Abstract
Dsk2p from Saccharomyces cerevisiae belongs to the class of proteins that contain a ubiquitin-like (UbL) domain at the N terminus together with a ubiquitin-associated (UBA) domain at the C terminus. We show here that the C-terminal UBA domain of Dsk2p binds to K48-linked polyubiquitin chains, and the N-terminal UbL domain of Dsk2p interacts with the proteasome. Overexpression of Dsk2p caused the accumulation of large amounts of polyubiquitin, and extragenic suppressors of the Dsk2p-mediated lethality proved to be temperature-sensitive mutations in two proteasome subunits, rpn1 and pre2. K48-linked ubiquitin-dependent degradation was impaired by disruption of the DSK2 gene. These results indicate that Dsk2p is K48-linked polyubiquitin-binding protein and also interacts with the proteasome. We discuss a possible role of adaptor function of Dsk2p via its UbL and UBA domains in the ubiquitin-proteasome pathway.
Keywords: ubiquitin-related protein‖polyubiquitin adaptor‖UBA domain‖ ubiquitin chains‖UbL domain
Dsk2p is a member of a protein family that is characterized by possession of an N-terminal ubiquitin (Ub)-like (UbL) domain and a C-terminal Ub-associated (UBA) domain (1). In yeast, DSK2 was isolated as a suppressor of kar1, which is defective in spindle pole duplication (2). Although DSK2 is not an essential gene, yeast strains deleted for both DSK2 and RAD23 are temperature-sensitive for growth because of a block of spindle pole body duplication. Dsk2p and Rad23p seems to perform overlapping functions, although the sequence of Dsk2p is hardly similar to that of Rad23p overall except that they show ≈30% identity in their N-terminal UbL domains (residues 1–76) and that both contain UBA domains at their C terminus. Recent work on yeast Rad23p indicates that it is connected with the Ub-proteasome pathway; the N-terminal UbL domain interacts with the proteasome (3). There are genetic interactions between RAD23 and the polyubiquitin-binding subunit of the proteasome, RPN10 (4), and Rad23p seems to inhibit polyubiquitin chain assembly in a cell-free assay system (5). We recently identified that Xenopus Dsk2-related protein, XDRP1, blocked Ub-dependent degradation of the cyclin A (6), and the human homologue of XDRP1, Plic1, is reported to form a link between the Ub machinery and the proteasome (7). Plic1 also is known as ubiquilin, independently isolated in a yeast two-hybrid screen by using presenilin as bait (8). In Dictostelium, a Dsk2p homologue known as SonA is involved in Ub-mediated proteolysis (9). These recent reports all suggest that Dsk2-related proteins play critical roles in protein degradation by the Ub-proteasome pathway.
Ub is a 76-residue polypeptide found in all eukaryotes, the sequence of which is highly conserved from yeast to human. Ub-mediated protein degradation plays a vital role in many eukaryotic cellular processes including protein quality control, cell cycle control, the regulation of transcription, and the response to a variety of external stress (10, 11). Proteins to be degraded are modified by the covalent attachment of the C-terminal glycine residue of Ub to certain of their lysine residues forming an isopeptide bond. In turn, the covalently linked Ubs themselves are ubiquitylated to form polyubiquitin chains that serve as recognition motifs for the proteasome, which degrades the target protein and recycles the Ub. The ubiquitylation process involves a Ub-activating enzyme (E1), a Ub-conjugating enzyme (E2), and a Ub ligase (E3) (11). The protein modified by the polyubiquitin chain is captured by interaction with receptors that are present in the 19S proteasomal cap unit and then degraded by the 20S proteasomal core (12). Compared with the well understood biochemistry of the ubiquitylation machinery and the molecular aspects of the proteasome, however, it is not completely clear yet how a targeted protein is delivered to the 26S proteasome; in particular, it has been puzzling how polyubiquitin chains are recognized by the proteasome. At least one component of the 19S proteasomal cap, Rpn10/S5a, seems capable of binding tightly to polyubiquitin (13, 14), but it is not an essential gene in yeast, raising the possibility that other factors are involved (15, 16). As we show in this paper, Dsk2p seems to be one of these previously unrecognized polyubiquitin-binding factors and uses its UbL and UBA domains to perform the recognition.
Also puzzling are a class of proteins that contain UbL domains at their N-terminal ends that display high homology to portions of the Ub molecule (1). There is no evidence that UbL domains embedded in their polypeptide are conjugated to other proteins as modifiers. Confusingly, these Ub-related proteins are involved in a wide range of cellular functions including protein degradation (5–7), DNA repair (17), spindle pole duplication (2), cell cycle arrest (18), chaperone functions (19), cell adhesion (20), and apoptosis (21). We show here that the UbL domain of Dsk2p is a binding site for the proteasome, which would imply that the wide range of cellular functions reflects the cases in which proteolysis underlies the effects. In this study we present evidence that Dsk2p binds to polyubiquitin, preferentially to K48-linked chain, via its C-terminal UBA domain. Dsk2p also interacts with the proteasome via its N-terminal UbL domain mediated by Rpn1 and Pre2 subunits. Based on the findings described here, we discuss that Dsk2p serves as an adaptor protein that participates in the delivery of polyubiquitylated proteins to the proteasome. In addition, we also discuss that the Dsk2p and Rad23p, both of which share similar domain structures, serve a similar and partially redundant function.
Materials and Methods
Strains, Media, and Two-Hybrid Assay.
The yeast strains used in this study were Saccharomyces cerevisiae YPH499 (MATa ade2-101 leu2-Δ1 trp1-Δ63 ura3-52 lys2-801 his3-Δ200) background (22). Yeast strains were grown in the standard culture medium. Yeast cells were transformed by the lithium-acetate method. For overexpression experiments, yeast cells transformed by galactose-inducible plasmid were cultured in minimal medium containing 2% raffinose at 30°C to ≈A600 = 0.8. Galactose (2%) then was added to the medium and incubated for 4 h. Escherichia coli DH5α strain was used for DNA manipulation, and E. coli BL21 (DE3) was used for the expression of glutathione S-transferase (GST)-Dsk2p protein.
In two-hybrid screening, yeast strain Y190 (MATa ade2-101 his3-Δ200 leu2-3,112 trp1-901 ura3-52 gal4Δ gal80Δ cyhr2 URA3∷Gal-lacZ LYS2∷GAL-HIS3) was used. DSK2 lacking the N terminus (72) was subcloned into the pAS404 (23) and used as bait in the two-hybrid screen by using yeast Gal4AD-cDNA library (a gift from Stephen Elledge, Houston). Among 1.5 × 105 transformants, 42 colonies that grew on the synthetic dextrose-His-Trp-Leu + 25 mM 3-aminotriazole plate was obtained and tested for sequence.
Construction of Plasmids and Mutants.
PCR-amplified DSK2 ORF (1.9-kb BamHI–ClaI fragment) was subcloned into the pRS316. For overexpression, DSK2 DNA was subcloned into galactose-inducible vectors, pGAL1-YEplac195 and pGAL1-YEplac112. For recombinant Dsk2p, DSK2 was subcloned into a XbaI–HindIII site in the pGEX-KG or pEG-KG vector. DSK2 deletion mutants were constructed by the PCR method and subcloned into pGEX-KG and pEG-KG similarly. To disrupt DSK2, a 480-bp BamHI–SacI fragment upstream of the DSK2 ORF was amplified by PCR (primer: 5′-ATAGAGCTCTATCGTGCTAGAATGATTCGG-3′ and 5′-CAAGGATCCCCGT AGCAGACGCGCTTCTTA-3′) and the 380-bp ClaI–ApaI fragment downstream of the DSK2 ORF was amplified by PCR (primer: 5′-TAAGGGCCCTTAAATAAACGTCTAGATGCC-3′ and 5′-TTTATCGATCTGAATCGGATACGGCAAGCC-3′). Then, these two fragments were subcloned into the pRS403. The resultant plasmid was digested by BamHI and used to transform into YPH499. Disruption of DSK2 was checked by PCR and immunoblotting.
Ub cDNA was cloned into pET21a and pET28a from UBI1 cDNA by the PCR method. Ub lysine mutants (K29R, K48R, K63R, K29R-K48R-K63R) were constructed from T7-tagged Ub in pET21a by PCR-based site-directed mutagenesis. T7-tagged Ub and its mutants in pET21a were subcloned into galactose-inducible YEpGAL112. His6-T7-tagged Ub in pET28a was subcloned into a GAL1-inducible yeast 2-μm vector, pNV7. The plasmids carrying the Ub-lacZ fusion gene, Ub-Leu-β-galactosidase (β-gal), and Ub-Ala-β-gal were provided originally from A. Varshavsky (24).
Immunoblotting, Pull-Down Assay, and Recombinant GST-Dsk2p.
Cell extracts were prepared in lysis buffer: 20 mM Hepes, pH 7.5, 100 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA, 1 mM p-amidinophenylmethanesulfonyl fluoride, and 1 μg/ml each of leupeptin, pepstatin, chymostatin (25). For GST pull-down, cell extracts were incubated with the glutathione-Sepharose 4B for 2 h. For His6-T7-Ub pull-down, cell extracts in lysis buffer containing 1 mM EGTA instead of EDTA were incubated and precipitated with Talon metal-affinity resin (CLONTECH). For proteasome-Dsk2p interaction, the cell lysate was prepared as described (26). Recombinant GST-Dsk2 protein was prepared from E. coli BL21 (DE3) transformed with pGEX-KG-DSK2 and used for pull-down assay as described (6).
Isolation of DSK2-Mediated Suppressor Mutant.
YPH499 cells harboring YEplac195-pGAL1-DSK2, YEplac112-pGAL1-DSK2, and pRS315-pGAL1-ADE2 were spread on synthetic medium containing 2% galactose minus uracil, tryptophan, leucine, and adenine, and colonies grown on plate were obtained. Of 400 candidates, nine temperature-sensitive mutants were isolated. Tetrad analysis showed that the suppression phenotype and the temperature sensitivity were caused by a single mutation. These mutants were transformed by the pRS200-based S. cerevisiae genomic library, and plasmid DNA that complemented the temperature sensitivity was sequenced. Mutated genes were cloned by the gap-repair method, and mutation sites were determined by sequencing.
Degradation Assay of N-End Rule Substrate.
The plasmid carrying the Ub-lacZ fusion gene (24) was transformed into strain YPH499. The transformant was grown at 30°C in raffinose medium minus uracil. Ub-Leu-β-gal and Ub-Ala-β-gal, a model substrate of N-end rule (27), were expressed in yeast under the control of the GAL1 promotor. Cells were collected once by centrifugation and transferred to galactose medium minus uracil. Incubation was continued for 2 h to induce Ub-Leu-β-gal or Ub-Ala-β-gal, and then dextrose was added to a final concentration of 2%. Cell extracts were prepared according to the method described (25) followed by immunoblotting with anti-β-gal antibody.
Antibodies and Tetra-Ub.
Anti-Dsk2p polyclonal antiserum was raised in rabbits by using purified Dsk2p prepared from thrombin-digested recombinant GST-Dsk2p. Anti-Ub (Santa Cruz Biotechnology), anti-GST (Santa Cruz Biotechnology), anti-T7 (Promega), anti-polyubiquitin (FK1, Nippon Bio-Test Laboratories, Tokyo), anti-Cdc28 (R49-4, a gift from Kim Nasmyth, Vienna, Austria), anti-20S proteasome “core” (Affiniti, Exeter, U.K.), anti-Rpt1 (Affiniti), anti-β-gal (Promega) antibodies, and tetra-Ub (Affiniti) were used in this study.
Results
Dsk2p Binds to Polyubiquitin.
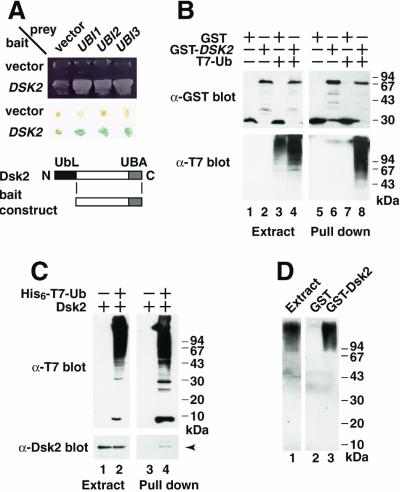
To investigate the function of Dsk2p in yeast, we screened a S. cerevisiae library for interacting factors with an N-terminal truncated Dsk2p in a two-hybrid system. Seventeen strong interacting clones were identified as Ub cDNAs fused with ribosomal proteins (13 UBI3, 3 UBI2, and 1 UBI1) (28). We first confirmed the interaction of Dsk2p with these proteins by using different two-hybrid reporters. Fig. 1A shows that all three Ub fusion proteins gave a strong interaction by using histidine-prototrophic growth (Top) and a β-gal assay (Middle). Next, GST-Dsk2p and T7-tagged Ub were expressed in yeast, and binding of GST-Dsk2p to Ub was tested. A significant fraction of the high molecular weight Ub was retained by GST-Dsk2p when detected by the anti-T7 antibody (Fig. 1B, lane 8). By using His6-T7-tagged Ub, we also showed that the Ub was coprecipitated with Dsk2p (Fig. 1C, an arrow in lane 4). The high molecular weight smear was proved to be polyubiquitin when recombinant GST-Dsk2p pull-downs from cell extracts were immunoblotted with an anti-polyubiquitin antibody (Fig. 1D, lane 3). No binding between monoubiquitin and Dsk2p was observed in any case (data not shown). Therefore, we conclude that Dsk2p binds to polyubiquitin but not to monoubiquitin.
Figure 1.
Dsk2p binds to polyubiquitin. (A) Interaction of Dsk2p with Ub was tested by two-hybrid assay. Among 1.5 × 105 transformants screened, 17 UBI1∼3 and 4 DSK2 interacting clones were obtained. Interaction of DSK2 with UBI1∼3 was tested by histidine-prototrophic growth (Top) and β-gal (Middle). N-terminal truncated Dsk2p (residues 72–373) was used as a bait (Bottom), because overexpression of full-length Dsk2p is toxic to yeast (2). (B) Binding of GST-Dsk2p to Ub. GST-Dsk2p (lanes 2 and 4) and T7-Ub (lanes 3 and 4) were expressed in yeast. GST-Dsk2p was pulled down by glutathione beads, and GST-bound materials were immunoblotted by anti-GST (Upper) and anti-T7 antibodies (Lower). Lanes 1–4, expression; lanes 5–8, GST pull-down. (C) Binding of Ub to Dsk2p. His6-T7-Ub (lanes 2 and 4) and Dsk2p (lanes 1–4) were expressed in yeast. His6-T7-Ub was pulled down by Talon resin, and the bound materials (lanes 3 and 4) were immunoblotted either by anti-T7 (Upper) or anti-Dsk2 (Lower) antibody. An arrow indicates Dsk2p (see lane 4). (D) Dsk2p binds to polyubiquitin. GST (lane 2) or GST-Dsk2p (lane 3) purified from E. coli BL21 (DE3) was incubated with yeast extracts for 2 h. GST pull-down materials were immunoblotted with anti-polyubiquitin (lanes 2 and 3). Endogenous polyubiquitin in the extracts is shown in lane 1.
The C-Terminal UBA Domain in Dsk2p Is Required for Its Binding to Polyubiquitin.
To define the domain of Dsk2p required for polyubiquitin binding, we made a number of deletion mutants of Dsk2p (Fig. 2B), the binding abilities of which for polyubiquitin were tested by immunoblotting. The N-terminal truncation (residues 78–373), which removes the N-terminal UbL domain, bound to polyubiquitin (Fig. 2A, lane 3). The C-terminal UBA domain alone (residues 328–373) bound polyubiquitin to the same extent as full-length Dsk2p (lane 4). By contrast, C-terminally truncated mutants (residues 1–327, 1–215, 1–77, and 78–335), all of which lack the UBA domain, did not bind to polyubiquitin (lanes 5–8). This result shows that the UBA domain at the C terminus of Dsk2p is necessary for its binding to polyubiquitin.
Figure 2.
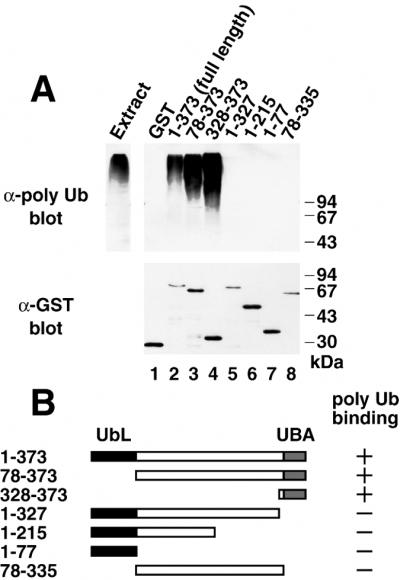
The C-terminal UBA domain of Dsk2p is required for its binding to polyubiquitin. (A) Binding assay of deletion mutants. Recombinant GST-Dsk2p deletions were incubated with yeast extract for 2 h, and GST-bound materials were immunoblotted either with the anti-polyubiquitin (Upper) or anti-GST (Lower) antibody. (B) A diagram of Dsk2p constructs. Binding abilities of the mutants are shown on the right.
Dsk2p Binds Directly to K48-Linked Chains via Its UBA Domain.
We next tested whether Dsk2p can bind to tetra-Ub as a model of a minimal polyubiquitin chain for proteasomal targeting (29) in the absence of any other cellular factors. K48-linked tetra-Ub was incubated with GST-Dsk2p followed by GST pull-down and immunoblotting with the anti-Ub antibody (Fig. 3A). Tetra-Ub was retained by the glutathione-Sepharose when full-length GST-Dsk2p (residues 1–373) (lane 6) or C-terminal UBA domain 328–373 (lane 8) were present but not with C-terminal truncated Dsk2p (residues 1–327) (lane 7). As a control GST alone did not retain tetra-Ub (lane 5). Therefore, the UBA domain of Dsk2p is able to bind directly to K48-linked tetra-Ub chain in vitro.
Figure 3.
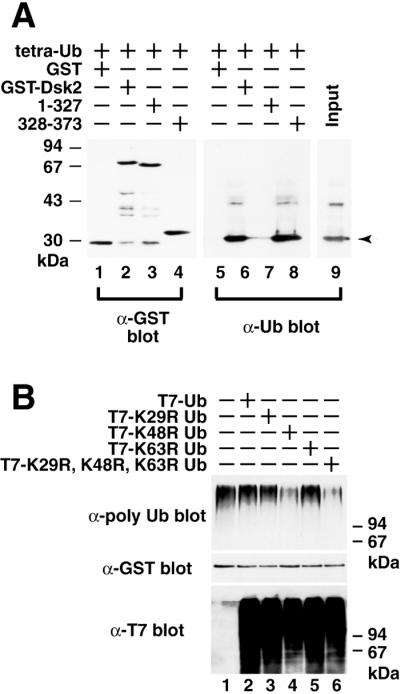
The UBA domain of Dsk2p binds directly to K48-linked polyubiquitin chains. (A) Direct binding of Dsk2p to tetra-Ub. K48-linked tetra-Ub was incubated in vitro with full-length GST-Dsk2p (residues 1–373), C-terminal truncated GST-Dsk2p (residues 1–327), and C-terminal UBA (residues 328–373). GST pull-downs were immunoblotted with anti-GST (lanes 1–4) and anti-Ub (lanes 5–8) antibodies. The arrow in lane 9 indicates the position of tetra-Ub. (B) Effects of Ub lysine mutants on binding between Dsk2p and polyubiquitin. T7-tagged lysine mutants of Ub (K29R, K48R, K63R, and K29R-K48R-K63R) were expressed in yeast (Bottom), and the cell extracts were incubated with recombinant GST-Dsk2p. GST-bound materials were immunoblotted with anti-polyubiquitin (Top) and anti-GST (Middle) antibodies for GST-Dsk2p.
Polyubiquitin chains can be formed in vivo by at least three different linkages of lysine residues in the Ub molecule, K29, K48, and K63 (30–33). Next, a series of Ub lysine mutants, K29R, K48R, K63R, or the triple substitution mutant, were made, and the effects of Ub lysine mutants on polyubiquitin-Dsk2p binding were examined (Fig. 3B). T7-tagged Ub mutants were expressed in yeast from galactose-inducible promoter, and the extracts of yeast cells were incubated with recombinant GST-Dsk2p followed by glutathione-Sepharose affinity chromatography and immunoblotting with anti-polyubiquitin antibody. The expression of K48R Ub reduced the amount of endogenous polyubiquitin bound to GST-Dsk2p (Fig. 3B Top, lane 4) compared with that of wild type (lane 2) and no additional Ub expression (lane 1), whereas expression of K29R (lane 3) or K63R (lane 5) had no effect on the recovery of polyubiquitin. Expression of the triple lysine mutant (K29R-K48R-K63R) decreased polyubiquitin binding to GST-Dsk2p to about the same extent as the K48R (lane 6). As a control, we checked that these T7-Ub mutants were expressed similarly in cells (Bottom) and that the level of GST-Dsk2p is similar in each sample when immunoblotted with anti-GST antibody (Middle). These results, together with domain analysis (Fig. 2), indicate that the UBA domain of Dsk2p binds to K48-linked polyubiquitin chains.
Accumulation of Polyubiquitin Chains Caused by Overexpression of Dsk2p in Cells.
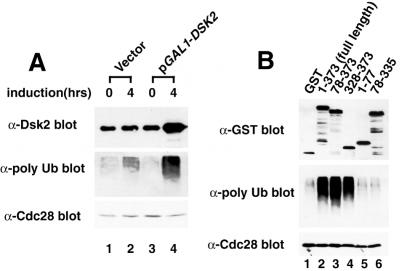
We noticed in the experiment shown in Fig. 1 that cells expressing GST-Dsk2p showed increased amounts of high molecular weight T7-tagged polyubiquitin (Fig. 1B, lanes 3 and 4). We therefore tested whether endogenous polyubiquitin accumulated in cells where Dsk2p was overexpressed (Fig. 4A). Dsk2p was expressed from pGAL1-DSK2 by the addition of galactose to the medium followed by immunoblotting with the anti-polyubiquitin antibody. Overexpression of Dsk2p led to the accumulation of a large amount polyubiquitin chains in cells (lane 4). We then determined which domain of Dsk2p was required for the increased polyubiquitin levels (Fig. 4B). Deletion of the N terminus of Dsk2p (residues 78–373 and 328–373) accumulated polyubiquitin (lanes 3 and 4), whereas expression of mutants lacking the C terminus (residues 1–77 and 78–335) did not (lanes 5 and 6). Therefore, the C-terminal UBA domain of Dsk2p is required for the increased accumulation of polyubiquitin seen when Dsk2p overexpression is induced. The accumulation of polyubiquitin was parallel to that obtained when polyubiquitin binding was measured (see Fig. 2B). When we tested the accumulation of polyubiquitin in cells expressing Ub lysine mutants, the expression of Ub K48R or the triple K29R-K48R-K63R inhibited the accumulation of polyubiquitin, whereas K29R or K63R was relatively less inhibited (data not shown). These results indicate that overexpression of Dsk2p caused the accumulation of K48-linked chains.
Figure 4.
Dsk2p causes the accumulation of polyubiquitin chains. (A) Accumulation of polyubiquitin by overexpression of Dsk2p. Yeast cells transformed by pGAL1-DSK2 were incubated in raffinose synthetic medium (lanes 1 and 3), and Dsk2p expression was induced in 4 h by adding galactose to the medium (lanes 2 and 4). Proteins from the extracts were immunoblotted with anti-Dsk2p (Top), anti-polyubiquitin (Middle), and anti-Cdc28 (Bottom) antibodies. (B) The C-terminal UBA domain of Dsk2p is required for accumulation of polyubiquitin. GST-Dsk2p and its deletions (see Fig. 2) were galactose-induced in yeast, and proteins from the extracts were immunoblotted with anti-GST (Top), anti-polyubiquitin (Middle), and anti-Cdc28 (Bottom) antibodies.
Suppressors That Restore the DSK2-Mediated Toxicity Are Identified as Mutations in the Subunit of Proteasome Rpn1 and Pre2.
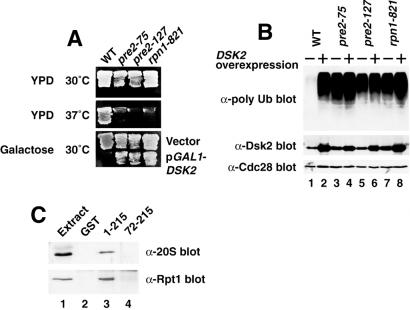
As originally observed (2), the overexpression of full-length Dsk2p inhibits the growth of yeast. Then, we looked for extragenic suppressors that could restore the DSK2-mediated growth arrest (Fig. 5). The procedures are essentially the same as described (34). Nine suppressor mutants that also conferred a temperature-sensitive growth were isolated, and three of them were identified as recessive mutations in subunits of the proteasome: pre2-75 (G75A), pre2-127 (C127F), and rpn1-821 (G821D), respectively (Fig. 5A). Rpn1p is found in the lid of the 19S regulatory subunit, and Pre2p is a β-core subunit (35). Even in the absence of DSK2 overexpression, these three mutants accumulated polyubiquitin (Fig. 5B, lanes 3, 5, and 7). We next investigated whether Dsk2p interacts physically with the proteasome by using pull-down of Dsk2p-GST followed by immunoblotting with anti-20S proteasome antibody. Fig. 5C shows that the Dsk2p fragment (1) bound to the 20S proteasome (lane 3, Upper), whereas the N-terminal deletion of the Dsk2p fragment (72) failed to bind (lane 4). The same result was obtained by immunoblotting with anti-Rpt1 antibody, a base component of the 26S proteasome (Lower). Thus, the N-terminal UbL domain of Dsk2p physically interacts with the proteasome. Both genetic and biochemical analysis shows an interaction between Dsk2p and the proteasome.
Figure 5.
Genetic and physical interactions between Dsk2p and the proteasomal subunits. (A) Identification of pre2 and rpn1. Suppressors were isolated as temperature-sensitive mutants that can suppress the Dsk2p-mediated growth arrest. The detailed procedure is described elsewhere (34). Mutation sites were determined by the gap-repair method as pre2-75, pre2-127, and rpn1-821. WT, wild type. (B) Polyubiquitin accumulates in pre2 and rpn1. DSK2 was overexpressed in pre2 and rpn1 at the permissive temperature (30°C), and cell extracts (lanes 2, 4, 6, and 8) were immunoblotted with anti-polyubiquitin (Top), anti-Dsk2p (Middle) and anti-Cdc28 (Bottom) antibodies. Lanes 1, 3, 5, and 7, no induction of Dsk2p. (C) Physical interaction of Dsk2p with the proteasome. GST-fused Dsk2p recombinant protein was incubated in the yeast cell extracts. GST pull-downs were immunoblotted with anti-20S (Upper) and anti-Rpt1 (Lower) antibodies. Lane 1, cell extracts; lane 2, GST control; lane 3, Dsk2p (residues 1–215); lane 4, N-terminal truncated Dsk2p (residues 72–215).
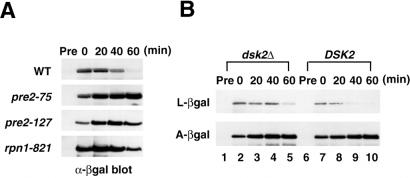
We next tested the effects of these mutations on protein degradation by using a model substrate of N-end rule for degradation, Leu-β-gal (27). Ub-Leu-β-gal was expressed in the mutant cells pre2 and rpn1 followed by extractions of proteins and immunoblotting with anti-β-gal antibody. We found that the degradation of Leu-β-gal was prevented in pre2-75, pre2-127, and rpn1-821 compared with that in the wild-type strain (Fig. 6A). These mutant phenotypes thus indicate that the proteasome activity is decreased by the mutation of either gene.
Figure 6.
Inhibition of protein degradation by the proteasome mutations pre2 and rpn1 and by DSK2 disruption. (A) Inhibition of Leu-β-gal degradation in pre2 and rpn1. Ub-Leu-β-gal, a model substrate of N-end rule (originally provided by A. Varshavsky), was induced by galactose induction in the wild-type, pre2, and prn1 strains followed by chasing with the addition of glucose to the medium. After the induction was shut off, cell extracts (0.1 A600 equivalents) were taken at 20-min intervals and immunoblotted with anti-β-gal antibody (Promega). Pre; before induction; WT, wild type. (B) Degradation of K48-linked N-end rule substrate is inhibited by DSK2 disruption. N-end rule substrates, Ub-Leu-β-gal and Ub-Ala-β-gal, were induced by galactose-induction in dsk2Δ (lanes 1–5) and the wild-type strain (lanes 6–10). Degradation of Leu-β-gal and Ala-β-gal was followed by immunoblotting as described for A.
Degradation of K48-Linked N-End Rule Substrate Is Impaired by Disruption of DSK2.
We further examined the effects of Dsk2p on protein degradation in the Ub-proteasome pathway by using Leu-β-gal and Ala-β-gal as model substrates. Leu-β-gal is degraded rapidly depending on K48-linked Ub chains, whereas Ala-β-gal is stable (36). Ub-Leu-β-gal or Ub-Ala-β-gal was expressed in the wild-type strain and dsk2Δ. When immunoblotting with the anti-β-gal antibody (Fig. 6B), degradation of Leu-β-gal is inhibited in dsk2Δ (Upper, lanes 2–5) compared with that in the wild-type strain (lanes 7–10). Unlike Leu-β-gal, the stability of Ala-β-gal was unaffected by Dsk2p depletion (Lower). These results suggest that Dsk2p participates in K48-linked protein degradation in the Ub-proteasome pathway.
Discussion
The Role of Dsk2p as an Adaptor Protein in Ub-Proteasome Pathway.
In this paper we provide evidence that the C-terminal UBA domain of Dsk2p binds to K48-linked polyubiquitin chains and that its N-terminal UbL domain binds to the proteasome. Overexpression of Dsk2p is toxic to the wild-type strain, allowing us to isolate the suppressor mutants rpn1 and pre2, components of the proteasome subunits (35). The reduced toxicity caused by overexpression of Dsk2p in these suppressors is explained presumably by the inability of the proteasome that contained the altered Rpn1p or Pre2p. Dsk2p binds to polyubiquitin but not to monoubiquitin, interestingly in contrast to a recent report in budding yeast Ddi1p (37). Taken together, these results indicate that Dsk2p might be a polyubiquitin-binding protein that helps to deliver polyubiquitin chains to the proteasome. This interpretation is supported by the recent report that Plic1, a human homologue of Dsk2p, associates with both the proteasome and Ub ligase in a large complex (7, 38). Moreover, disruption of Dsk2p inhibited the degradation of K48-linked substrate Leu-β-gal (Fig. 6); therefore, we suspect that Dsk2p binds to K48 polyubiquitin chains attached to target proteins. Here we propose that Dsk2p serves as an adaptor protein that participates in the delivery of polyubiquitylated substrates to the proteasome; Dsk2p binds to polyubiquitin chains via the C-terminal UBA domain and to the proteasome via the N-terminal UbL domain. K48-linked polyubiquitylated substrate captured by Dsk2p may be delivered to the receptor site onto the proteasome.
Furthermore, we observe a remarkable increase in the total amount of K48-linked polyubiquitin under the condition of Dsk2p excess in yeast. Polyubiquitin chains can be edited also by the activities of specific de-ubiquitylating enzymes (16), the activity of which is rather critical for the persistence of long Ub chains (39, 40). In the condition of Dsk2p excess, therefore, either the activation of ubiquitylation or the inhibition of de-ubiquitylation might be induced by excess Dsk2p via its UBA domain, and excess Dsk2p may cause an increase in flux through the proteasome.
In vertebrates, consistent with a positive effect of Dsk2p on Ub-proteasome pathway, a Xenopus homolog of Dsk2p, XDRP1, promotes the degradation of cyclin B in egg extracts (unpublished result). On the other hand, we originally observed that the degradation of cyclin A is inhibited by XDRP1 (6). Such a distinct response for cyclin degradation may be caused by different XDRP1-related proteins, because several XDRP1-related proteins are present in Xenopus eggs (data not shown).
Possible Interactions of Dsk2p Adaptor with Other Polyubiquitin Receptors.
Several polyubiquitin receptors seem to be involved in the recognition of polyubiquitin chains. The proteasome subunit Rpn10/S5a was shown to be a Ub receptor that binds to polyubiquitin chains (13, 14). However, it was puzzling at the time that a deletion of RPN10 is viable and the rpn10 mutant lacking the ability to bind polyubiquitin chains did not inhibit protein degradation in vivo (41), because no other polyubiquitin-binding protein had been identified. It has since become clear that several 19S subunits, Rpn1 and Rpn10, are involved in the recognition of polyubiquitinated substrates (15, 38, 42). For example, rpn10 exerts synthetic effects with rad23, indicating that Rpn10p receptor and Rad23p have overlapping functions via binding to the same proteasome with different sites (4). Taken together with our data described here, Dsk2p binds to K48-linked polyubiquitin, and the N-terminal UbL domain of Dsk2p also interacts with the 26S proteasome, mediated by Rpn1 and/or Pre2 subunits (Fig. 5). We therefore can imagine that Dsk2p may act by passing distinct polyubiquitin K48 chains on to Rpn1p of the proteasome.
Functional Redundancy of Dsk2p and Rad23p.
Dsk2p has functional redundancy with another N-terminal Ub-related protein, Rad23p. Although DSK2 is not an essential gene (2), we also confirmed that a double mutant, dsk2Δrad23Δ, is synthetically lethal at 35°C. The sequence of Rad23p is hardly similar to Dsk2p overall, but it contains an N-terminal UbL and two UBA domains (17). A recent paper suggested that Rad23p is connected with the Ub-proteasome pathway (3–5). Rad23p is shown to negatively regulate the polyubiquitin pathway by inhibition of ubiquitylated chain assembly factor E4 (5, 43), whereas Dsk2p seems to function positively in polyubiquitylation. Overexpression of RAD23 stabilizes mutant Pds1p (44), and thus we tested whether, in contrast, overexpressed Dsk2p could stimulate Leu-β-gal degradation. However, we cannot observe distinct effects of Leu-β-gal degradation (data not shown), probably because an expressed Dsk2p to a single Leu-β-gal substrate was titrated out by many other substrates present in cells. Furthermore, it is possible also that Dsk2p and Rad23p each could recognize different lysine-linked chains by virtue of their abilities to bind polyubiquitin chains. It has been published recently that fission yeast Dph1p and Rhp23p (Dsk2p and Rad23p homologs) bind to multi-Ub chains (45). In this paper we show that budding yeast Dsk2p binds to polyubiquitin. We therefore suspect that both Dsk2p and Rad23p function as adaptors, but they might work on some subset of polyubiquitylated substrates. Our data suggests that Dsk2p mediates the delivery of K48-polyubiquitin and the proteasome, whereas Rad23p-related function may link to different lysine chains (5, 16). Overlapping but distinct chain specificity of Dsk2p and Rad23p might play a regulatory role in the fail-safe delivery system of the Ub-proteasome pathway. Further work on functional redundancy of these proteins is required to clarify this point.
Acknowledgments
We would like to acknowledge Dr. Tim Hunt for critical reading of the manuscript and Dr. Mark Carrington for valuable comments. We also thank Dr. Randolph Hampton for yeast strains and Dr. Mark Rose for Dsk2p antibody in the initial stage of the work. This work was supported in part by Grants-in-Aid from the Ministry of Education, Science, Sports, and Technology of Japan. M.F. was supported by a research fellowship of the Japan Society for the Promotion of Science.
Abbreviations
- Ub
ubiquitin
- UbL
ubiquitin-like
- UBA
ubiquitin-associated
- GST
glutathione S-transferase
- β-gal
β-galactosidase
References
- 1.Jentsch S, Pyrowlakis G. Trends Cell Biol. 2000;10:335–342. doi: 10.1016/s0962-8924(00)01785-2. [DOI] [PubMed] [Google Scholar]
- 2.Biggins S, Ivanovska I, Rose M D. J Cell Biol. 1996;133:1331–1346. doi: 10.1083/jcb.133.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K. Nature (London) 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 4.Lambertson D, Chen L, Madura K. Genetics. 1999;153:69–79. doi: 10.1093/genetics/153.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortolan T G, Tongaonkar P, Lambertson D, Chen L, Schauber C, Madura K. Nat Cell Biol. 2000;2:601–608. doi: 10.1038/35023547. [DOI] [PubMed] [Google Scholar]
- 6.Funakoshi M, Geley S, Hunt T, Nishimoto T, Kobayashi H. EMBO J. 1999;18:5009–5018. doi: 10.1093/emboj/18.18.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleijnen M F, Shih A H, Zhou P, Kumar S, Soccio R E, Kedersha N L, Gill G, Howley P M. Mol Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- 8.Mah A L, Perry G, Smith M A, Monteiro M J. J Cell Biol. 2000;151:847–862. doi: 10.1083/jcb.151.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pukatzki S, Ennis H L, Kessin R H. Biochem Biophys Acta. 2000;1499:154–163. doi: 10.1016/s0167-4889(00)00124-5. [DOI] [PubMed] [Google Scholar]
- 10.Hershko A. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- 11.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 12.Baumeister W, Walz J, Zuhl F, Seemuller E. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 13.Konimami K, Okura N, Kawamura M, DeMartino G N, Slaughter C A, Shimbara N, Chung C H, Fujimuro M, Yokosawa H, Shimizu Y, et al. Mol Biol Cell. 1997;8:171–187. doi: 10.1091/mbc.8.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young P, Deveraux Q, Beal R E, Pickart C M, Rechsteiner M. J Biol Chem. 1998;273:5461–5467. doi: 10.1074/jbc.273.10.5461. [DOI] [PubMed] [Google Scholar]
- 15.Wilkinson C R M, Ferrell K, Penney M, Wallace M, Dubiel W, Gordon C. J Biol Chem. 2000;275:15182–15192. doi: 10.1074/jbc.275.20.15182. [DOI] [PubMed] [Google Scholar]
- 16.Pickart C M. Trends Biochem Sci. 2000;25:544–548. doi: 10.1016/s0968-0004(00)01681-9. [DOI] [PubMed] [Google Scholar]
- 17.Watkins J F, Sung P, Prakash L, Prakash S. MolCellBiol. 1993;13:7757–7765. doi: 10.1128/mcb.13.12.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He X, Jones M H, Winey M, Sazer S. J Cell Sci. 1998;111:1635–1647. doi: 10.1242/jcs.111.12.1635. [DOI] [PubMed] [Google Scholar]
- 19.Kaye F J, Modi S, Ivanovska I, Koonin E V, Thress K, Kubo A, Kornbluth S, Rose M D. FEBS lett. 2000;467:348–352. doi: 10.1016/s0014-5793(00)01135-2. [DOI] [PubMed] [Google Scholar]
- 20.Wu A-L, Wang J, Zheleznyak A, Brown E J. Mol Cell. 1999;4:619–625. doi: 10.1016/s1097-2765(00)80212-9. [DOI] [PubMed] [Google Scholar]
- 21.Thress K, Henzel W, Shillinglaw W, Kornbluth S. EMBO J. 1998;17:6135–6143. doi: 10.1093/emboj/17.21.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakashima N, Noguchi E, Nishimoto T. Genetics. 1999;152:853–867. doi: 10.1093/genetics/152.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bachmair A, Finley D, Varshavsky A. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 25.Rudner A D, Hardwick K G, Murray A W. J Cell Biol. 2000;149:1361–1376. doi: 10.1083/jcb.149.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies R J. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varshavsky A. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozkaynak E, Finley D, Solomon M J, Varshavsky A. EMBO J. 1987;6:1429–1439. doi: 10.1002/j.1460-2075.1987.tb02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thrower J S, Hoffman L, Rechsteiner M, Pickart C M. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnason T, Ellison M J. Mol Cell Biol. 1994;14:7876–7883. doi: 10.1128/mcb.14.12.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mastrandrea L D, You J, Niles E G, Pickart C M. J Biol Chem. 1999;274:27299–27306. doi: 10.1074/jbc.274.38.27299. [DOI] [PubMed] [Google Scholar]
- 32.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen Z J. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 33.Spence J, Gali R R, Dittmar G, Sherman F, Karin M, Finley D. Cell. 2000;102:67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- 34.Funakoshi M, Sikder H, Ebihara H, Sugimoto K, Matsumoto K, Hunt T, Nishimoto T, Kobayashi H. Genes Cells. 1997;2:329–343. doi: 10.1046/j.1365-2443.1997.1230320.x. [DOI] [PubMed] [Google Scholar]
- 35.Glickman M H, Rubin D M, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried V A, Finley D. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 36.Johnson E S, Ma P C M, Ota I M, Varshavsky A. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 37.Bertolaet B L, Clarke D J, Wolff M, Watson M H, Henze M, Divita G, Reed S I. Nat Struct Biol. 2001;8:417–422. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- 38.Xie Y, Varshavsky A. Proc Natl Acd Sci USA. 2000;97:2497–2502. doi: 10.1073/pnas.060025497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lam Y A, Xu W, DeMartino G N, Cohen R E. Nature (London) 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 40.Swaminathan S, Amerik A Y, Hochstrasser M. Mol Biol Cell. 1999;10:2583–2594. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Nocker S, Sadis S, Rubin D M, Glickman M, Fu H, Coux O, Wefes I, Finley D, Vierstra R D. Mol Cell Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeuchi J, Fujimuro M, Yokosawa H, Tanaka K, Toh-e A. Mol Cell Biol. 1999;19:6575–6584. doi: 10.1128/mcb.19.10.6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koegl M, Hoppe T, Schlenker S, Ulrich H D, Mayer T U, Jentsch S. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 44.Clarke D J, Mondesert G, Segal M, Bertolaet B L, Jensen S, Wolff M, Henze M, Reed S I. Mol Cell Biol. 2001;21:1997–2007. doi: 10.1128/MCB.21.6.1997-2007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilkinson C R M, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]