Abstract
Metazoan genomes contain vast tracts of cis-regulatory DNA that have been identified typically through tedious functional assays. As a result, it has not been possible to uncover a cis-regulatory code that links primary DNA sequences to gene expression patterns. In an initial effort to determine whether coordinately regulated genes share a common “grammar,” we have examined the distribution of Dorsal recognition sequences in the Drosophila genome. Dorsal is one of the best-characterized sequence-specific transcription factors in Drosophila. The homeobox gene zerknullt (zen) is repressed directly by Dorsal, and this repression is mediated by a 600-bp silencer, the ventral repression element (VRE), which contains four optimal Dorsal binding sites. The arrangement and sequence of the Dorsal recognition sequences in the VRE were used to develop a computational algorithm to search the Drosophila genome for clusters of optimal Dorsal binding sites. There are 15 regions in the genome that contain three or more optimal sites within a span of 400 bp or less. Three of these regions are associated with known Dorsal target genes: sog, zen, and Brinker. The Dorsal binding cluster in sog is shown to mediate lateral stripes of gene expression in response to low levels of the Dorsal gradient. Two of the remaining 12 clusters are shown to be associated with genes that exhibit asymmetric patterns of expression across the dorsoventral axis. These results suggest that bioinformatics can be used to identify novel target genes and associated regulatory DNAs in a gene network.
The recent revelation that the human genome contains only ≈30,000 genes underscores the importance of gene regulation in generating organismal diversity (1, 2). Cis-regulatory DNAs account for up to 50% of the total DNA that comprises metazoan genomes. However, there is no obvious “code” that links primary DNA sequence with gene expression patterns. Here we describe a whole-genome analysis of the binding sites for Dorsal (Dl), one of the best-characterized sequence-specific transcription factors in Drosophila (3, 4).
The Dl protein is distributed in a broad concentration gradient across the dorsoventral axis of the early embryo (5–7). This gradient initiates the differentiation of several embryonic tissues by regulating various target genes in a concentration-dependent manner (8). Peak levels of Dl activate snail and twist expression in ventral regions that form mesoderm (9–12), whereas lower levels activate neurogenic genes such as sim and rhomboid in lateral regions (13, 14). The Dl gradient also functions as a context-dependent repressor that restricts the expression of zen, dpp, and tolloid to dorsal regions that will form derivatives of the dorsal ectoderm (15–17).
Altogether, the Dl gradient generates five different thresholds of gene activity across the dorsoventral axis of the early embryo. These thresholds are interpreted by complex enhancers that are 300 bp–1 kb in length and contain clustered binding sites for Dl. Target enhancers that contain low-affinity Dl binding sites such as those present in the 180-bp twist proximal enhancer are activated only in the ventral mesoderm in response to peak levels of the Dl gradient (18). In contrast, enhancers that contain high-affinity sites, as seen in the 300-bp rhomboid neurogenic ectoderm enhancer (NEE) enhancer, can be activated in the lateral neurogenic ectoderm in response to low levels of the Dl gradient (14, 18).
Cis-regulatory DNAs have been characterized for seven different Dl target genes that initiate the differentiation of the mesoderm (twist and snail; refs. 9–12), mesectoderm (sim; ref. 13), neurogenic ectoderm (rhomboid; ref. 14), and dorsal ectoderm (zen, dpp, and tolloid; refs. 19–21). zen, dpp, and tolloid are repressed by low levels of the Dl gradient that are insufficient to activate the rhomboid NEE. There is only one putative Dl target gene, sog, that is activated by the same low levels of the Dl gradient. sog is expressed in broad lateral stripes that encompass the entire presumptive neurogenic ectoderm (22). The sog stripes are broader than the rhomboid stripes and represent a distinct threshold readout of the Dl gradient. Because zen and sog exhibit similar sensitivity to the Dl gradient, it is possible that their cis-regulatory DNAs share some common features. The zen 5′ cis-regulatory region includes a distal silencer, the ventral repression element (VRE), which contains four high-affinity Dl binding sites (4). Dl is inherently a transcriptional activator, but facilitates the binding of “corepressor” proteins to neighboring AT-rich sequence motifs within the VRE (19, 20).
To identify cis-regulatory DNAs and associated genes that are regulated by Dl, a bioinformatics approach was used to identify clusters of high-affinity Dl binding sites in the Drosophila genome. The search was based on the observation that the zen VRE contains a cluster of four optimal Dl binding sites (4, 15, 19). There are only 15 sequences in the entire genome, including the VRE, that contain three or more optimal sites within a stretch of 400 bp or less. One of the optimal clusters is located within the first intron of the sog gene, ≈700 bp downstream of the transcription start site. This sog intronic region contains four optimal Dl binding sites within a 263-bp interval. A 393-bp DNA fragment that encompasses the cluster was inserted into a lacZ P-element transformation vector and analyzed in transgenic embryos. The resulting sog-lacZ fusion gene exhibits broad lateral stripes of expression in early embryos similar to the endogenous sog pattern. The expression pattern is broader than the lateral stripes seen for rhomboid and indicates that the sog intronic enhancer is activated by low levels of the Dl gradient. Thus, the zen VRE and sog intronic enhancer share a similar code that permitted the de novo identification of the sog enhancer in a whole-genome search of optimal Dl binding clusters.
Computational methods can be used to identify target genes in a regulatory network. Three of the 15 optimal clusters are associated with known Dl target genes: Brinker (21, 23), sog (22), and zen (19). Most of the remaining clusters are linked to genes that are not known to be involved in dorsoventral patterning. In situ localization assays were done to determine whether any of these genes might represent previously uncharacterized Dl targets. Two of the genes, Phm (24) and Ady (25), are shown to be up-regulated in the presumptive mesoderm of early embryos. The Ady gene exhibits a pattern of expression that is consistent with activation by intermediate levels of the Dl gradient. Thus, the search for optimal Dl binding sites led to the identification of putative target genes and cis-regulatory DNAs. We discuss the application of this method to other systems and compare the efficacy of this strategy with computational search algorithms that employ multiple classes of regulatory factors.
Materials and Methods
Fly Stocks.
yw67 was used for P-element transformations and in situ hybridizations. Males carrying the sog B/lacZ transgene were mated with females homozygous for a null mutation in gastrulation defective (obtained from the Bloomington Stock Center line, v1 gd7/TM3, BL stock 3109).
Cloning and Injection of DNA Fragments Containing Dl Binding Clusters.
Genomic DNA was prepared from a single anesthetized yw male as described (26). sog A, B, and C fragments were amplified from genomic DNA with the following primer pairs: Sog A, 102 (5′-TTTGCCTGCAACATGTTGCTGCCGATCGTTAGATGT-3′) and 103 (5′-GCTTTATGGTCCATGGTCCATACCACCCAATGGTCT-3′); Sog B, 101 (5′-GTTGCCAATGCCATTGCGCATACGCCGTGTCGTCTA-3′) and 103; and Sog C, 101 and 105 (5′-CTGGGTATATGCAGGGACAGAGCGAAGTGCCTTTGA-3′). PCR products were purified with the Qiagen (Chatsworth, CA) QiaQuick PCR purification kit and cloned directly into the Promega pGEM T-Easy vector. The fragments were excised from the pGem vector by NotI digestion, gel-purified with the Qiagen QiaQuick gel extraction kit, and cloned into a unique NotI site upstream of the eve minimal promoter–lacZ reporter within a modified pCaSpeR vector (27), E2G. E2G contains SuHw insulator elements flanking the NotI insertion + eve-lacZ portion of the vector. These pCaSpeR constructs were introduced into the Drosophila germline by the standard injection protocol (28). Between three and nine independent transformed lines were obtained for each construct.
In Situ Hybridization.
Embryos were hybridized with digoxigenin-labeled antisense RNA probes as described (11). An antisense lacZ RNA probe (11) was used to examine the staining patterns of each of the lines obtained for the sog A, B, and C transformants. To examine the pattern of endogenous Phm, the expressed sequence tag clone SD13745 was obtained from Research Genetics (Huntsville, AL), linearized with EcoRI, and transcribed with SP6. To examine the expression pattern of the endogenous Ady gene, the following primers were used to obtain a 992-bp fragment from exon 3 of the coding region: 5′-CTCTCGGTGCCTTACAAACCCGGGTGGCT-3′ and 5′-TGAAATGGTGGACAAATGTTTGACTAAGCCT-3′. This fragment was cloned into pGem T-Easy, linearized with NcoI, and transcribed with SP6. sog and zen antisense probes were prepared from cDNA containing plasmids.
Genome-Wide Scanning.
Bioinformatic screens of Dl binding clusters used the Berkeley Drosophila Genome Project database Release I of the Drosophila genome (29), which contains 114 megabases (ignoring indeterminate positions encoded with Ns). The program FLY ENHANCER was developed and used in this study to scan the genome for clusters of binding sites. A manuscript describing the program is in preparation. To use FLY ENHANCER or for more information about our bioinformatics approach, visit www.insilicolabs.com/flyenhancer.
Statistics.
Our computations take into account the observation that the Drosophila genome is AT-rich, with A and T each occurring with probability 0.287 and G and C each occurring with probability 0.213. Given the A/T bias in the Drosophila genome, the probability of finding one of the 208 “optimal” Dl sequences searched (encoded by GGGWWWWCCM or GGGWDWWWCCM and their reverse complements) at any point in the genome is 0.0000691. We found 8,789 instances of optimal Dl sequences (the expected number was 7,898). The expected number of k clusters was estimated by computing the probability of finding (k − 1) or more instances of optimal sequence patterns in a window of length n, following each of the 8,789 Dl sites, using the binomial distribution as an approximation.
Results
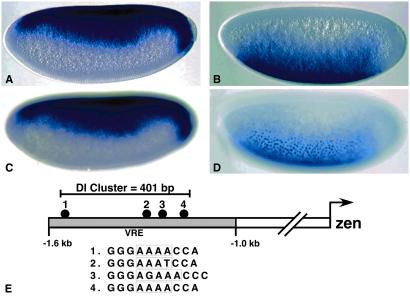
Previous studies have shown that zen is an immediate target of the maternal Dl gradient (4, 15, 19). The gene is activated initially at nuclear cleavage cycle 11-12 within 1 h after the Dl gradient is formed (30). zen initially exhibits a broad pattern of expression in the presumptive dorsal ectoderm and at the termini (Fig. 1 A and C). As discussed earlier, high and low levels of the Dl gradient keep zen off in ventral and lateral regions. sog exhibits a complementary pattern of expression because it is activated by Dl, whereas zen is repressed (Fig. 1 B and D). As seen for zen, sog expression is detected shortly after the formation of the Dl gradient (22).
Figure 1.
zen and sog expression patterns. Precellular embryos are oriented with anterior to the left and dorsal up. A and C were hybridized with a digoxigenin-labeled zen antisense RNA probe, and B and D were hybridized with a sog probe. The staining patterns were visualized with anti-digoxigenin antibodies and histochemical staining. (A and C) Parasagittal and surface views of the same embryo. (B and D) Different planes of focus through a single embryo. Note that sog RNAs are detected in nuclei (D). (E) Diagram of the zen 5′ regulatory region showing distribution of the four Dl binding sites in the VRE.
The zen VRE contains four optimal Dl recognition sequences within a span of 400 bp (ref. 4; Fig. 1E). Three of the four Dl binding sites contained within the zen VRE conform to the following consensus sequence for high-affinity Dl binding sites: GGG(W)nCCM (where W = A or T, M = C or A, and n corresponds to either four or five W residues). The fourth recognition sequence (binding site 3 within the VRE) contains a G residue in the AT-rich central region and is represented by the optimal consensus sequence GGGWDWWWCCM (where D = A, T, or G). To determine whether a similar density of optimal Dl sites might account for the regulation of sog, we scanned the entire Drosophila genome for clusters of any of the 208 unique Dl sequences that conform (either directly or by reverse complement) to the two degenerate sequences discussed above: GGG(W)4CCM and GGGWDWWWCCM.
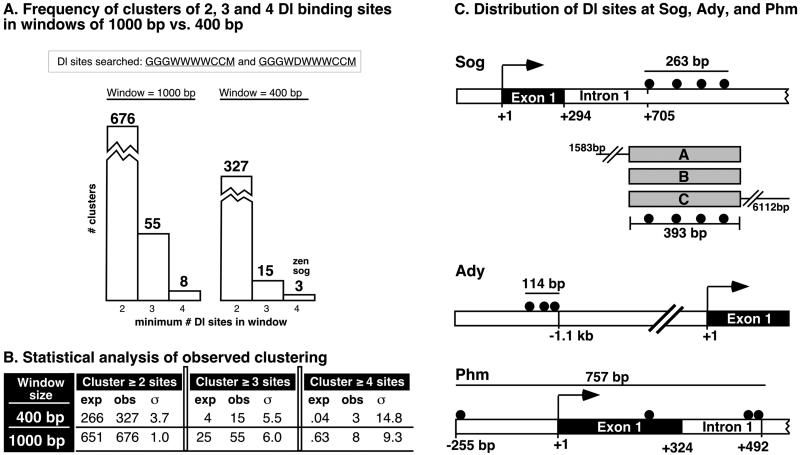
We scanned the genome for clusters of Dl binding sites in windows of 400 bp, the interval that the sites are clustered in the zen VRE, and also for clustering in windows of 1,000 bp because the operational size of enhancers can generally be thought of as about 1,000 bp. Although the genome-wide occurrence of 676 clusters of two or more optimal Dl sites in 1,000 bp is not statistically significant (Fig. 2 A and B), the occurrence of 55 clusters with at least three sites and of eight clusters containing four sites is enriched beyond what one would expect from random chance. However, none of the clusters within 1,000 bp identified known Dl targets that were missed by our more stringent screen for clustering within 400 bp. Therefore, as discussed below, we focused this study on the results from the more stringent screen.
Figure 2.
Distribution of Dl clusters. (A) Frequency of clusters in genome containing a minimum of two, three, or four Dl binding sites in intervals of 1,000 or 400 bp. The Dl sequences searched are represented by the degenerate sequences GGGWWWWCCM and GGGWDWWWCCM, which encode a total of 208 unique sequences. Of the three clusters found to contain four sites in 400 bp, one is associated with zen and another with sog. (B) Statistical analysis of the expected (exp) vs. observed (obs) numbers of clusters with two, three, and four Dl sites found in windows of 1,000 and 400 bp. The number of observed clusters of three and four sites are many standard deviations (σ) from their expected frequencies, suggesting that their occurrence at the observed frequencies is not a random event. See Materials and Methods for details. (C) Distribution of Dl binding sites associated with sog, Ady, and Phm. Illustrated below the sog cluster are the three DNA fragments (sog A, B, and C) that were tested for regulatory activities in transgenic embryos.
As expected, the occurrence of 400-bp windows containing at least two sites (327 clusters; Fig. 2A) is much greater than the occurrence of 400-bp windows containing at least three sites (15 clusters) or four sites (3 clusters). However, the statistical significance of the clusters increases with their rarity (Fig. 2B). For example, the occurrence of 15 clusters with three or more Dl sites is 6 standard deviations from expected, making the probability of finding 15 clusters by random chance less than one in a million. The probability of finding three 400-bp clusters with at least four Dl sites is less than 10−49. Remarkably, two of the clusters in this rarest class are associated with the sog and zen genes (Fig. 2A), which exhibit the most sensitive response to the Dl gradient. Of the remaining 13 clusters containing three or more Dl sites, one is associated with the Brinker gene, which is expressed in lateral stripes and probably is a direct target of the Dl gradient (ref. 21; see Table 1). The other remaining 12 clusters neighbor genes that were not known previously to be involved in dorsoventral patterning. In the remainder of this report we examine the expression patterns of two of these genes, Phm and Ady, and also investigate the regulatory activities of the “sog cluster.”
Table 1.
Possible Dorsal targets: genes neighboring clusters of three or more Dorsal sites in a window of 400 bp
| Gene | Position of DI sites in cluster | Position of cluster relative to gene | Putative or known function | |
|---|---|---|---|---|
| Cluster of 3 in 100 | CG12444 | 0,41,64 | 5.5 kb 5′ of txn start | No info |
| ? none | 0,73,86 | 30 kb from Ppv; 57 kb from CG3367 | – | |
| CG1412 | 0,115,146,185 | 9 kb 5′ of CG1412 txn start | Contains PDZ domain | |
| CG1812 | 11.5 kb 5′ of CG1812 txn start | Contains BTB/POZ domain | ||
| Cluster of 3 in 200 | Ady43A | 0,73,104 | 1.1 kb 5′ of Ady txn start | Adenosine kinase |
| CG11086 | 3 kb 5′ of CG11086 txn start | No info | ||
| Fas-3 | 0,15,175,(811) | 11 kb 5′ of Fas-3 txn start | “Fasciclin 3”; cell adhesion, axon guidance | |
| Acp36DE | 12.5 kb 5′ of Acp36D txn start | “Accessory Gland Peptide”; peptide hormone | ||
| Zen | 0,68,118,392 | 1.5 kb 5′ of txn start | “Zerknullt”; transcription factor | |
| Sog | 0,79,162,253 | 750 bp 3′ of txn start, in intron 1 | “Short-gastrulation”; dpp antagonist/agonist | |
| Cluster of 3 in 300 | CG5549 | 0,154,215 | 1.3 kb 5′ of txn start | Glycine:sodium symporter |
| CG4763 | 4 kb 5′ of txn start | No info | ||
| Runt | 0,143,230 | 300 bp 3′ of txn stop | “Runt”; transcription factor | |
| Kr-h2 | 0,67,249 | 2.7 kb 3′ of kr-h2 txn stop | “Krupple homolog 2” | |
| CG9162 | 2.2 kb 5′ of CG9162 txn start | No info | ||
| Phm | 0,24,262,(747) | 272 bp 5′ txn start to +457 bp in Intron 1 | “Peptidylglycine-α-hydroxylating monooxygenase” | |
| Cluster of 3 in 400 | CG9595 | 0,191,306 | 2 kb 3′ txn stop | Kinesin motor |
| eya | 18 kb 5′ txn start | “Eyes absent”; involved in transcription | ||
| CG1924 | 1,115,330 | 15 kb 3′ of CG1924 txn stop | Chaperone | |
| PpD5 | 0,175,362,(926) | 1 kb bp 5′ of txn start | “Protein phosphatase D5” | |
| CG13500 | 12.5 kb 3′ txn stop | No info | ||
| Brk | 0,158,346 | 10.5 kb 5′ of brk txn start | “Brinker”; transcriptional repressor | |
| Unc-119 | 5.7 kb 5′ of unc-119 txn start | Neuronal protein |
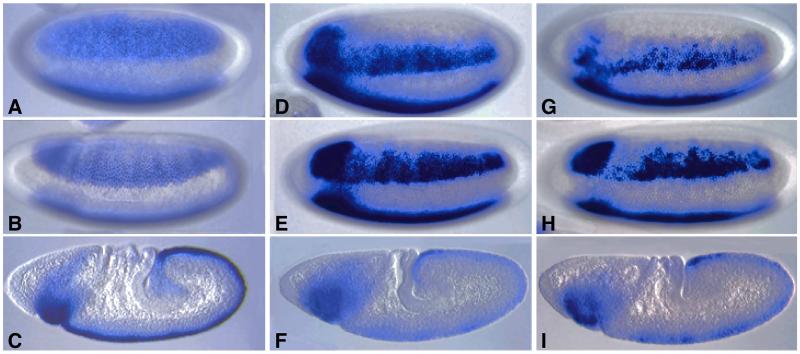
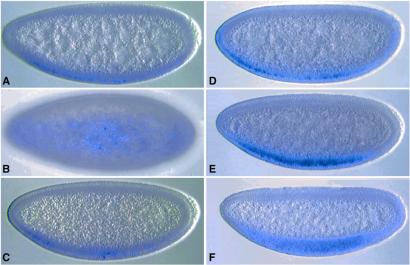
sog is expressed initially in broad lateral stripes that encompass the entire presumptive neurogenic ectoderm (ref. 20; Fig. 3A). In this lateral view, only one of the stripes is observed; the other stripe is out of the plane of focus. Staining persists in these lateral stripes during cellularization and the onset of gastrulation (Fig. 3B) but quickly refines within the mesectoderm at the ventral midline of elongating embryos (Fig. 3C). There are four optimal Dl binding sites located within a 263-bp region of sog intron 1 (Fig. 2C). Three different DNA fragments that encompass this region of the sog gene were placed 5′ of a lacZ reporter gene and expressed in transgenic embryos. The largest fragment is 6 kb in length and includes two-thirds of intron 1, whereas the smallest is just 393 bp and centered around the cluster of Dl binding sites. We also tested a 1.5-kb fragment that extends into the 5′-flanking region (Fig. 2C). All three fragments direct lateral stripes of lacZ expression that are similar to those seen for the endogenous gene (Fig. 3 D–I). These broad stripes persist until gastrulation (Fig. 3 E and H) and then refine within the mesectoderm (Fig. 3 F and I). Midline staining becomes weak and erratic in older embryos (data not shown). The 393-bp sog fragment directs essentially the same staining pattern (Fig. 3 G–I) as those obtained with the 6-kb DNA fragment (Fig. 3 D–F) as well as the 1.5-kb fragment (data not shown). These results suggest that the 393-bp fragment (hereafter called the sog lateral stripe enhancer) contains most of the cis elements responsible for regulating the early sog pattern.
Figure 3.
The sog lateral stripe enhancer. Wild-type and transgenic embryos are oriented with anterior to the left and dorsal up. A–C were hybridized with a sog antisense RNA probe, and D–I were hybridized with a lacZ probe to monitor the activities of different sog-lacZ transgenes. (A–C) Endogenous sog expression pattern in precellular (A), gastrulating (B), and elongating (C) embryos. Staining is detected initially in broad lateral stripes (A and B) but is restricted to the mesectoderm during germ band elongation (C). (D–F) sog-lacZ transgene that contains a 6-kb region of sog intron 1. Staining is detected in broad lateral stripes before (D) and after (E) cellularization but is restricted to the mesectoderm in elongating embryos (F). The staining pattern is similar to the normal sog expression pattern except that there is progressive loss of staining in the mesectoderm (compare C with F; data not shown). (G–I) sog-lacZ transgene that contains a 393-bp fragment from sog intron 1, which encompasses all four high-affinity Dl binding sites. The lacZ expression pattern is similar to that obtained with the 6-kb sog DNA fragment except that staining may be somewhat weaker and mottled.
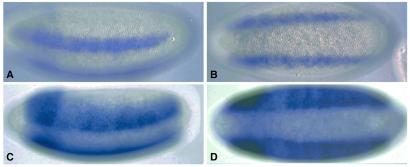
The sog lateral stripe enhancer shares a number of similarities with the previously characterized rhomboid NEE (14), which also mediates gene expression in the neurogenic ectoderm (Fig. 4 A and B). However, the NEE stripes are narrower than those generated by the sog enhancer (Fig. 4 C and D), suggesting that the sog enhancer responds to lower levels of the Dl gradient than the NEE. This difference might be due, at least in part, to the quality or organization of the Dl binding sites in the two enhancers. For example, only two of the four Dl binding sites contained in the NEE are optimal sites, whereas all four sites are optimal in the sog enhancer. The NEE contains four binding sites for the zinc finger snail repressor (14), which is expressed selectively in the ventral mesoderm and thereby restricts rhomboid expression to lateral regions (Fig. 4 A and B). The sog lateral stripe enhancer contains two potential snail repressor sites (CACCT) that might be responsible for attenuated staining in ventral regions (e.g., Fig. 4D).
Figure 4.
Comparison of the rhomboid NEE and sog lateral stripe enhancer. Transgenic, cellularizing embryos were hybridized with a lacZ RNA probe to visualize the activities of the 300-bp rhomboid NEE (A and B) and 393-bp sog lateral stripe enhancer (C and D). The lateral stripes generated by the NEE encompass 6–8 cells in the ventral half of the presumptive neurogenic ectoderm (A). In contrast, the stripes produced by the sog enhancer encompass 12–14 cells and include the entire neurogenic ectoderm (C). Both enhancers are inactive or attenuated in the ventral mesoderm (B and D). The NEE has been shown to be repressed by snail. The sog enhancer contains two potential snail repressor sites.
The preceding results indicate that computational methods can identify cis-regulatory DNAs. To determine whether these methods also identify target genes, we examined the expression profiles of two of the genes that contain optimal Dl clusters: Phm and Ady (see Table 1). Digoxigenin-labeled antisense RNA probes were prepared for each gene and hybridized to fixed embryos. Both genes exhibit expression in ventral regions of early embryos, although Ady appears to be expressed earlier than Phm (Fig. 5 A and D). Staining persists in the developing mesoderm during cellularization (Fig. 5 C and E) and gastrulation (Fig. 5F). The Ady pattern may be somewhat broader than the Phm pattern and may extend into ventral regions of the presumptive neurogenic ectoderm (Fig. 5B). These results suggest that one or both genes represent direct targets of the Dl gradient.
Figure 5.
Expression patterns of putative target genes. Normal embryos were hybridized with either an Ady (A–C) or Phm (D–F) RNA probe. Both genes are expressed in ventral regions of precellular embryos (A and D), although Ady may be activated slightly earlier than Phm. Both genes are expressed in cellularized embryos (C and E), and expression persists during gastrulation (F; data not shown). The Ady gene exhibits broad expression in ventral and ventrolateral regions (B). This pattern appears to include the entire presumptive mesoderm and extends into the ventral-most regions of the neurogenic ectoderm.
Discussion
An essential finding of this study is that the zen VRE and sog lateral stripe enhancer share a common code that could be identified accurately by using a genome-wide computational method. Only three regions in the entire genome contain four optimal Dl binding sites in intervals of 400 bp or less. One corresponds to the zen VRE, and evidence was presented that a second cluster corresponds to the sog lateral stripe enhancer. The VRE and sog enhancer are regulated by low levels of the Dl gradient that are insufficient to activate other target genes such as snail (12), sim (13), rhomboid (14), or Brinker (21). The third optimal Dl binding cluster is located between two divergently transcribed genes of unknown function. Preliminary in situ hybridization assays suggest that neither gene is regulated by the Dl gradient (see below).
There are 12 Dl binding clusters that contain exactly three optimal sites in 400 bp or less (see Table 1). One of these is located ≈10 kb 5′ of the Brinker transcription start site. Brinker probably is a direct target of the Dl gradient in that it exhibits lateral stripes of expression that are similar to those observed for rhomboid (21). The remaining 11 clusters generally are associated with genes that are not known to be involved in dorsoventral patterning. The expression patterns of two of these genes, Phm and Ady, were determined by in situ hybridization. Both genes exhibit expression in the ventral mesoderm of early embryos, suggesting that they might be activated by high levels of the Dl gradient. Moreover, the Ady pattern appears to include ventral regions of the neurogenic ectoderm. Thus, Ady expression might be activated by both high and intermediate levels of the Dl gradient. It seems that the computational genome-wide search for optimal Dl clusters successfully identified the full spectrum of Dl gradient thresholds including genes that are activated by high (Phm), intermediate (Ady), and low (sog) levels of the Dl gradient. Perhaps the exact sequence, arrangement, and density of the binding sites help to determine the response to these different thresholds. For example, the Dl binding sites in zen and sog appear to be helically phased, occurring every 60–80 bp. This phasing may lead to cooperative protein–protein interactions. The Phm gene might respond to only peak levels of the Dl gradient, because the binding sites exhibit a dispersed organization (see Fig. 2C). Of course, it is likely that these thresholds also depend on the binding of additional regulatory factors within the different target enhancers.
We can account for five of the 15 optimal Dl clusters in the Drosophila genome (sog, zen, Brinker, Phm, and Ady). These five genes seem directly regulated by the Dl gradient. It is unclear whether any of the remaining 10 clusters are associated also with Dl target genes. As discussed earlier, one of the very best clusters, containing four optimal sites within a span of 400 bp, is located near two genes that do not exhibit asymmetric patterns of expression across the dorsoventral axis. Moreover, another cluster is associated with the Runt gene, which is involved in segmentation and almost certainly is not a target of the Dl gradient (31, 32). However, Runt is a member of the AML family of transcription factors, which have been implicated in mammalian hematopoiesis (33). It is conceivable that the cluster of Dl binding sites located 3′ of the Runt transcription unit are recognized by another Rel-containing transcription factor in Drosophila, Dif or Relish, which mediates immune responses (34).
The identification of target genes and associated cis-regulatory elements based on the clustering of binding sites should be applicable generally to other systems, because clustering is a common feature of virtually all metazoan enhancers (35). For example, the eve stripe 2 enhancer contains five Bicoid activator sites within a 480-bp interval (27). Moreover, the mouse κ light chain enhancer contains clustered binding sites for two different classes of sequence-specific transcription factors, NF-κB and E12/E47 basic helix–loop–helix (bHLH) activators (36, 37). One way to improve the computational identification of cis-regulatory DNAs is to search for clustering of two or more different classes of recognition sequences. The rhomboid NEE was not identified because two of the four Dl sites possess low binding affinities and do not conform to the optimal consensus sequence used in this study. However, like the κ enhancer, the NEE contains clustered binding sites for Dl (which is related to NF-κB) as well as bHLH activators. The NEE also contains several snail repressor binding sites (14), thus different combinations of Dl, bHLH, and snail recognition sequences could be used in more sophisticated searches. Unfortunately, such searches depend on prior knowledge about a gene regulatory cascade, which generally is not available in most systems. This study indicates that a single transcription factor (and associated recognition sequences) is sufficient for the identification of at least a subset of cis-regulatory DNAs and target genes.
Acknowledgments
We thank Yutaka Nibu for helpful discussions, Ka-Ping Yee for creating a web interface for FLY ENHANCER, Hilary Ashe for the E2G transformation vector, and John Cowden for the NEE-lacZ stainings shown in Fig. 4. This work was supported by National Institutes of Health Grant GM46638.
Abbreviations
- NEE
neurogenic ectoderm enhancer
- VRE
ventral repression element
Footnotes
See commentary on page 546.
References
- 1.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, Fitzhaugh W, et al. Nature (London) 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Steward R. Science. 1987;238:692–694. doi: 10.1126/science.3118464. [DOI] [PubMed] [Google Scholar]
- 4.Ip Y T, Kraut R, Levine M, Rushlow C. Cell. 1991;64:439–446. doi: 10.1016/0092-8674(91)90651-e. [DOI] [PubMed] [Google Scholar]
- 5.Rushlow C, Han K, Manley J L, Levine M. Cell. 1989;59:1165–1177. doi: 10.1016/0092-8674(89)90772-1. [DOI] [PubMed] [Google Scholar]
- 6.Steward R. Cell. 1989;59:1179–1188. doi: 10.1016/0092-8674(89)90773-3. [DOI] [PubMed] [Google Scholar]
- 7.Roth S, Stein D, Nusslein-Volhard C. Cell. 1989;59:1189–1202. doi: 10.1016/0092-8674(89)90774-5. [DOI] [PubMed] [Google Scholar]
- 8.Huang A, Rusch J, Levine M. Genes Dev. 1997;11:1963–1973. doi: 10.1101/gad.11.15.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thisse C, Perrin-Schmitt F, Stoetzel C, Thisse B. Cell. 1991;65:1191–1201. doi: 10.1016/0092-8674(91)90014-p. [DOI] [PubMed] [Google Scholar]
- 10.Pan D J, Huang J D, Courey A J. Genes Dev. 1991;5:1892–1901. doi: 10.1101/gad.5.10.1892. [DOI] [PubMed] [Google Scholar]
- 11.Jiang J, Kosman D, Ip Y T, Levine M. Genes Dev. 1991;5:1881–1891. doi: 10.1101/gad.5.10.1881. [DOI] [PubMed] [Google Scholar]
- 12.Ip Y T, Park R, Kosman D, Yazdanbakhsh K, Levine M. Genes Dev. 1992;6:1518–1530. doi: 10.1101/gad.6.8.1518. [DOI] [PubMed] [Google Scholar]
- 13.Kasai Y, Stahl S, Crews S. Gene Expression. 1998;7:171–189. [PMC free article] [PubMed] [Google Scholar]
- 14.Ip Y T, Park R, Kosman D, Bier E, Levine M. Genes Dev. 1992;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- 15.Jiang J, Cai H, Zhou Q, Levine M. EMBO J. 1993;12:3201–3209. doi: 10.1002/j.1460-2075.1993.tb05989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirov N, Zhelnin L, Shah J, Rushlow C. EMBO J. 1993;12:3193–3199. doi: 10.1002/j.1460-2075.1993.tb05988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J D, Schwyter D H, Shirokawa J M, Courey A J. Genes Dev. 1993;7:694–704. doi: 10.1101/gad.7.4.694. [DOI] [PubMed] [Google Scholar]
- 18.Jiang J, Levine M. Cell. 1993;72:741–752. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- 19.Cai H N, Arnosti D N, Levine M. Proc Natl Acad Sci USA. 1996;93:9309–9314. doi: 10.1073/pnas.93.18.9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valentine S A, Chen G, Shandala T, Fernandez J, Mische S, Saint R, Courey A J. Mol Cell Biol. 1998;18:6584–6594. doi: 10.1128/mcb.18.11.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jazwinska A, Rushlow C, Roth S. Development (Cambridge, UK) 1999;126:3323–3334. doi: 10.1242/dev.126.15.3323. [DOI] [PubMed] [Google Scholar]
- 22.Francois V, Solloway M, O'Neill J W, Emery J, Bier E. Genes Dev. 1994;8:2602–2616. doi: 10.1101/gad.8.21.2602. [DOI] [PubMed] [Google Scholar]
- 23.Campbell G, Tomlinson A. Cell. 1999;96:553–562. doi: 10.1016/s0092-8674(00)80659-5. [DOI] [PubMed] [Google Scholar]
- 24.Jiang N, Kolhekar A S, Jacobs P S, Mains R E, Eipper B A, Taghert P H. Dev Biol. 2000;226:118–136. doi: 10.1006/dbio.2000.9832. [DOI] [PubMed] [Google Scholar]
- 25.Singh B, Lin A, Wu Z G, Gupta R S. DNA Cell Biol. 2001;20:53–65. doi: 10.1089/10445490150504693. [DOI] [PubMed] [Google Scholar]
- 26.Gloor G B, Preston C R, Johnson-Schiltz D M, Nassif N A, Phillis R W, Benz W K, Robertson H M, Engels W R. Genetics. 1993;135:81–95. doi: 10.1093/genetics/135.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Small S, Blair A, Levine M. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubin G, Spradling A. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 29.Adams MD, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 30.Doyle H, Harding K, Hoey T, Levine M. Nature (London) 1986;323:76–79. doi: 10.1038/323076a0. [DOI] [PubMed] [Google Scholar]
- 31.Klingler M, Soong J, Butler B, Gergen J P. Dev Biol. 1996;177:73–84. doi: 10.1006/dbio.1996.0146. [DOI] [PubMed] [Google Scholar]
- 32.Li L H, Gergen J P. Development (Cambridge, UK) 1999;126:3313–3322. doi: 10.1242/dev.126.15.3313. [DOI] [PubMed] [Google Scholar]
- 33.Tracey W D, Jr, Pepling M E, Horb M E, Thomsen G H, Gergen J P. Development (Cambridge, UK) 1998;125:1371–1380. doi: 10.1242/dev.125.8.1371. [DOI] [PubMed] [Google Scholar]
- 34.Imler J L, Hoffmann J A. Curr Opin Microbiol. 2000;3:16–22. doi: 10.1016/s1369-5274(99)00045-4. [DOI] [PubMed] [Google Scholar]
- 35.Davidson E H. Genomic Regulatory Systems. New York: Academic; 2000. [Google Scholar]
- 36.Picard D, Schaffner W. Nature (London) 1984;307:80–82. doi: 10.1038/307080a0. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh S, Gifford A M, Riviere L R, Tempst P, Nolan G P, Baltimore D. Cell. 1990;62:1019–1029. doi: 10.1016/0092-8674(90)90276-k. [DOI] [PubMed] [Google Scholar]