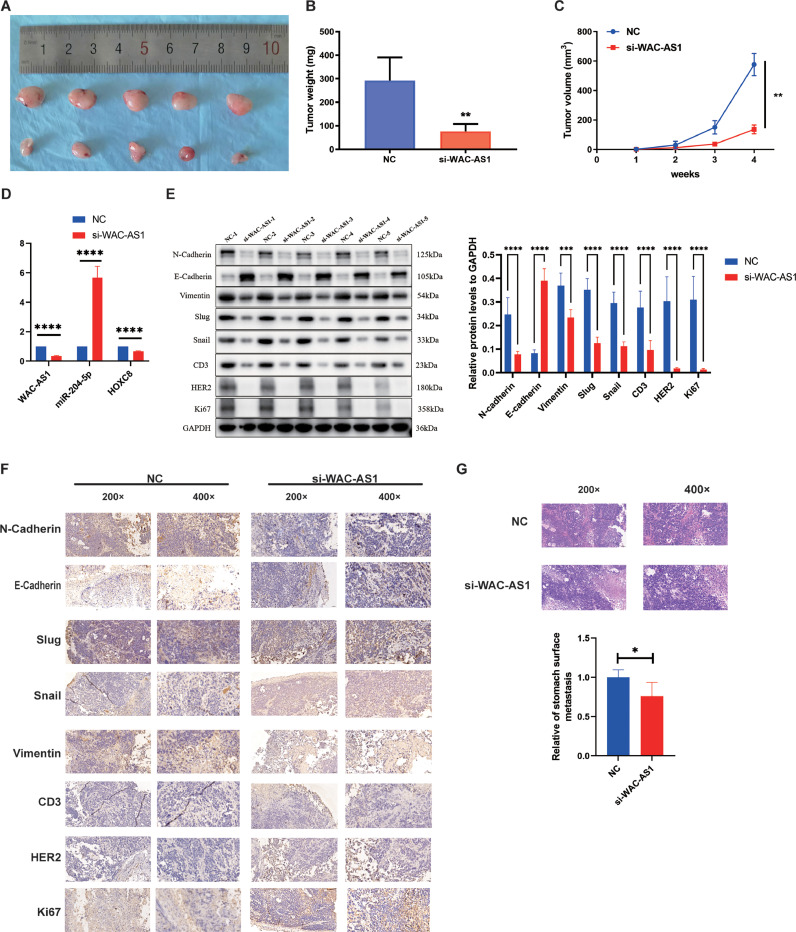
Knockdown WAC-AS1 suppressed the growth of GC in vivo. A. Image of the xenograft tumors from BALB/c nude mice in WAC-AS1 downregulated group and control group. B. The relative weights of tumors were evaluated. C. The tumor volume of mice was measured every week. D. Relative expression levels of WAC-AS1, miR-204-5p and HOXC8 were observed in subcutaneous tumor tissues by qRT-PCR. E. Relative expression levels of N-cadherin, E-cadherin, Vimentin, Slug, Snail, CD3, HER2, and Ki-67 were observed in subcutaneous tumor tissues by western blot assays. F. Relative expression levels of N-cadherin, E-cadherin, Vimentin, Slug, Snail, CD3, HER2, and Ki-67 were observed in subcutaneous tumor tissues by IHC. G. Representative images in HE staining of the gastric metastasis of HGC-27 cells. Data were showed as mean ± SD. *P < 0.05, **P < 0.01, ***P & ****P < 0.001.
Knockdown WAC-AS1 suppressed growth and GC gastric metastasis
To further investigate the effect of WAC-AS1 on tumor growth and gastric metastasis in vivo, HGC-27 cells stably transfected with si-WAC-AS1 or control vector were subcutaneously injected into BALB/c nude mice. Tumor volume was measured every 7 days. The tumor weight was measured after 28 days. Compared with the control group, WAC-AS1 knockdown reduced tumor weight and volume (Fig. 7A-C). These subcutaneous tumor tissues were subjected to qRT-PCR, WB, and IHC. The qRT-PCR results showed that the expression of HOXC8 was down-regulated in the WAC-AS1 knockdown group (Fig. 7D), and the WB results presented that N-cadherin, Vimentin, Slug, Snail, CD3, HER2, and Ki-67 expression levels were down-regulated. Upregulation of E-cadherin expression was observed in the WAC-AS1 knockdown group (Fig. 7E).
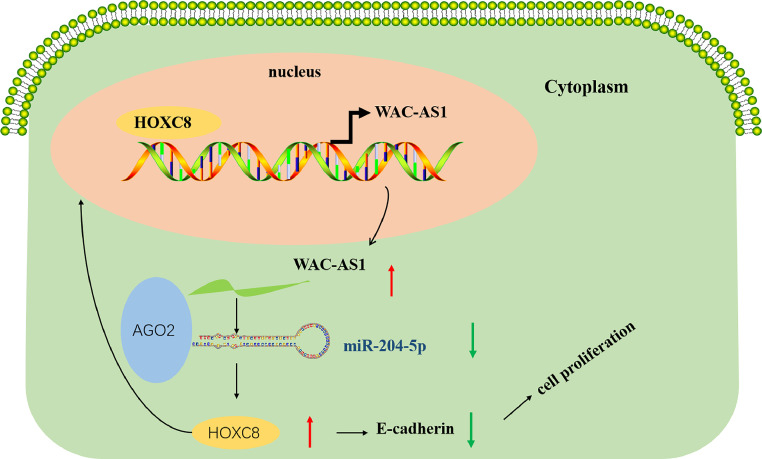
E-cadherin expression was increased, and N-cadherin, Vimentin, Slug, Snail, CD3, HER2, as well as Ki-67 expression levels were reduced in the transplanted tumor tissues by IHC assays (Fig. 7F). The role of WAC-AS1 in tumor metastasis was also investigated. Comparing with the control, the WAC-AS1 knockdown group had fewer gastric metastases (Fig. 7G). In summary, these findings together with the above evidence demonstrate that WAC-AS1 acts as a miR-204–5p sponge to promote GC progression by enhancing HOXC8 expression (Fig. 8).
Fig. 8.
The schematic diagram shows the mechanism underlying lncRNA WAC-AS1 as a ceRNA for miR-204-5p to regulate HOXC8 expression in the progression of gastric cancer.
Discussion
LncRNAs play a major role in genome transcription, translation, and post-translational modifications [34]. Some lncRNAs have been identified as biomarkers indicating the development of malignant tumors and predicting patient prognosis. For instance, RMST served as both a prognostic biomarker and tumor promoter by modulating miR-204–5p [24]. Similarly, lncRNA PVT1 was a key player in tumorigenesis and therapeutic in lung cancer [35]. Additionally, lncRNA CARMN acts as a tumor suppressor in the metastasis and a potential biomarker for progression in early-stage BC [36]. WAC-AS1 is an antisense RNA head-to-head of WAC [16] and related to ovarian cancer as well as hepatocellular carcinoma [17,37]. In present study, we confirmed WAC-AS1 mechanisms in GC. The DE WAC-AS1 in GC tissues was obtained through multiple bioinformatics analyses. In the microarray data, WAC-AS1 was up-regulated in GC tissues. It was related to GC patients’ poor prognosis. WAC-AS1 promoted the proliferation, migration, and invasion and inhibited GC cells’ apoptosis, suggesting that WAC-AS1 was a factor that induced the development of GC.
GSE99415, RNA22 and lncRNASNP2 could predict the target miRNA of WAC-AS1. Then, WAC-AS1 and miR-204–5p were determined to function together in the cytoplasm of GC cell, and WAC-AS1 targets miR-204–5p through dual luciferase reporter, RIP, and RNA pull down assay. MiR-204–5p is regulated by various lncRNAs, thereby affecting the tumor progression of human cancer. For example, lncSLCO1C1 promotes GC progression by enhancing cell growth and preventing DNA damage via interacting and scaffolding the SSRP1/H2A/H2b complex and absorbing both miR-211-5p and miR-204-5p to increase SSRP1 expression [38]. LncRNA NORAD mediates KMT2D expression by targeting miR-204–5p and affects the growth of gastric cancer [39]. Subsequently, WAC-AS1 reversed the oncogenic result of miR-204–5p confirmed by rescue experiments. MiR-204–5p is down-regulated in GC and promotes the invasion and metastasis of GC [[25], [40],41]. Furthermore, the downregulation of miR-204–5p expression level endorsed GC cell WAC-AS1 activation, indicating that miR-204–5p is the key regulator of WAC-AS1. WAC-AS1 provides a therapeutic target for GC patients. Suggesting, miR-204–5p was predicted the downstream miRNA of WAC-AS1, and significantly down-regulated in GC cells and tissues, and was related to the OS of patients and GC cell metastasis and proliferation [25].
WAC-AS1 can counteract the promoting effect of the HOXC8 gene on GC progression by binding to miR-204–5p found by further experiments. The active role of lncRNAs has been reported to occur mainly in the cytoplasm [14,42]. Most lncRNAs contain MREs, and the ceRNA mechanism is the main pathway for lncRNAs exertion of their functions [[12], [14], [31], [43]]. For example, lncRNA-MIAT endorses thyroid cancer development by targeting miR-150–5p and functions as a ceRNA on EZH2 [44]. There are many more studies on the mechanism of lncRNA sponges.
Homologous box (HOX) genes are a highly conserved homeobox superfamily subgroup, consisting of 39 transcription factors, including HOXA, HOXB, HOXC and HOXD, which significantly affect various cellular processes such as apoptosis, proliferation, cell shape, cell migration and angiogenesis genes are a highly conserved homologous box superfamily subgroup, consisting of 39 transcription factors, including HOXA, HOXB, HOXC and HOXD, which significantly affect various cellular processes such as apoptosis, proliferation, cell shape, cell migration and angiogenesis [45,46]. Large amount evidence have highlighted the potential role of HOX genes in the progression and metastasis of several tumors and resistance to therapy [47]. As a member of the HOX family, HOXC8 can regulate genes related to proliferation, adhesion and migration, and is considered to be a global regulator of human cell growth and differentiation [48]. HOXC8 has been reported to participate in the growth and migration of breast cancer cells by promoting the epithelial-mesenchymal transition (EMT) of tumor cells [49]. In addition, upregulated HOXC8 expression was also observed in prostate, cervical,and lung, and gastric cancers, promoting the metastasis and progression of tumor cells [[50], [51], [52], [53], [54]]. In our research, we also found HOXC8 expression was up-regulated through comprehensive bioinformatics analysis. Furthermore, HOXC8 is the target gene of miR-204-5p by RIP and dual luciferase reporter assays. HOXC8 endorsed the GC occurrence and progression and was related to poor prognosis. How WAC-AS1 and miR-204-5p interact with HOXC8 to affect the progression of GC has also been further studied. lncRNA WC-AS1 interacted with miR-204-5p to inhibit HOXC8 expression. WAC-AS1/miR-204-5p/HOXC8 axis is a new mechanism in GC development.
EMT factors, HER2, CD3, and ki67, are related to GC [4,55,56]. WAC-AS1 promoted the mesenchymal transition of GC cells in this study while inhibiting the expression of HER2, CD3, and ki67 through the miR-204-5p/HOXC8 axis.
However, our findings have several limitations. First of all, the clinical samples in our study are small and heterogeneous, which cannot fully explain the role of WAC-AS1 in GC, and more clinical samples are needed to verify its function. Secondly, our preliminary study showed that WAC-AS1 was significantly upregulated in GC. Therefore, the intensive study of WAC-AS1 in clinical samples makes it possible to become an ideal biomarker and therapeutic target for GC. Thirdly, our study demonstrated the binding ability of WAC-AS1 to miR-204-5p. However, there may be other miRNAs that bind to WAC-AS1 to regulate the development of GC that were not predicted by bioinformatics analysis. Fourthly, whether WAC-AS1 regulates GC development through other mechanisms, such as interaction with RNA-binding proteins and sponge trans-acting elements, requires further investigation. Therefore, a deeper understanding of the therapeutic potential of WAC-AS1 in GC needs to be further explored.
Conclusion
WAC-AS1 serves as a tumor inducer and a prognostic biomarker in GC. In addition, WAC-AS1 acts in the blockage of miR-204-5p activity, thus contributing in HOXC8-induced pathways and promoting the GC cell invasion and proliferation abilities. This study offers new targets for GC patients.
Ethics statement
All experiments in this study were endorsed by the Ethics Committee of China-Japan Union Hospital of Jilin University and complied with the Declaration of Helsinki. The Ethics Committee of China-Japan Union Hospital of Jilin University specifically approved that no informed consent was required, because data was analyzed anonymously.
Funding statement
This work was supported by Wu Jieping Medical Foundation clinical research project (YDZJ202201ZYTS069).
Data availability statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
CRediT authorship contribution statement
Yan Liu: Writing – review & editing, Writing – original draft, Data curation. Kaixuan Li: Writing – original draft, Software. Yongjian Gao: Writing – review & editing, Writing – original draft, Investigation. Ye Feng: Writing – original draft, Formal analysis. Xiaoling Zhao: Writing – review & editing, Writing – original draft, Data curation. Ruizhi Hou: Writing – review & editing, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare no competing interests
Acknowledgments
No applicable.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2024.102139.
Contributor Information
Xiaoling Zhao, Email: xiaoling.12356@163.com.
Ruizhi Hou, Email: hourz@jlu.edu.cn.
Appendix. Supplementary materials
References
- 1.Zheng R.S., Chen R., Han B.F., Wang S.M., Li L., Sun K.X., Zeng H.M., Wei W.W., He J. [Cancer incidence and mortality in China, 2022] Zhonghua Zhong Liu Za Zhi. 2024;46:221–231. doi: 10.3760/cma.j.cn112152-20240119-00035. [DOI] [PubMed] [Google Scholar]
- 2.Alsina M., Arrazubi V., Diez M., Tabernero J. Current developments in gastric cancer: from molecular profiling to treatment strategy. Nat. Rev. Gastroenterol. Hepatol. 2023;20:155–170. doi: 10.1038/s41575-022-00703-w. [DOI] [PubMed] [Google Scholar]
- 3.Norwood D.A., Montalvan-Sanchez E., Dominguez R.L., Morgan D.R. Gastric cancer: emerging trends in prevention, diagnosis, and treatment. Gastroenterol. Clin. North Am. 2022;51:501–518. doi: 10.1016/j.gtc.2022.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Hogner A., Moehler M. Immunotherapy in gastric cancer. Curr. Oncol. 2022;29:1559–1574. doi: 10.3390/curroncol29030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan W.L., He Y., Xu R.H. Gastric cancer treatment: recent progress and future perspectives. J. Hematol. Oncol. 2023;16:57. doi: 10.1186/s13045-023-01451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashrafizaveh S., Ashrafizadeh M., Zarrabi A., Husmandi K., Zabolian A., Shahinozzaman M., Aref A.R., Hamblin M.R., Nabavi N., Crea F., et al. Long non-coding RNAs in the doxorubicin resistance of cancer cells. Cancer Lett. 2021;508:104–114. doi: 10.1016/j.canlet.2021.03.018. [DOI] [PubMed] [Google Scholar]
- 7.Mirzaei S., Gholami M.H., Hushmandi K., Hashemi F., Zabolian A., Canadas I., Zarrabi A., Nabavi N., Aref A.R., Crea F., et al. The long and short non-coding RNAs modulating EZH2 signaling in cancer. J. Hematol. Oncol. 2022;15:18. doi: 10.1186/s13045-022-01235-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirzaei S., Paskeh M.D.A., Okina E., Gholami M.H., Hushmandi K., Hashemi M., Kalu A., Zarrabi A., Nabavi N., Rabiee N., et al. Molecular landscape of LncRNAs in prostate cancer: a focus on pathways and therapeutic targets for intervention. J. Exp. Clin. Cancer Res. 2022;41:214. doi: 10.1186/s13046-022-02406-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mirzaei S., Zarrabi A., Hashemi F., Zabolian A., Saleki H., Ranjbar A., Seyed Saleh S.H., Bagherian M., Sharifzadeh S.O., Hushmandi K., et al. Regulation of Nuclear Factor-KappaB (NF-kappaB) signaling pathway by non-coding RNAs in cancer: inhibiting or promoting carcinogenesis? Cancer Lett. 2021;509:63–80. doi: 10.1016/j.canlet.2021.03.025. [DOI] [PubMed] [Google Scholar]
- 10.Bhan A., Soleimani M., Mandal S.S. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Statello L., Guo C.J., Chen L.L., Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021;22:96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan H., Bu P. Non-coding RNA in cancer. Essays Biochem. 2021;65:625–639. doi: 10.1042/EBC20200032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan J.J., Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi X., Zhang D.H., Wu N., Xiao J.H., Wang X., Ma W. ceRNA in cancer: possible functions and clinical implications. J. Med. Genet. 2015;52:710–718. doi: 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 15.Yuan L., Xu Z.Y., Ruan S.M., Mo S., Qin J.J., Cheng X.D. Long non-coding RNAs towards precision medicine in gastric cancer: early diagnosis, treatment, and drug resistance. Mol. Cancer. 2020;19:96. doi: 10.1186/s12943-020-01219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y., Gong H., Cao Y. LncRNA WAC-AS1 expression in human tumors correlates with immune infiltration and affects prognosis. Hereditas. 2023;160:26. doi: 10.1186/s41065-023-00290-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai J., Qiu J., Wang H., Sun J., Ji Y. Identification of potential biomarkers in ovarian carcinoma and an evaluation of their prognostic value. Ann. Transl. Med. 2021;9:1472. doi: 10.21037/atm-21-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng J., Zhou Z., Qiu Y., Wang M., Yu H., Wu Z., Wang X., Jiang X. A prognostic ferroptosis-related lncRNAs signature associated with immune landscape and radiotherapy response in glioma. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.675555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia X., Zhang H., Xia P., Zhu Y., Liu J., Xu K., Yuan Y. Identification of glycolysis-related lncRNAs and the novel lncRNA WAC-AS1 promotes glycolysis and tumor progression in hepatocellular carcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.733595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao M., Yuan D., Jiang H., Zhou G., Chen C., Han G. Long non-coding RNA WAC antisense RNA 1 mediates hepatitis B virus replication <em>in vitro</em>by reinforcing miR-192-5p/ATG7-induced autophagy. Eur. J. Histochem. 2022;66 doi: 10.4081/ejh.2022.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He H., Chen K., Wang F., Zhao L., Wan X., Wang L., Mo Z. miR-204-5p promotes the adipogenic differentiation of human adipose-derived mesenchymal stem cells by modulating DVL3 expression and suppressing Wnt/beta-catenin signaling. Int. J. Mol. Med. 2015;35:1587–1595. doi: 10.3892/ijmm.2015.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F., Bian Z., Xu P., Sun S., Huang Z. MicroRNA-204-5p: a pivotal tumor suppressor. Cancer Med. 2023;12:3185–3200. doi: 10.1002/cam4.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 24.Cai H., Li C., Wu Z. lncRNA RMST is associated with the progression and prognosis of gastric cancer via miR-204-5p. Cell Div. 2024;19:12. doi: 10.1186/s13008-024-00117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang S., Chen B., Zhang B., Li C., Qiu Y., Yang H., Huang Z. miR‑204‑5p promotes apoptosis and inhibits migration of gastric cancer cells by targeting HER‑2. Mol. Med. Rep. 2020;22:2645–2654. doi: 10.3892/mmr.2020.11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Min S., Zhang L., Zhang L., Liu F., Liu M. LncRNA MIR100HG affects the proliferation and metastasis of lung cancer cells through mediating the microRNA-5590-3p/DCBLD2 axis. Immun. Inflamm. Dis. 2024;12:e1223. doi: 10.1002/iid3.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu L., Zhu Y., Han S., Chen M., Song P., Dai D., Xu W., Jiang T., Feng L., Shin V.Y., et al. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death. Dis. 2019;10:383. doi: 10.1038/s41419-019-1585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Y., Zhou B., Hu X., Ying S., Zhou Q., Xu W., Feng L., Hou T., Wang X., Zhu L., et al. LncRNA LINC00942 promotes chemoresistance in gastric cancer by suppressing MSI2 degradation to enhance c-Myc mRNA stability. Clin. Transl. Med. 2022;12:e703. doi: 10.1002/ctm2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian J., Cheng L., Kong E., Gu W., Jiang Y., Hao Q., Kong B., Sun L. linc00958/miR-185-5p/RSF-1 modulates cisplatin resistance and angiogenesis through AKT1/GSK3beta/VEGFA pathway in cervical cancer. Reprod. Biol. Endocrinol. 2022;20:132. doi: 10.1186/s12958-022-00995-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Zheng W., Zhang L., Gu Y., Zhu L., Huang Y. LncRNA FBXO18-AS promotes gastric cancer progression by TGF-beta1/Smad signaling. Eur. J. Histochem. 2023;67 doi: 10.4081/ejh.2023.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ala U. Competing endogenous RNAs and cancer: how coding and non-coding molecules cross-talk can impinge on disease. Int. J. Biochem. Cell Biol. 2021;130 doi: 10.1016/j.biocel.2020.105874. [DOI] [PubMed] [Google Scholar]
- 32.Su K., Wang N., Shao Q., Liu H., Zhao B., Ma S. The role of a ceRNA regulatory network based on lncRNA MALAT1 site in cancer progression. Biomed. Pharmacother. 2021;137:111389. doi: 10.1016/j.biopha.2021.111389. [DOI] [PubMed] [Google Scholar]
- 33.Xue S.T., et al. Long non-coding RNA LINC00680 functions as a ceRNA to promote esophageal squamous cell carcinoma progression through the miR-423-5p/PAK6 axis. Mol. Cancer. 2022;21(1):69. doi: 10.1186/s12943-022-01539-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang G., Xiang Z., Wu H., He Q., Dou R., Lin Z., Yang C., Huang S., Song J., Di Z., et al. The lncRNA BDNF-AS/WDR5/FBXW7 axis mediates ferroptosis in gastric cancer peritoneal metastasis by regulating VDAC3 ubiquitination. Int. J. Biol. Sci. 2022;18:1415–1433. doi: 10.7150/ijbs.69454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hakami M.A., Hazazi A., Khan F.R., Abdulaziz O., Alshaghdali K., Abalkhail A., Nassar S.A., Omar B.I.A., Almarshadi F., Gupta G., et al. PVT1 lncRNA in lung cancer: a key player in tumorigenesis and therapeutic opportunities. Pathol. Res. Pract. 2024;253 doi: 10.1016/j.prp.2023.155019. [DOI] [PubMed] [Google Scholar]
- 36.Liao H., Wang H., Zheng R., Yu Y., Zhang Y., Lv L., Zhang B., Chen J. LncRNA CARMN suppresses EMT through inhibiting transcription of MMP2 activated by DHX9 in breast cancer. Cell Signal. 2024;113 doi: 10.1016/j.cellsig.2023.110943. [DOI] [PubMed] [Google Scholar]
- 37.Wang S., Bai H., Fei S., Miao B. A cuproptosis-related LncRNA risk model for predicting prognosis and immunotherapeutic efficacy in patients with hepatocellular carcinoma. Biochem. Genet. 2023 doi: 10.1007/s10528-023-10539-x. [DOI] [PubMed] [Google Scholar]
- 38.Xiao Y.F., Li B.S., Liu J.J., Wang S.M., Liu J., Yang H., Hu Y.Y., Gong C.L., Li J.L., Yang S.M. Role of lncSLCO1C1 in gastric cancer progression and resistance to oxaliplatin therapy. Clin. Transl. Med. 2022;12:e691. doi: 10.1002/ctm2.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X., Pan T., Guo G., Chen G., Cai Y., Tang Y., Wang Y., Wang Y., Deng Z., Li L., et al. LncRNA NORAD mediates KMT2D expression by targeting miR-204-5p and affects the growth of gastric cancer. J. Gastrointest. Oncol. 2022;13:2832–2844. doi: 10.21037/jgo-22-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang Y., Zhang C.D., Zhang C., Dai D.Q. DLX6-AS1/miR-204-5p/OCT1 positive feedback loop promotes tumor progression and epithelial-mesenchymal transition in gastric cancer. Gastric. Cancer. 2020;23:212–227. doi: 10.1007/s10120-019-01002-1. [DOI] [PubMed] [Google Scholar]
- 41.Wang S., Zhu W., Qiu J., Chen F. lncRNA SNHG4 promotes cell proliferation, migration, invasion and the epithelial-mesenchymal transition process via sponging miR-204-5p in gastric cancer. Mol. Med. Rep. 2021;23 doi: 10.3892/mmr.2020.11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W., Min L., Qiu X., Wu X., Liu C., Ma J., Zhang D., Zhu L. Biological function of long non-coding RNA (LncRNA) Xist. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.645647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill M., Tran N. miRNA interplay: mechanisms and consequences in cancer. Dis. Model. Mech. 2021;14 doi: 10.1242/dmm.047662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo K., Qian K., Shi Y., Sun T., Wang Z. LncRNA-MIAT promotes thyroid cancer progression and function as ceRNA to target EZH2 by sponging miR-150-5p. Cell Death. Dis. 2021;12:1097. doi: 10.1038/s41419-021-04386-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mallo M. Reassessing the role of Hox genes during vertebrate development and evolution. Trends. Genet. 2018;34:209–217. doi: 10.1016/j.tig.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Shah N., Sukumar S. The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 47.Wu S., Zhu D., Feng H., Li Y., Zhou J., Li Y., Hou T. Comprehensive analysis of HOXC8 associated with tumor microenvironment characteristics in colorectal cancer. Heliyon. 2023;9:e21346. doi: 10.1016/j.heliyon.2023.e21346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lei H., Wang H., Juan A.H., Ruddle F.H. The identification of Hoxc8 target genes. Proc. Natl. Acad. Sci. U S. A. 2005;102:2420–2424. doi: 10.1073/pnas.0409700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gong C., Zou J., Zhang M., Zhang J., Xu S., Zhu S., Yang M., Li D., Wang Y., Shi J., et al. Upregulation of MGP by HOXC8 promotes the proliferation, migration, and EMT processes of triple-negative breast cancer. Mol. Carcinog. 2019;58:1863–1875. doi: 10.1002/mc.23079. [DOI] [PubMed] [Google Scholar]
- 50.Alami Y., Castronovo V., Belotti D., Flagiello D., Clausse N. HOXC5 and HOXC8 expression are selectively turned on in human cervical cancer cells compared to normal keratinocytes. Biochem. Biophys. Res. Commun. 1999;257:738–745. doi: 10.1006/bbrc.1999.0516. [DOI] [PubMed] [Google Scholar]
- 51.Axlund S.D., Lambert J.R., Nordeen S.K. HOXC8 inhibits androgen receptor signaling in human prostate cancer cells by inhibiting SRC-3 recruitment to direct androgen target genes. Mol. Cancer Res. 2010;8:1643–1655. doi: 10.1158/1541-7786.MCR-10-0111. [DOI] [PubMed] [Google Scholar]
- 52.Gu H., Zhong Y., Liu J., Shen Q., Wei R., Zhu H., Zhang X., Xia X., Yao M., Ni M. The role of miR-4256/HOXC8 signaling axis in the gastric cancer progression: evidence from lncRNA-miRNA-mRNA network analysis. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.793678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu H., Zhang M., Xu S., Zhang J., Zou J., Yang C., Zhang Y., Gong C., Kai Y., Li Y. HOXC8 promotes proliferation and migration through transcriptional up-regulation of TGFbeta1 in non-small cell lung cancer. Oncogenesis. 2018;7:1. doi: 10.1038/s41389-017-0016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai C.Y., Liao J.B., Lee Y.C., Yang Y.F. HOXC8 mediates osteopontin expression in gastric cancer cells. J. Cancer. 2023;14:2552–2561. doi: 10.7150/jca.84460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li D., Xia L., Huang P., Wang Z., Guo Q., Huang C., Leng W., Qin S. Heterogeneity and plasticity of epithelial-mesenchymal transition (EMT) in cancer metastasis: focusing on partial EMT and regulatory mechanisms. Cell Prolif. 2023;56:e13423. doi: 10.1111/cpr.13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu Y., Zhu X., Wei X., Tang C., Zhang W. HER2-targeted therapies in gastric cancer. Biochim. Biophys. Acta Rev. Cancer. 2021;1876 doi: 10.1016/j.bbcan.2021.188549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.