Abstract
Early embryonic cells in Caenorhabditis elegans embryos interact through a signaling pathway closely related to the Notch signaling pathway in Drosophila and vertebrates.Components of this pathway include a ligand, receptor, the presenilin proteins, and a novel protein, APH-2, that is related to the Nicastrin protein in humans. Here we identify the aph-1 gene as a new component of the Notch pathway in Caenorhabditis elegans. aph-1 is predicted to encode a novel, highly conserved multipass membrane protein. We show that aph-1 and the presenilin genes share a similar function in that they are both required for proper cell-surface localization of APH-2/Nicastrin.
The Notch signaling pathway mediates a variety of cell interactions that control cell fate specification during metazoan development. Components of the Notch signaling pathway are highly conserved among vertebrates and invertebrates (1–3). It is now clear that the presenilin proteins implicated in Alzheimer's disease in humans have a role in normal development in the Notch signaling pathway. In response to its ligand, Notch receptor undergoes a series of cleavage events culminating in a final presenilin-mediated intramembranous cleavage that releases the Notch intracellular domain (4). This fragment is then free to travel to the nucleus, where it influences transcriptional regulation (1–3). The role of presenilins in processing of the Notch receptor is related to their role in processing amyloid precursor protein, the protein that accumulates in Alzheimer's disease patients. Recent studies have shown that the human presenilins exist in a complex with a protein called Nicastrin (5). The Caenorhabditis elegans orthologue of Nicastrin, APH-2, is essential for Notch signaling during early embryogenesis (6); however, the exact role of APH-2/Nicastrin is not known.
The first requirements for the Notch signaling pathway in C. elegans development occur during the 4-cell and 12-cell stages of embryogenesis. Cell interactions at these stages are mediated by GLP-1, a receptor closely related to Drosophila Notch. The 12-cell interaction is required for the development of the anterior half of the pharynx, and embryos defective in this interaction have an Aph (anterior pharynx defective) phenotype (6). Although the posterior half of the pharynx normally contains many of the same differentiated cell types as the anterior half, it develops through a pathway that is independent of Notch signaling. Mutations in genes, such as pha-1 or pha-4, that control the differentiation of the various cell types within the pharynx affect the development of both the anterior and posterior halves of the pharynx (7, 8). In contrast, removal of Notch pathway components, such as GLP-1, LAG-1, APH-2, or the two presenilin proteins SEL-12 and HOP-1, causes an Aph phenotype (6, 9–12). GLP-1 and a second receptor related to Notch called LIN-12 function in multiple cell interactions during subsequent embryonic and postembryonic development (2, 3). These include GLP-1-mediated interactions that are required for germline proliferation and LIN-12-mediated interactions that are required for the development of the egg-laying system.
We have identified aph-1 as a gene that is involved in Notch signaling in the early C. elegans embryo, and show that closely related genes exist in Drosophila, mice, and humans. Mutations in aph-1 cause the embryonic Aph phenotype, indicative of defective Notch signaling at the 4- and 12-cell stages of embryogenesis. At these stages, the aph-1 mutant embryos show a marked defect in the ability to localize the Notch pathway component APH-2/Nicastrin. We show that inactivation of the presenilin genes causes a very similar mislocalization of APH-2/Nicastrin.
Materials and Methods
Strains and Genetic Analysis.
Bristol strain N2 was used as the standard wild-type strain, and culture and genetic analysis was as described by Brenner (13). The following genetic markers were used: unc-13(e1091), unc-29(e1072), aph-1(zu123), aph-1(zu147), aph-1(or28), lin-11(n566), sel-12(ty11) (14), and the balancer: hT2[bli-4(e937) let-?(h661)] (I;III). zu123 and zu147 were isolated as described (15). Hermaphrodites heterozygous for aph-1(zu123) and the chromosomal deficiency nDf25 are viable and lay dead embryos that resemble aph-1(zu123) embryos (100% inviable, n = 1,149 from six heterozygous mothers).
Analysis of Embryos.
Embryos were collected and prepared for microscopy as described (6). The following antisera/markers were used: pharyngeal cells, mAb 3NB12 (9), hypodermal cell boundaries, jam-1∷GFP fusion construct, jcIs1 (16), anti-APX-1 polyclonal serum (17), anti-GLP-1 affinity-purified polyclonal serum (kindly provided by S. Crittenden and J. Kimble; ref. 18), anti-APH-2 affinity-purified polyclonal serum (6). For quantification of embryo survival, individual homozygous mothers were allowed to lay 30–100 eggs, and the number of unhatched eggs was scored 15–25 h later. Egg laying and sterility were scored on individual L4 worms monitored daily for 4–5 days. Worms were scored as egg laying defective if eggs developed inside the parent instead of being laid, or scored as sterile if no eggs were produced. Sterile worms were found to contain sperm, a few undifferentiated germ cells, and very few or no mature oocytes. Anchor cells were scored in L3 worms by light microscopy.
Molecular Analysis of aph-1.
Transformation rescue was analyzed in unc-29 aph-1(zu123)/unc-13 lin-11 animals by using the transformation marker pRF4 (19). A 1.7 kb rescuing genomic fragment, generated by PCR from the rescuing yeast artificial chromosome Y62H1, contained 163 bp of upstream untranslated sequence and 248 bp of downstream untranslated sequence; this fragment rescued the embryonic lethality of aph-1(zu123) and aph-1(or28) as well as the egg-laying defect of aph-1(or28). The sequence of aph-1 mutants was obtained by amplifying the 1.7-kb genomic region directly from single aph-1 homozygous worms (Advantage high-fidelity PCR kit, CLONTECH) and obtaining double-stranded sequence for the complete 1.7-kb fragment. Sequencing was performed by the University of Massachusetts Automated DNA Sequencing Facility. Sequence differences between mutant and wild-type fragments were verified on independent PCR products. To obtain aph-1 cDNAs, total worm RNA was extracted from wild-type N2 worms by using Ultraspec (Biotecx Laboratories, Houston) and used with Moloney murine leukemia virus (M-MuLV) reverse transcriptase (New England Biolabs) and poly(T) oligomers to make cDNA. Primers specific for the C. elegans SL1 or SL2 trans-splicing leaders (20) or for the 5′ end of the predicted aph-1 gene were coupled with a primer specific for the predicted 3′ untranslated end of aph-1 and used to amplify aph-1 cDNA products (Advantage cDNA PCR kit, CLONTECH). More than 10 independent cDNA amplifications were carried out, and all products matched the expected splicing prediction for VF36H2L.1 with the addition of either SL1 or SL2 sequences at the 5′ end. Five of these products were fully sequenced. The 3′ end of the gene was also confirmed by the sequence reported for the independently isolated 3′ partial cDNA, CEESY69 (GenBank Accession number T02051). Searches of public database were performed by National Center for Biotechnology Information blast (http://www.ncbi.nlm.nih.gov/blast) and alignments were performed by using clustalw, courtesy of the Human Genome Sequencing Center at Baylor College of Medicine. Transmembrane regions were predicted according to Kyte and Doolittle (21) and according to Sonnhammer et al. (ref. 22, http://www.cbs.dtu.dk/services/TMHMM).
RNA Interference.
A complete aph-1 cDNA was cloned into pBluescript plasmid vector (Stratagene) and transcribed with the Ampliscribe kit (Epicentre Technologies, Madison, WI). Double-stranded RNA at a concentration of 300–500 ng/μl was injected into N2 worms at the L4 or young adult stage. Eight hours after injection the injected animals were placed on fresh plates for 12-h intervals, and their progeny were analyzed as described above.
Analysis of hop-1; sel-12 Double Mutants.
The hop-1; sel-12 double mutants were generated by injecting homozygous sel-12 L4-stage worms with double-stranded RNA transcribed as above from a hop-1 cDNA clone (11) (kindly provided by Iva Greenwald, Columbia University, New York). Injected worms were kept at 20°C; 18–20 h after injection, embryos were collected from parents and prepared for immunofluorescence as described above. Of all 2- to 12-cell stage embryos in which APH-2 could be detected, 71% showed perinuclear localization of APH-2 (n = 42 from >30 injected mothers). To assess the efficiency of hop-1 RNA-mediated interference (RNAi), embryos from five injected animals were allowed to develop instead of being processed for immunofluorescence; 78% became arrested in embryogenesis with Aph-like characteristics (n = 191).
Results
Isolation and Phenotypic Characterization of aph-1 Mutants.
We used a genetic screen described in ref. 6 to identify Aph mutants. In addition to several alleles of glp-1 and aph-2, we isolated two noncomplementing mutations that mapped to a position on chromosome I that did not correspond to the positions of any known component of the Notch pathway. We designate these mutations, zu123 and zu147, as alleles of a previously uncharacterized gene, aph-1. A third allele of aph-1, or28, was identified in an independent screen by Bruce Bowerman (personal communication). Each of the aph-1 alleles is a recessive, maternal-effect mutation. All (100%) of the embryos produced by homozygous zu123 and or28 mothers are inviable (n > 3000). The zu147 mutation appears to be slightly weaker, with 95.8% embryonic lethality (n = 3,394).
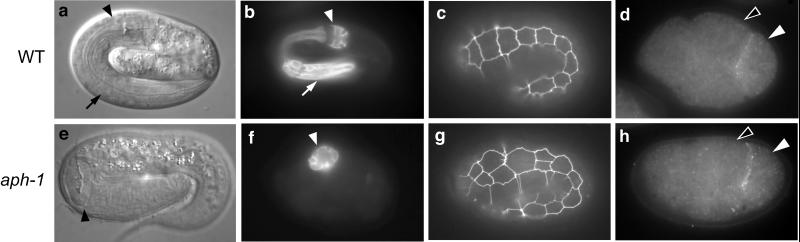
As expected for a Notch pathway component, aph-1 mutant embryos lack the anterior pharynx but can develop a relatively normal posterior pharynx (Fig. 1 e and f). The posterior pharynx can form a specialized cuticular structure called the grinder that normally differentiates late in embryogenesis, and the muscles in the posterior pharynx can contract. A second hallmark of a defect in the Notch-mediated 12-cell interaction is the development of supernumerary lateral skin cells; these extra cells are apparent in embryos homozygous for weak glp-1 and aph-2 alleles that disrupt primarily the 12-cell interaction (6, 15, 23, 24) (Fig. 1g; see also ref. 15). We observed a very similar pattern of ectopic skin cells in embryos from mothers homozygous for the weak allele aph-1(zu147) (Fig. 1g). Strong mutations in Notch pathway components prevent the 4-cell interaction in addition to preventing the 12-cell interaction (6, 15, 24, 25). In addition to the Aph phenotype, such mutations cause transformations in cell fate that prevent morphogenesis and cause skin cells to clump together on the dorsal surface (6, 15). We found that the strong alleles aph-1(zu123) and aph-1(or28) prevented body enclosure with the characteristic dorsal clump of skin cells (99.7%, n = 743 and 100%, n = 289 respectively). Although aph-1 mutants show defects associated with Notch signaling at the 4-cell stage, they do not show defects in concurrent interactions that are mediated by a separate, wingless Wnt-like signaling pathway. During the 4-cell stage of wild-type embryogenesis, the P2 blastomere provides separate signals for the Notch-mediated and Wnt-mediated interactions. Defects in the Wnt-mediated interactions can prevent the specification of intestinal cells (26); however, intestinal cells were present in nearly all aph-1 mutant embryos (zu123: 99.2%, n = 498; or28: 99.6%, n = 289; zu147: 99.1%, n = 340).
Figure 1.
aph-1 and wild-type embryos. (a) Living wild-type embryo viewed by light microscopy; the arrow indicates the anterior half of the pharynx and the arrowhead indicates the posterior half. (b) Immunostaining of pharyngeal cells; markers as in a. (c) Hypodermal cell boundaries delineated by green fluorescent protein expression; 10 cells are visible in the lateral row. (d) Four-cell embryo showing expression of the ligand APX-1. APX-1 is visible in the posterior-most cell (P2; white arrowhead) as a line contacting one of the GLP-1-expressing cells (ABp; black arrowhead); embryos oriented with the anterior to the left. (e–h) aph-1 mutant embryos staged, prepared, and labeled as above. Note the extra hypodermal cells at the first, second, and fourth positions in g. e and g are aph-1(zu147), and f and h are aph-1(zu123).
We addressed whether aph-1 participates in postembryonic cell interactions that are mediated by the Notch pathway. In wild-type larvae, the proliferation of germ-line cells requires GLP-1-mediated interactions with a somatic cell called the distal tip cell (27). We found that RNAi (28) of the aph-1 gene in wild-type adults caused most of the embryos to die with Aph terminal phenotypes (data not shown). However, the surviving embryos developed into adults that were sterile (94% sterility, n = 1,106). These adults appeared to have underproliferated germ lines; the gonads lacked oocytes, and sperm were present in the place of mitotic germ cells. Although we have not investigated the cellular defect responsible for sterility, these features are characteristic of adults lacking either glp-1 function (27) or the functions of the presenilins hop-1 and sel-12 (11, 12). Because homozygous aph-1 mutant adults that are produced by the self-fertilization of heterozygous hermaphrodites do not show a similar sterility, we propose that maternally provided aph-1(+) function is sufficient for germ-line proliferation. Development of the egg-laying system requires LIN-12 function to specify a cell called the anchor cell, and to mediate additional signaling events that occur after the anchor cell is specified (2, 29, 30). We found that the aph-1(or28) mutation caused a recessive, fully penetrant egg-laying defect (100%, n > 600). The aph-1(or28) larvae correctly specified a single anchor cell (n = 43). Thus aph-1(or28) appears to disrupt the development of the egg-laying system after the anchor cell is specified. Certain alleles of the Notch pathway genes lin-12, sel-12, and aph-2 also cause an egg-laying defect after anchor cell specification, and these phenotypes have been demonstrated to result from defects in uterine cell specification (6, 14, 29, 31, 32).
Identification of the aph-1 Gene Product.
The position of aph-1 on chromosome I was determined by mapping relative to visible markers and DNA polymorphisms (data available from the Caenorhabditis Genetics Center, University of Minnesota, St. Paul). Transformation rescue of aph-1 mutants was obtained with two overlapping C. elegans/yeast artificial chromosome DNAs, Y62H1 and YG434, and a 1.7-kb genomic fragment from the overlap region. This fragment contains a single predicted gene, VF36H2L.1, hereafter called aph-1 (GenBank accession no. AL021466). We cloned and sequenced five independent aph-1 cDNAs by reverse transcription PCR of mixed stage wild-type worm RNA and found their sequences to correspond exactly to that predicted for VF36H2L.1 by Gene Finder as part of the C. elegans Genome Project. The aph-1 cDNAs contain each of the four predicted exons and a C. elegans leader sequence, SL1 or SL2 (20), trans-spliced 2 bp before the start codon. The predicted initiator methionine codon is mutated to an isoleucine codon in the aph-1(zu123) allele.
The predicted APH-1 protein is not related to any known proteins. However, database searches identified DNA sequences in humans, mice, and Drosophila that could encode proteins closely related to APH-1 (Fig. 2a). Hydropathy plot analysis showed that APH-1 and related proteins are largely hydrophobic, with a common pattern of seven hydrophobic regions that are predicted to be membrane-spanning regions (Fig. 2b). None of the proteins contain predicted glycosylation sites, consistent with the idea that the majority of the protein is embedded within a cellular membrane. There are conserved, hydrophilic residues within the hydrophobic domains (Cys-9, Ser-45, Ser-50, Ser-130, His-183, Ser-213, His-214, Ser-257), suggesting that these residues may be critical for protein structure or interactions with other proteins. The aph-1(or28) mutation is predicted to replace a glycine residue with an aspartic acid residue within the fourth hydrophobic domain, and may therefore disrupt the topography of APH-1 in the membrane. The C. elegans APH-1 protein is unique in containing a hydrophilic 40-aa “tail” at the C terminus. The aph-1(zu147) mutation results in a premature stop codon near the beginning of this hydrophilic tail. Because aph-1(zu147) appears to be a hypomorphic allele that retains some aph-1(+) activity, the nonconserved C-terminal tail may not be essential for APH-1 function. We note that the only other multipass membrane proteins known to act in C. elegans Notch pathways are the presenilin proteins.
Figure 2.
(a) alignment of the predicted APH-1 protein of C. elegans with related proteins in Drosophila (33% identity, GenBank accession no. AAF51212) and humans (33% and 34% identity, GenBank accession nos. AAD34072 and AL136671, respectively). Each of the two human genes also has a highly related mouse gene, which is not shown here (GenBank accession nos. AC015932.1 and AK002310). Identical amino acids are black and conserved amino acids are gray; predicted transmembrane domains are overlined. Asterisks correspond to positions of aph-1 mutations as follows. zu123, Met to Ile; or28, Gly to Asp; zu147, Arg to opal stop codon. (b) APH-1 hydrophobicity plots (21); alignments as shown in a; predicted membrane-spanning regions are numbered above the plots.
APH-1 and Presenilins Are Required for Cell-Surface Localization of APH-2.
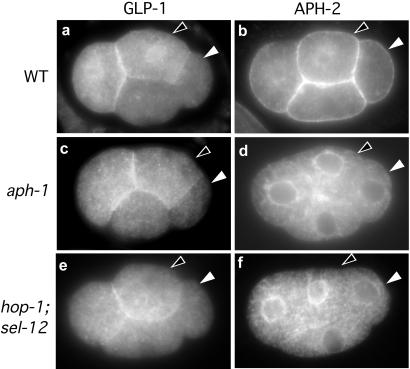
Because of its characteristics as a membrane protein, we asked whether the APH-1 protein might influence the expression or localization of cell-surface components of the Notch pathway in the early embryo. In wild-type 4-cell stage embryos, the ligand APX-1 is localized to the surface membrane of the posterior-most cell (Fig. 1d), where it contacts one of the two cells that express the receptor GLP-1 (17, 18) (Fig. 3a). The APH-2 protein is associated with the surface membranes of all four cells (Fig. 3b) (6). We found that APX-1 and GLP-1 had the wild-type localization pattern in aph-1 mutant embryos (Figs. 1h and 3c); however, the APH-2 protein was mislocalized in all aph-1 mutant embryos examined (n > 200 embryos between the 2-cell and 28-cell stages). In aph-1 mutant embryos, APH-2 was not detectable on the cell surface, and was instead prominent in the cytoplasm of all four cells, where it was concentrated around the nucleus in a pattern characteristic of the endoplasmic reticulum (Fig. 3d). These results suggest that APH-1 facilitates the translocation of APH-2 to the cell surface.
Figure 3.
GLP-1 and APH-2 localization in 4-cell stage embryos. The signaling cell (P2; white arrowhead) and responding cell (ABp; black arrowhead) are indicated in all panels. (a) Immunostaining of GLP-1 in wild type. GLP-1 is expressed on the surface of the anterior-most cell (Left) and its sister (black arrowhead). (b) Immunostaining of APH-2 in wild type. APH-2 is visible on the surface membranes of all four cells. (c and d) aph-1(zu123) embryos labeled as above. (e and f) Embryos deficient in presenilins [sel-12(ty11); hop-1(RNAi)] labeled as above. The level of APH-2 immunostaining is highest when cells are mid-interphase; in the images shown, the GLP-1-expressing cells appear to express a higher level of APH-2 because they are slightly more advanced in the cell cycle than the other two cells.
Presenilin proteins have been localized to the endoplasmic reticulum in some systems, and are essential for Notch signaling (4, 11, 12, 31). Because the human orthologue of APH-2, Nicastrin, has been shown to associate with presenilins (5), we asked whether presenilin function was required for APH-2 localization. Remarkably, presenilin-deficient embryos showed an abnormal accumulation of APH-2 around the nucleus in an apparently identical pattern to that of aph-1 mutant embryos (Fig. 3f); these embryos had the wild-type pattern of GLP-1 localization (Fig. 3e). These results suggest that the presenilins and APH-1 may function together in an event that is a prerequisite for APH-2 to localize to the cell surface.
Discussion
We have shown that the phenotypes of aph-1 mutant embryos are similar in all respects to the embryonic phenotypes associated previously with mutations in the C. elegans Notch pathway components. In addition, we have shown that APH-1 is required for the proper cell surface localization of the Notch pathway component APH-2/Nicastrin. We do not believe that APH-1 has a general role in protein secretion, because localization of the transmembrane receptor GLP-1 and transmembrane ligand APX-1 both appear normal in aph-1 mutant embryos. Moreover, interactions at the 4-cell stage that are mediated by secreted and transmembrane components of a separate, Wnt-like, signaling pathway appear to occur normally in aph-1 mutant embryos. We conclude from these several lines of evidence that the aph-1 gene is a component of the Notch signaling pathway in C. elegans. The observation that the Drosophila and human genomes contain predicted proteins that are closely related to APH-1 protein raises the possibility that these proteins have similar roles in Notch signaling.
Homozygous aph-1 mutant embryos produced from heterozygous parents do not exhibit at least some of the late embryonic defects described previously in mutants lacking both of the receptors encoded by glp-1 and lin-12 (the “Lag” phenotype; ref. 33). Although it is possible that certain Notch-mediated interactions have no, or different, requirements for aph-1 function, a simple explanation for this difference is that maternally contributed aph-1 is sufficient to mediate late embryonic cell-fate decisions. For example, maternal hop-1 activity appears to prevent hop-1; sel-12 double mutants from displaying the Lag phenotype (12). We have shown that maternally provided aph-1 appears to play a role in postembryonic germ-line proliferation, and previous studies have demonstrated that maternally provided sel-12 can rescue the germ-line proliferation defects of hop-1; sel-12 double mutants (12).
Human presenilins have been found to exist in high molecular weight complexes that include the APH-2/Nicastrin protein. Presenilins appear to function at the cell surface in the intramembranous cleavage of Notch in response to ligand stimulation, and recent studies suggest that APH-2/Nicastrin may directly influence this cleavage event (5). We have shown that embryos lacking presenilin function and aph-1 mutant embryos both show a similar defect in APH-2/Nicastrin localization. The APH-2 localization defect is observed in all cells of the 4-cell stage embryo, including the anterior-most cell (ABa), which does not contact the ligand-expressing cell (P2). Thus, cells have at least one requirement for aph-1 and presenilin function that is independent of ligand-stimulated Notch processing. The observation that presenilins have a role in APH-2/Nicastrin localization raises the question of whether a similar requirement for presenilin function contributes to Alzheimer's disease in humans.
Future studies are needed to clarify whether cells have additional requirements for aph-1 upon ligand stimulation of Notch. For example, formation of an APH-1–APH-2–presenilin complex could be a prerequisite for transport of these proteins to the cell surface, where the tripartite complex subsequently mediates the intramembranous cleavage of ligand-activated Notch. Thus, APH-1 may prove to be a functional component of the large presenilin complex presumed to mediate intramembranous cleavage events. Alternatively, it is possible that APH-1 function is limited to the endoplasmic reticulum, where it promotes the assembly or modification of an APH-2–presenilin complex. It will be interesting in future studies to determine whether the human APH-1 orthologues have an essential role in the presenilin-mediated processing of amyloid precursor protein, the process that is defective in patients with Alzheimer's disease, in addition to potential roles in Notch signaling.
Acknowledgments
We are grateful to Bruce Bowerman, Sarah Crittenden, Judith Kimble, Anna Newman, and Iva Greenwald for valuable reagents; to Katie Mickey, David Wynne, Craig Mello, and Peter Pryciak for assistance and suggestions; and to the C. elegans Genetics Stock Center and the Genetics Tool Kit for supplying nematode strains. J.R.P. is an Investigator of the Howard Hughes Medical Institute. This work was supported by grants from the National Institutes of Health (to J.R.P.) and the National Science Foundation (to C.G.).
Abbreviation
- RNAi
RNA-mediated interference
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Artavanis-Tsakonas S, Rand M D, Lake R J. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Greenwald I. Genes Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 3.Kimble J, Simpson P. Annu Rev Cell Dev Biol. 1997;13:333–361. doi: 10.1146/annurev.cellbio.13.1.333. [DOI] [PubMed] [Google Scholar]
- 4.Kopan R, Goate A. Genes Dev. 2000;14:2799–2806. doi: 10.1101/gad.836900. [DOI] [PubMed] [Google Scholar]
- 5.Yu G, Nishimura M, Arawaka S, Levitan D, Zhang L, Tandon A, Song Y Q, Rogaeva E, Chen F, Kawarai T, et al. Nature (London) 2000;407:48–54. doi: 10.1038/35024009. [DOI] [PubMed] [Google Scholar]
- 6.Goutte C, Hepler W, Mickey K M, Priess J R. Development (Cambridge, UK) 2000;127:2481–2492. doi: 10.1242/dev.127.11.2481. [DOI] [PubMed] [Google Scholar]
- 7.Mango S E, Lambie E J, Kimble J. Development (Cambridge, UK) 1994;120:3019–3031. doi: 10.1242/dev.120.10.3019. [DOI] [PubMed] [Google Scholar]
- 8.Schnabel H, Schnabel R. Science. 1990;250:686–688. doi: 10.1126/science.250.4981.686. [DOI] [PubMed] [Google Scholar]
- 9.Priess J R, Thomson J N. Cell. 1987;48:241–250. doi: 10.1016/0092-8674(87)90427-2. [DOI] [PubMed] [Google Scholar]
- 10.Christensen S, Kodoyianni V, Bosenberg M, Friedman L, Kimble J. Development (Cambridge, UK) 1996;122:1373–1383. doi: 10.1242/dev.122.5.1373. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Greenwald I. Proc Natl Acad Sci USA. 1997;94:12204–12209. doi: 10.1073/pnas.94.22.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westlund B, Parry D, Clover R, Basson M, Johnson C D. Proc Natl Acad Sci USA. 1999;96:2497–2502. doi: 10.1073/pnas.96.5.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cinar H N, Sweet K L, Hosemann K E, Earley K, Newman A P. Dev Biol. 2001;237:173–182. doi: 10.1006/dbio.2001.0374. [DOI] [PubMed] [Google Scholar]
- 15.Priess J R, Schnabel H, Schnabel R. Cell. 1987;51:601–611. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- 16.Mohler W A, Simske J S, Williams-Masson E M, Hardin J D, White J G. Curr Biol. 1998;8:1087–1090. doi: 10.1016/s0960-9822(98)70447-6. [DOI] [PubMed] [Google Scholar]
- 17.Mickey K M, Mello C C, Montgomery M K, Fire A, Priess J R. Development (Cambridge, UK) 1996;122:1791–1798. doi: 10.1242/dev.122.6.1791. [DOI] [PubMed] [Google Scholar]
- 18.Evans T C, Crittenden S L, Kodoyianni V, Kimble J. Cell. 1994;77:183–194. doi: 10.1016/0092-8674(94)90311-5. [DOI] [PubMed] [Google Scholar]
- 19.Mello C C, Kramer J M, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spieth J, Brooke G, Kuersten S, Lea K, Blumenthal T. Cell. 1993;73:521–532. doi: 10.1016/0092-8674(93)90139-h. [DOI] [PubMed] [Google Scholar]
- 21.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 22.Sonnhammer E L, von Heijne G, Krogh A. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 23.Mello C C, Draper B W, Priess J R. Cell. 1994;77:95–106. doi: 10.1016/0092-8674(94)90238-0. [DOI] [PubMed] [Google Scholar]
- 24.Hutter H, Schnabel R. Development (Cambridge, UK) 1994;120:2051–2064. doi: 10.1242/dev.120.7.2051. [DOI] [PubMed] [Google Scholar]
- 25.Moskowitz I P, Gendreau S B, Rothman J H. Development (Cambridge, UK) 1994;120:3325–3338. doi: 10.1242/dev.120.11.3325. [DOI] [PubMed] [Google Scholar]
- 26.Thorpe C J, Schlesinger A, Bowerman B. Trends Cell Biol. 2000;10:10–17. doi: 10.1016/s0962-8924(99)01672-4. [DOI] [PubMed] [Google Scholar]
- 27.Austin J, Kimble J. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- 28.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 29.Newman A P, White J G, Sternberg P W. Development (Cambridge, UK) 1995;121:263–271. doi: 10.1242/dev.121.2.263. [DOI] [PubMed] [Google Scholar]
- 30.Sundaram M, Greenwald I. Genetics. 1993;135:755–763. doi: 10.1093/genetics/135.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levitan D, Greenwald I. Nature (London) 1995;377:351–354. doi: 10.1038/377351a0. [DOI] [PubMed] [Google Scholar]
- 32.Levitan G Y, St. George Hyslop P, Goutte C. Dev Biol. 2001;240:654–661. doi: 10.1006/dbio.2001.0486. [DOI] [PubMed] [Google Scholar]
- 33.Lambie E J, Kimble J. Development (Cambridge, UK) 1991;112:231–240. doi: 10.1242/dev.112.1.231. [DOI] [PubMed] [Google Scholar]