Abstract
A major challenge in interpreting genome sequences is understanding how the genome encodes the information that specifies when and where a gene will be expressed. The first step in this process is the identification of regions of the genome that contain regulatory information. In higher eukaryotes, this cis-regulatory information is organized into modular units [cis-regulatory modules (CRMs)] of a few hundred base pairs. A common feature of these cis-regulatory modules is the presence of multiple binding sites for multiple transcription factors. Here, we evaluate the extent to which the tendency for transcription factor binding sites to be clustered can be used as the basis for the computational identification of cis-regulatory modules. By using published DNA binding specificity data for five transcription factors active in the early Drosophila embryo, we identified genomic regions containing unusually high concentrations of predicted binding sites for these factors. A significant fraction of these binding site clusters overlap known CRMs that are regulated by these factors. In addition, many of the remaining clusters are adjacent to genes expressed in a pattern characteristic of genes regulated by these factors. We tested one of the newly identified clusters, mapping upstream of the gap gene giant (gt), and show that it acts as an enhancer that recapitulates the posterior expression pattern of gt.
The development of multicellular organisms is, to a large extent, dictated by a carefully choreographed progression of domain- and tissue-specific gene expression. To understand development, it is therefore necessary to understand the logic and mechanisms of this transcriptional network. Much of the information that determines when and where genes will be expressed is encoded in an organism's genome sequence. Although we now have genome sequences for many important metazoans, our understanding of how this information is encoded is extremely limited. Cracking this “cis-regulatory code” is a major problem in biology.
A paradigmatic model for studying transcriptional control of development is the early Drosophila embryo. Most of the important players have been identified by exhaustive genetic analysis, and there are sophisticated tools for characterizing the sequence features controlling the transcriptional network organized by these key developmental regulators. Although the early Drosophila embryo is relatively simple, many of the genes involved in early development of the fly are known to control development in other animals (1). Thus, it is likely that an understanding of the developmental cis-regulatory code in Drosophila will be applicable to other higher eukaryotes, including humans.
Careful genetic and biochemical dissection of numerous genes involved in Drosophila development suggests some general principles for how cis-regulatory regions are organized. For example, the cis-regulatory region of the pair-rule gene even-skipped (eve)—expressed in seven stripes in the blastoderm embryo—is organized into a series of discrete sequence regions of roughly 500 bp in length, each of which controls a distinct component of eve's expression pattern (2–7). This modular organization of cis-regulatory regions is observed in many developmental genes in Drosophila, and in other organisms (8). In general, several transcription factors bind to each of these cis-regulatory modules (CRMs), and there are often multiple binding sites for each of these factors (8). Presumably, multiple bound transcription factors act combinatorially to confer specific transcriptional activity. For example, the enhancer controlling expression of eve's second stripe contains at least three binding sites for Hunchback (Hb) and Giant (Gt), five for Bicoid (Bcd), and six for Krüppel (Kr) (4, 9).
It has been proposed that the high local density of transcription factor binding sites required for the proper function of these CRMs could be used as the basis for identifying novel CRMs (10–13). Here, we demonstrate the utility of this approach by examining the genome-wide distribution of binding sites for five transcription factors known to act together in the early Drosophila embryo.
Materials and Methods
Collection and Alignment of Transcription Factor Binding Sites.
Bcd, Cad, Hb, Kr, and Kni binding sequences determined by in vitro DNase protection assays were compiled from a previous study (14) and additional sources. These sequences and their sources are listed in Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org.
Binding site sequences for Bcd, Hb, and Kr were aligned by using the pattern discovery tool meme (v 3.0; ref. 15), with the following command line settings “-mod zoops -revcomp -dna.” The “-minsites” parameter was set to 80% of the total number of sites collected for each transcription factor. This setting allowed for up to 20% of binding site sequences that aligned poorly to be omitted as potential sources of experimental error. For Bcd, 51/51 sites were aligned; for Hb, 93/93 sites were aligned; for Kr, 29/37 sites were aligned. A -bfile or background model file was used, which included mono-nucleotide, di-nucleotide, and tri-nucleotide frequencies determined from the intergenic Drosophila melanogaster genomic sequence, as annotated in Berkeley Drosophila Genome Project (BDGP)/Celera Release 1 (Rel 1.; ref. 16). Individual binding site sequences for Cad and Kni were aligned manually.
Construction of Position Weight Matrices (PWMs) and Searching.
patser (v. 3b; ref. 17) was used to construct PWMs from sequences as aligned as described above, and to search genomic sequence for matches to the PWM. patser was run with the following command line options: “-c -d2 -l 4.” An “alphabet” file (specified with the command line parameter “-a”) was used to provide the following background frequencies: A/T = 0.297, G/C = 0.203. These frequencies were determined from the intergenic D. melanogaster genomic sequence as annotated in Rel. 1.
patser was run on Rel. 1 genomic sequence, and cis-analyst was used to identify all potential binding sites with P value < site_p. cis-analyst examines sequence windows of length wind_size, retaining only those containing at least min_sites binding sites. cis-analyst then collapses all overlapping windows into a single “cluster.”
Collection of CRMs.
Test CRM boundaries were determined as described in the studies listed in Table 1, which is published as supporting information on the PNAS web site. If the CRM had been sequenced as part of a prior study, we aligned this sequence with Rel. 1 genomic sequence and used the aligned segment from BDGP/Celera sequence (all sequences matched perfectly or with greater than 99% identity). If the CRM element had not been previously sequenced, we identified the restriction sites bordering the element, and extracted the genomic sequence occurring between these sites.
Test CRM Independent Matrices.
In analyzing the overlap between binding site clusters and our test CRMs, we sought to avoid evaluating a particular CRM with PWMs built by using binding sites from that CRM. For each CRM, we constructed a separate set of PWMs that excluded binding sites derived from that CRM and used these PWMs to determine whether the CRM overlapped a binding site cluster. The sole exception was the Kni PWM for the eve stripe 3/7 CRM, because all Kni example binding sequences were derived from the eve stripe 3/7 CRM.
Genome-Wide Searches.
cis-analyst was used to search 93 Mb of noncoding DNA from Rel. 1 for clusters of Cad, Bcd, Hb, Kr, and Kni by using the parameters site_p = 0.0003 and wind_size = 700, and values of min_sites from 12 to 18. cis-analyst was also used to search for clusters of Bcd, Hb, Kr, and Kni by using the parameters site_p = 0.0003, wind_size = 700, and min_sites = 13. The set of 28 clusters in Table 2 (which is published as supporting information on the PNAS web site) represents the union of the results for searches for Cad, Bcd, Hb, Kr, and Kni with min_sites = 15 and Bcd, Hb, Kr and Kni with min_sites = 13.
Whole-Mount in Situ Hybridizations and DNA Microarray Hybridizations.
Embryonic whole-mount in situ RNA hybridizations and DNA microarray hybridizations were performed as described on the Berkeley Drosophila Genome Project web site (http://www.fruitfly.org/).
Giant Transgenics and Mutant Embryos.
A 1.1-kb DNA fragment located upstream of the transcription start site of gt (from −2.7 to −1.6 kb) was amplified from y w fly genomic DNA by PCR by using two primers containing synthetic AscI and NotI restriction sites: ttaggcgcgccagaaacttaccatcacttcg, attgcggccgccccattcagggggattggg. The PCR product was digested with AscI and NotI, and inserted in their native orientation into the AscI-NotI site of a modified CaSpeR-AUG-bgal transformation vector (18) containing the eve basal promoter, starting at −42 bp and continuing through codon 22 fused in-frame with lacZ (19). The P-element transformation vectors were injected into yw embryos, as described previously (19, 20).
Results
The transcription factors Bicoid (Bcd), Caudal (Cad), Hunchback (Hb), Krüppel (Kr), and Knirps (Kni) act at very early stages of Drosophila development to define the anterior–posterior axis of the embryo (reviewed in ref. 21). Bcd (22) and Cad (23–25) are maternal activators broadly distributed in the anterior and posterior portions of the embryo, respectively. Hb, Kr, and Kni are zinc-finger gap proteins that act primarily as repressors in specific embryonic domains (reviewed in ref. 26). Aided greatly by a prior study (14), we collected sequences of previously described binding sites for these five factors present in the cis-regulatory regions of known target genes. We aligned the binding sequences for each factor by using the motif-assembly program meme (15), and modeled the binding specificities of each factor with a PWM. PWMs are a useful way to represent binding specificities and provide a statistical framework for searching for novel instances of the motif in genome sequences (27, 28). The sequences used and PWMs produced are shown in Fig. 5.
We used the freely available program patser (17) to search the genome for sequences that matched these PWMs, and developed a web-based visualization tool, cis-analyst (http://www.fruitfly.org/cis-analyst/) to display the location of predicted binding sites along with genome annotations in selected genomic regions. patser assigns a score to each potential site that reflects the agreement between the site and the corresponding PWM. These scores approximates the free energy of binding between the factor and site (27, 29), and cis-analyst uses a user-defined cutoff parameter (site_p) to eliminate predicted low-affinity sites.
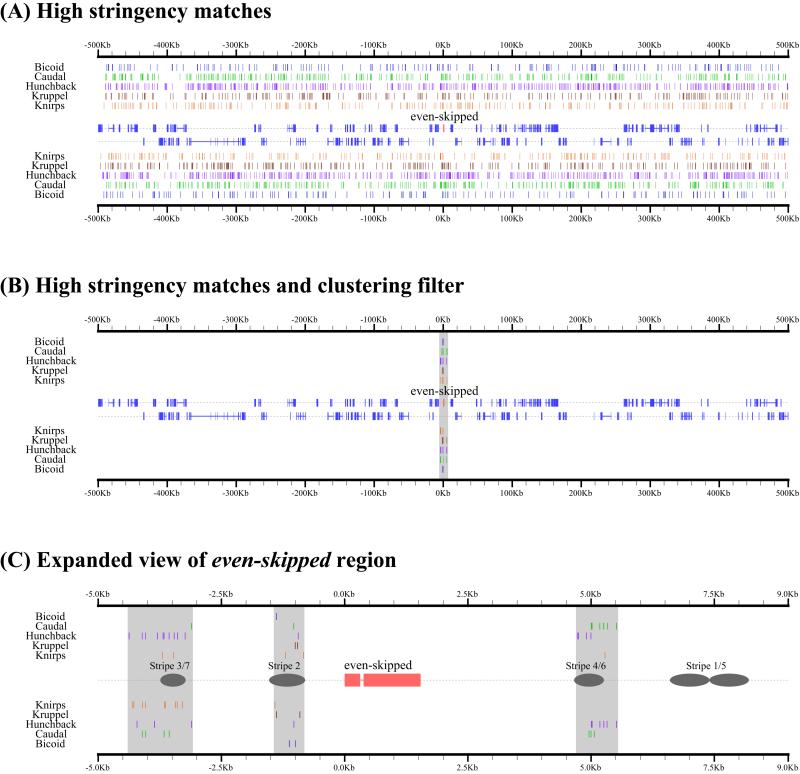
Using cis-analyst, we examined the distribution of Bcd, Cad, Hb, Kr, and Kni binding sites in a 1-Mb genomic region surrounding the well-characterized eve locus at a site_p value of 0.0003 (Fig. 1). At this relatively high-stringency value, most experimentally verified binding sites are retained; at more restrictive values, many of these sites would be lost. Fig. 1A shows all predicted binding sites for all five factors and reveals that binding sites for these factors are densely and widely distributed across this region of the genome.
Figure 1.
Distribution of predicted transcription factor binding sites and binding site clusters in the vicinity of eve. (A) Predicted high-affinity (P < 0.0003) binding sites for the transcription factors Bcd, Cad, Hb, Kr, and Kni in 1 Mb of genomic sequence surrounding the gene even-skipped (eve) are displayed as colored boxes. Blue boxes in the center of the panel represent positions of annotated exons, with eve highlighted in red. Binding sites and genes shown above the midline map to the forward DNA strand; those below the midline map to the reverse strand. (B) Sites from A that occur in 700-bp windows containing at least 13 predicted binding sites. (C) Expanded view of region containing all clusters in B, with positions of known eve enhancers marked with gray ellipses.
To investigate whether binding site clustering could help to explain the specificity of these factors for eve, we incorporated a simple notion of binding site clustering into cis-analyst, allowing searches for segments of a specified length containing a minimum number of predicted binding sites. When we searched the 1-Mb region surrounding eve for dense clusters of predicted high-affinity sites (at least 13 Bcd, Cad, Hb, Kr, or Kni sites in a 700-bp window), three discrete regions were identified (Fig. 1 B and C). Strikingly, these three clusters were all adjacent to eve, and overlapped the previously characterized stripe 2, stripe 3 + 7, and stripe 4 + 6 enhancers.
To generalize and quantify these promising results, we compiled a broader collection of 19 well-defined CRMs from 9 Drosophila genes known to be required for proper embryonic development (see Table 1). Each of these CRMs is sufficient to direct the expression of a distinct anterior–posterior pattern in early embryos, and genetic evidence suggests that each is regulated by at least one of Bcd, Cad, Hb, Kr, and Kni. Mutation and in vitro DNA binding studies completed on a subset of the CRMs provide evidence for a direct regulatory relationship. The same clustering criteria that were successful for identifying CRMs in eve (700-bp regions with at least 13 predicted binding sites) identified clusters overlapping 14 of these 19 known CRMs (binding site plots for each of these CRMs are shown in Fig. 6, which is published as supporting information on the PNAS web site).
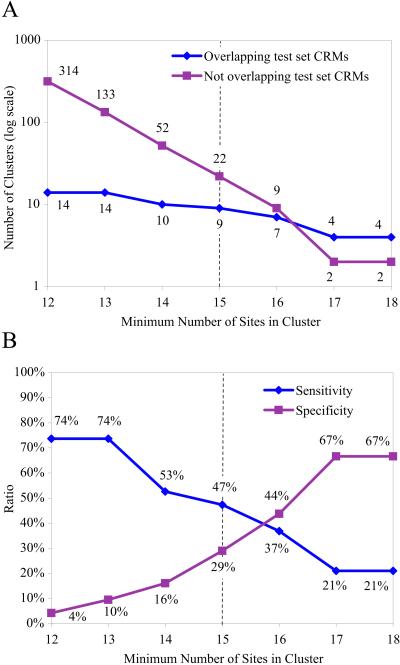
A search of the entire genome for 700-bp windows containing at least 13 predicted binding sites identified 133 clusters in addition to the 19 described above, or ≈1 per 700 kb of noncoding sequence. As expected, when more stringent clustering criteria are used, both the number of known CRMs recovered and the number of novel clusters identified decrease (see Fig. 2). We chose to examine further the novel clusters identified with a density of at least 15 binding sites per 700 bp, a level at which half of the known CRMs are still recovered. Binding site plots for the 22 novel clusters identified at this high stringency condition, and 6 additional novel clusters identified with an equally stringent search by using only Bcd, Hb, Kr, and Kni (see Materials and Methods) are shown in Fig. 7 (which is published as supporting information on the PNAS web site).
Figure 2.
Binding site clusters identified as a function of binding site density. (A) Number of binding site clusters in 93 Mb of noncoding genomic DNA at varying densities. Number of clusters overlapping test CRMs is shown in blue. Number of additional clusters is shown in pink. (B) Sensitivity (test set CRMs recovered divided by total number of test set CRMs) is shown in blue. Specificity (test set CRMs recovered divided by total number of clusters identified) is shown in pink. It is important to note that these sensitivity and specificity measures are computed assuming that only previously known CRMs are true positives. Because there are almost certainly additional bona fide CRMs in this set, the actual specificities and sensitivities of the method are expected to be better. Dotted line indicates density level chosen for the exploration of novel clusters described in the text.
Twenty-three of these 28 clusters fall in regions between genes, whereas the remaining 5 fall in introns. There are therefore 49 genes that either contain a novel cluster of binding sites or flank an intergenic region that does. We examined the expression patterns of these 49 genes in early embryos by whole-mount RNA in situ hybridization and DNA microarray hybridization. The locations of these clusters and details and expression patterns of these adjacent and flanking genes are presented in Table 2. At least 10 of the 28 clusters were adjacent to a gene that showed localized anterior–posterior expression in the syncitial or cellular blastoderm stages (see Fig. 3), consistent with early regulation by maternal effect or gap transcription factors. Although the numbers are small, this is significantly more than the 1 or 2 expected if the positions of clusters had been chosen at random.‖
Figure 3.
Expression patterns of selected genes flanking novel binding site clusters. We examined the expression patterns of 49 genes adjacent to one of the 28 novel binding site clusters described in Table 2 in syncytial and cellular blastoderm embryos (whole mount RNA in situ images are available in Table 2 (which is published as supporting information on the PNAS web site) and on the Berkeley Drosophila Genome Project website (http://www.fruitfly.org/). Eleven of these genes representing 10 clusters had early embryonic expression patterns characteristic of genes regulated by maternal and gap transcription factors and are shown here. §, References for flanking genes are as follows: gt (25, 30, 37–40), otd (41–43), btd (44, 45), pdm1 (46), pdm2 (46), Dfd (47–49), Antp (49, 50), ftz (51–53), odd (54), and psq (55)
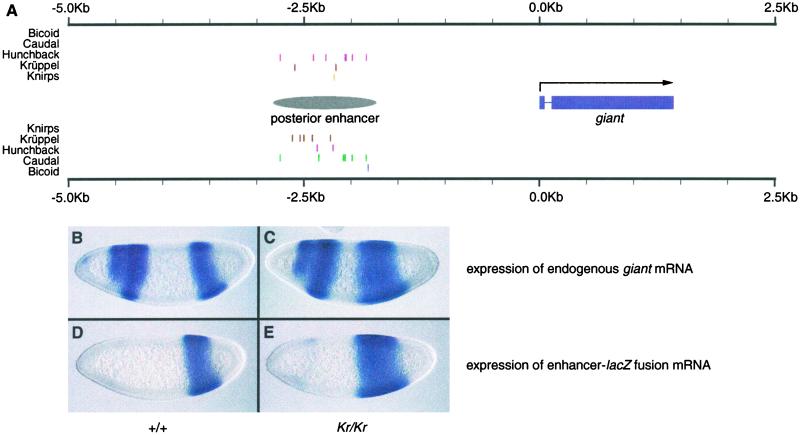
One of these clusters is located ≈2 kb upstream of the gap gene giant (gt; Fig. 4A). During cellularization, gt is expressed in two broad domains, one in the anterior and one in the posterior portion of the embryo (Fig. 4B). The pattern of expression of the posterior expression domain is known from a genetic analysis to be determined by the activities of Cad, Hb, and Kr (30). However, the cis-regulatory sequence controlling this posterior expression pattern has not been precisely identified. We sought to evaluate whether this cluster of binding sites might be the gt posterior enhancer. A 1.1-kb fragment containing this cluster was placed in a reporter construct containing the eve minimal promoter fused to a lacZ reporter gene. As shown in Fig. 4D, the expression pattern of this construct largely recapitulates the early expression pattern of the gt posterior expression domain. In the absence of Kr function, the anterior border of the gt posterior domain shifts anteriorly, indicating repression by Kr (Fig. 4C and ref. 30). The construct containing our gt posterior enhancer exhibits a similar shift in the absence of Kr (Fig. 4E).
Figure 4.
Identification of a novel enhancer controlling posterior expression of giant. (A) Cluster of binding sites found between 2.9 Kb and 1.8 Kb upstream of giant. The DNA segment surrounding the cluster (labeled “posterior enhancer”) was cloned into a lacZ fusion construct and introduced into the genome via germline transformation as described in Materials and Methods. (B and C) Expression of giant in syncitial blastoderm stage embryos as determined by RNA in situ hybridization. B shows a wild-type embryo, and C shows a Kr1/Kr1 embryo lacking Krüppel (Kr) function. Without repression by Kr, the anterior border of the posterior expression domain shifts anteriorly. (D and E) Expression of lac Z in embryos containing construct from A. D shows a wild-type embryo, and E shows a Kr1/Kr1 embryo. Expression of the lacZ construct in the mutant embryo shows similar expansion to that seen in gt.
Discussion
A central conundrum in understanding transcriptional regulation is that exquisite specificity in where and when genes are expressed is achieved through the action of sequence-specific DNA binding proteins whose sequence specificities are often not highly specific. Many transcription factors, like those examined here, bind DNA as monomers and recognize relatively short and degenerate sequences. Multiple predicted binding sites for each of the five transcription factors we examined are found adjacent to virtually every gene in the genome. Yet, only a handful of genes are regulated by these factors. The most striking result of this study is that variation in the local density of these binding sites appears sufficient to account for much of the specificity in their activities. At least a third, and likely more, of high-density clusters of maternal effect and gap transcription factor binding sites examined here correspond to bona fide cis-regulatory modules active in the early embryo. Although we did not observe patterned transcriptional activity in the early embryo for genes associated with a number of these clusters, Bcd, Cad, Hb, Kr, and Kni are also active later in development, and some clusters may represent regulatory elements active in these later processes. For instance, Hb and Kr are expressed in developing neuroblast cells much later in embryogenesis, and both are thought to contribute to fate determination of these cell populations (31).
Although it has been previously proposed that binding site clustering could be used to identify cis-regulatory modules (12), it is striking just how effective we have found this approach to be. The identification of functionally significant noncoding regions, especially those that control transcription, is one of the major challenges in understanding genome sequences, and we are optimistic that approaches like the one presented here will generalize to later stages of Drosophila development and to other organisms. Many of the transcription factors active in early Drosophila embryogenesis are also active in other organisms. In addition, binding site clustering has been observed in CRMs involved in processes later in Drosophila development and in the development of other organisms.
It is likely that improved methods for identifying binding site clusters will yield even better results than those presented here. The 700-bp window that we used is not appropriate for all CRMs. We are currently implementing a statistical model that will identify significant clusters of binding sites in windows of arbitrary size. This model will also consider the degree of agreement between prospective binding sites and the motif model, with increased significance assigned to clusters containing predicted high-affinity sites. Our binding site models will also be improved to account for any positional dependence occurring within transcription factor binding sites themselves; the current PWM model assumes that each base is independent. Models that incorporate some of these features have been successfully applied to other systems (10, 11, 13, 32). Additional functional exploration of computationally identified clusters will provide new examples that will permit further refinement of these methods.
Although binding data exist for some additional Drosophila transcription factors (33), a major roadblock to further evaluating and developing this approach is the paucity of available binding data for most transcription factors. We have initiated a project to gather accurate sequence specificity information for all of the ≈120 transcription factors believed to act in early Drosophila embryogenesis. With these data, it will be possible to search the genome for significant clusters of binding sites for additional combinations of factors. However, we cannot simply look for clusters of any combination of factors. Although high-density clusters of binding sites for particular combinations of factors are relatively rare, we expect every region of the genome to contain significant clusters of binding sites for some combinations of these 120 transcription factors. The analysis presented here worked in large part because we already knew that Bcd, Cad, Hb, Kr, and Kni act together. We plan to systematically identify additional sets of coacting factors by analyzing the expression patterns of transcription factors and through further genetic studies. The imminent availability of the Drosophila pseudoobscura genome sequence will provide additional means for distinguishing biologically relevant clusters from those that occur by chance, because only functionally significant clusters should be found in both genomes. In addition, the identification of blocks of noncoding DNA conserved between D. melanogaster and D. pseudoobscura will be useful in subsequent studies because recent analyses in many species suggest that such sequences are significantly enriched for functional transcription factor binding sites and CRMs (34, 35).
The grammar of the cis-regulatory code is clearly more complex than simply the density of transcription factor binding sites. The relative positioning of sites within cis-regulatory modules has been demonstrated to be significant in many cases. This result is to be expected, because we know that there are often important protein–protein interactions between bound factors that can influence CRM function. However, some plasticity in the positioning of binding sites is tolerated in some situations (14, 36). The analysis of orthologous CRMs in multiple species should help to further elucidate the rules governing CRM structure (as in ref. 14). Ultimately, we would like to incorporate these rules into our methods for identifying CRMs. However, to achieve a sufficient understanding of the architecture of cis-regulatory modules, we need to expand the number of identified and characterized CRMs. We believe that CRM detectors based on binding site clustering are a useful first step along this path.
Supplementary Material
Acknowledgments
We thank Amy Beaton, Elaine Kwan, Steven Richards, Richard Weiszman, Joseph Nunoo, and Todd Laverty for assistance with RNA in situ hybridizations; Chris Mungall for assistance with genome annotations; and Dmitri Papatsenko for sharing unpublished data and ideas. The binding site plots included in this work were created by using the GFF2PS (http://www1.imim.es/software/gfftools/GFF2PS.html), and we thank the author, Josep Francesco Abril Ferrando, for his personal assistance. B.P.B. wishes to thank Ed Lewis, Michael Brodsky, Erwin Frise, and Derek Chiang for helpful discussions. We also thank Casey Bergman for his critical reading of the manuscript. The cis-analyst tool, and all sequences and other data used in this work are available at http://www.fruitfly.org/cis-analyst. This work was supported by National Institutes of Health Grants GM34431 (to M.S.L.), P50 HG00750 (to G.M.R.), and HL667201 (to M.B.E.); Department of Energy contract DE-AC03-76SF00098 (to M.B.E.); and by the Howard Hughes Medical Institute. M.B.E. is a Pew Scholar in the Biomedical Sciences.
Abbreviations
- CRM
cis-regulatory module
- PWM
position weight matrix
Footnotes
See commentary on page 546.
Only 15 of 828 whole mount in situ hybridizations performed on randomly selected genes by the Berkeley Drosophila Genome Project (http://fruitfly.berkeley.edu) show localized anterior/posterior expression during the blastoderm stage. Based on this result, and accounting for the proportion of noncoding DNA in intergenic regions and introns, we estimate that a randomly selected 700-bp fragment has a 3.2% chance of being adjacent to a gene expressed in such a pattern.
References
- 1.Carroll S B, Grenier J K, Weatherbee S D. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. Oxford: Blackwell Scientific; 2001. [Google Scholar]
- 2.Harding K, Hoey T, Warrior R, Levine M. EMBO J. 1989;8:1205–1212. doi: 10.1002/j.1460-2075.1989.tb03493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goto T, Macdonald P, Maniatis T. Cell. 1989;57:413–422. doi: 10.1016/0092-8674(89)90916-1. [DOI] [PubMed] [Google Scholar]
- 4.Stanojevic D, Small S, Levine M. Science. 1991;254:1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- 5.Small S, Blair A, Levine M. Dev Biol. 1996;175:314–324. doi: 10.1006/dbio.1996.0117. [DOI] [PubMed] [Google Scholar]
- 6.Sackerson C, Fujioka M, Goto T. Dev Biol. 1999;211:39–52. doi: 10.1006/dbio.1999.9301. [DOI] [PubMed] [Google Scholar]
- 7.Fujioka M, Emi-Sarker Y, Yusibova G L, Goto T, Jaynes J B. Development (Cambridge, UK) 1999;126:2527–2538. doi: 10.1242/dev.126.11.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson E H. Genomic Regulatory Systems: Development and Evolution. San Diego: Academic; 2001. [Google Scholar]
- 9.Ludwig M Z, Patel N H, Kreitman M. Development (Cambridge, UK) 1998;125:949–958. doi: 10.1242/dev.125.5.949. [DOI] [PubMed] [Google Scholar]
- 10.Crowley E M, Roeder K, Bina M. J Mol Biol. 1997;268:8–14. doi: 10.1006/jmbi.1997.0965. [DOI] [PubMed] [Google Scholar]
- 11.Wasserman W W, Fickett J W. J Mol Biol. 1998;278:167–181. doi: 10.1006/jmbi.1998.1700. [DOI] [PubMed] [Google Scholar]
- 12.Wagner A. Bioinformatics. 1999;15:776–784. doi: 10.1093/bioinformatics/15.10.776. [DOI] [PubMed] [Google Scholar]
- 13.Frith M C, Hansen U, Weng Z. Bioinformatics. 2001;17:878–889. doi: 10.1093/bioinformatics/17.10.878. [DOI] [PubMed] [Google Scholar]
- 14.Ludwig M Z, Bergman C, Patel N H, Kreitman M. Nature (London) 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- 15.Bailey T L, Elkan C. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 16.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 17.Hertz G Z, Stormo G D. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 18.Thummel C S, Boulet A M, Lipshitz H D. Gene. 1988;74:445–456. doi: 10.1016/0378-1119(88)90177-1. [DOI] [PubMed] [Google Scholar]
- 19.Small S, Blair A, Levine M. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosman D, Small S. Development (Cambridge, UK) 1997;124:1343–1354. doi: 10.1242/dev.124.7.1343. [DOI] [PubMed] [Google Scholar]
- 21.Niessing D, Rivera-Pomar R, La Rosee A, Hader T, Schock F, Purnell B A, Jackle H. J Cell Physiol. 1997;173:162–167. doi: 10.1002/(SICI)1097-4652(199711)173:2<162::AID-JCP15>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 22.St Johnston D, Nusslein-Volhard C. Cell. 1992;68:201–219. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- 23.Macdonald P M, Struhl G. Nature (London) 1986;324:537–545. doi: 10.1038/324537a0. [DOI] [PubMed] [Google Scholar]
- 24.Mlodzik M, Gehring W J. Cell. 1987;48:465–478. doi: 10.1016/0092-8674(87)90197-8. [DOI] [PubMed] [Google Scholar]
- 25.Rivera-Pomar R, Lu X, Perrimon N, Taubert H, Jackle H. Nature (London) 1995;376:253–256. doi: 10.1038/376253a0. [DOI] [PubMed] [Google Scholar]
- 26.Sauer F, Rivera-Pomar R, Hoch M, Jackle H. Philos Trans R Soc London B. 1996;351:579–587. doi: 10.1098/rstb.1996.0057. [DOI] [PubMed] [Google Scholar]
- 27.Berg O G, von Hippel P H. J Mol Biol. 1987;193:723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- 28.Stormo G D. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 29.Stormo G D, Fields D S. Trends Biochem Sci. 1998;23:109–113. doi: 10.1016/s0968-0004(98)01187-6. [DOI] [PubMed] [Google Scholar]
- 30.Kraut R, Levine M. Development (Cambridge, UK) 1991;111:601–609. doi: 10.1242/dev.111.2.601. [DOI] [PubMed] [Google Scholar]
- 31.Isshiki T, Pearson B, Holbrook S, Doe C Q. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- 32.Krivan W, Wasserman W W. Genome Res. 2001;11:1559–1566. doi: 10.1101/gr.180601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wingender E, Chen X, Fricke E, Geffers R, Hehl R, Liebich I, Krull M, Matys V, Michael H, Ohnhauser R, et al. Nucleic Acids Res. 2001;29:281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pennacchio L A, Rubin E M. Nat Rev Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- 35.Wasserman W W, Palumbo M, Thompson W, Fickett J W, Lawrence C E. Nat Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- 36.Arnosti D N, Barolo S, Levine M, Small S. Development (Cambridge, UK) 1996;122:205–214. doi: 10.1242/dev.122.1.205. [DOI] [PubMed] [Google Scholar]
- 37.Mohler J, Eldon E D, Pirrotta V. EMBO J. 1989;8:1539–1548. doi: 10.1002/j.1460-2075.1989.tb03538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraut R, Levine M. Development (Cambridge, UK) 1991;111:611–621. doi: 10.1242/dev.111.2.611. [DOI] [PubMed] [Google Scholar]
- 39.Eldon E D, Pirrotta V. Development (Cambridge, UK) 1991;111:367–378. doi: 10.1242/dev.111.2.367. [DOI] [PubMed] [Google Scholar]
- 40.Capovilla M, Eldon E D, Pirrotta V. Development (Cambridge, UK) 1992;114:99–112. doi: 10.1242/dev.114.1.99. [DOI] [PubMed] [Google Scholar]
- 41.Finkelstein R, Perrimon N. Nature (London) 1990;346:485–488. doi: 10.1038/346485a0. [DOI] [PubMed] [Google Scholar]
- 42.Gao Q, Wang Y, Finkelstein R. Mech Dev. 1996;56:3–15. doi: 10.1016/0925-4773(96)00504-7. [DOI] [PubMed] [Google Scholar]
- 43.Gao Q, Finkelstein R. Development (Cambridge, UK) 1998;125:4185–4193. doi: 10.1242/dev.125.21.4185. [DOI] [PubMed] [Google Scholar]
- 44.Wimmer E A, Jackle H, Pfeifle C, Cohen S M. Nature (London) 1993;366:690–694. doi: 10.1038/366690a0. [DOI] [PubMed] [Google Scholar]
- 45.Wimmer E A, Simpson-Brose M, Cohen S M, Desplan C, Jackle H. Mech Dev. 1995;53:235–245. doi: 10.1016/0925-4773(95)00439-8. [DOI] [PubMed] [Google Scholar]
- 46.Cockerill K A, Billin A N, Poole S J. Mech Dev. 1993;41:139–153. doi: 10.1016/0925-4773(93)90044-x. [DOI] [PubMed] [Google Scholar]
- 47.Martinez-Arias A, Ingham P W, Scott M P, Akam M E. Development (Cambridge, UK) 1987;100:673–683. doi: 10.1242/dev.100.4.673. [DOI] [PubMed] [Google Scholar]
- 48.Jack T, McGinnis W. EMBO J. 1990;9:1187–1198. doi: 10.1002/j.1460-2075.1990.tb08226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reinitz J, Levine M. Dev Biol. 1990;140:57–72. doi: 10.1016/0012-1606(90)90053-l. [DOI] [PubMed] [Google Scholar]
- 50.Harding K, Levine M. EMBO J. 1988;7:205–214. doi: 10.1002/j.1460-2075.1988.tb02801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dearolf C R, Topol J, Parker C S. Genes Dev. 1989;3:384–398. doi: 10.1101/gad.3.3.384. [DOI] [PubMed] [Google Scholar]
- 52.Dearolf C R, Topol J, Parker C S. Nature (London) 1989;341:340–343. doi: 10.1038/341340a0. [DOI] [PubMed] [Google Scholar]
- 53.Topol J, Dearolf C R, Prakash K, Parker C S. Genes Dev. 1991;5:855–867. doi: 10.1101/gad.5.5.855. [DOI] [PubMed] [Google Scholar]
- 54.Coulter D E, Swaykus E A, Beran-Koehn M A, Goldberg D, Wieschaus E, Schedl P. EMBO J. 1990;9:3795–3804. doi: 10.1002/j.1460-2075.1990.tb07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lehmann M, Siegmund T, Lintermann K G, Korge G. J Biol Chem. 1998;273:28504–28509. doi: 10.1074/jbc.273.43.28504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.