Abstract
Xeroderma pigmentosum variant (XP-V) cells are deficient in their ability to synthesize intact daughter DNA strands after UV irradiation. This deficiency results from mutations in the gene encoding DNA polymerase η, which is required for effecting translesion synthesis (TLS) past UV photoproducts. We have developed a simple cellular procedure to identify XP-V cell strains, and have subsequently analyzed the mutations in 21 patients with XP-V. The 16 mutations that we have identified fall into three categories. Many of them result in severe truncations of the protein and are effectively null alleles. However, we have also identified five missense mutations located in the conserved catalytic domain of the protein. Extracts of cells falling into these two categories are defective in the ability to carry out TLS past sites of DNA damage. Three mutations cause truncations at the C terminus such that the catalytic domains are intact, and extracts from these cells are able to carry out TLS. From our previous work, however, we anticipate that protein in these cells will not be localized in the nucleus nor will it be relocalized into replication foci during DNA replication. The spectrum of both missense and truncating mutations is markedly skewed toward the N-terminal half of the protein. Two of the missense mutations are predicted to affect the interaction with DNA, the others are likely to disrupt the three-dimensional structure of the protein. There is a wide variability in clinical features among patients, which is not obviously related to the site or type of mutation.
Xeroderma pigmentosum (XP) is a rare autosomal recessive disorder characterized by extreme sensitivity of the skin to sunlight-induced changes (1). In particular, affected individuals have a 1,000-fold increase in the incidence of sunlight-induced skin cancers. Genetically, XP is complex and in the majority of patients the clinical features result from a defect in one of seven genes, XPA–G, controlling nucleotide excision repair (NER). The products of these XP genes are involved in one or other of the steps in the NER process (2, 3). Approximately 20% of patients with XP are so-called XP variants (XP-V), which are defined by their normal levels of NER. Cultured fibroblasts from XP-V donors are only mildly sensitive to the killing effects of UV light, in contrast to the NER-defective XP cells, which have very marked hypersensitivity. Both NER-defective XP and XP-V cells are hypersensitive to the mutagenic effects of UV irradiation (4–6).
The defect in XP-V cells is a reduced ability to replicate DNA after irradiation (also referred to as postreplication repair). XP-V cells are unable to synthesize intact daughter DNA strands on UV-irradiated templates (7). In vitro studies showed that this deficiency resulted from an inability to carry out translesion synthesis (TLS), the synthesis of DNA directly past damaged sites (8). These and other in vitro studies led to the cloning of the gene deficient in patients with XP-V (9, 10), and the demonstration that the gene product is DNA polymerase η (polη). Polη is a member of a newly discovered family of DNA polymerases, designated the polymerase Y family, which are related in sequence to each other but not to other DNA polymerase families (11). The polymerase Y family has in common low fidelity using undamaged templates, poor processivity, and the ability to carry out TLS past different types of DNA lesions (12). Thus, polη can replicate efficiently past cyclobutane pyrimidine dimers, the most common lesion generated in DNA by UV, and in most cases is able to insert the “correct” bases opposite this lesion (13). It is also able to carry out TLS past other lesions such as acetylaminofluorene (AAF) adducts (13). A recent study has implicated polη in somatic hypermutation of Ig variable genes (14), findings that are in accord with the mutational specificity of polη (15).
The N-terminal 350 amino acids of polη are highly conserved in the polymerase Y family and form the active site (see Discussion). We have shown previously that the C-terminal 120 amino acids of polη are required for nuclear localization and for accumulation into replication foci after irradiation (16). The function of the central 240 amino acids is not yet known.
The two groups who cloned the polη gene identified mutations in this gene in a few patients (9, 10). In the majority of those examined, the mutations resulted in severely truncated proteins, proving that polη was indeed the XP-V gene product. These mutations were, however, of limited use for understanding structure–function relationships in polη. In the current work, we have identified mutations in 21 donors with XP-V, from several different European and North African countries. We also correlate the sites of the mutations with the ability of cell extracts to carry out TLS.
Materials and Methods
Cells.
The XP-V primary fibroblast cell strains that we have analyzed are listed in Table 2. We have also used the cell strains 1BR3 (normal), XP1BR (XP-D), and the immortalized XP-V cell strain CTAg (17) for the TLS studies.
Table 2.
Details of patients with XP-V, cell strains, and mutations
| Cell strain | Age of patient | Country of origin | Skin tumors* | Mutation | Amino acid change† | Location | TLS‡ |
|---|---|---|---|---|---|---|---|
| XP51VI | 22 | Algeria | ++ | Del exon 5 | Fs 163 (hom) | Exon 5 | — |
| XP56RO | 25 | Belgium | − | G890A | Trp-297stop | Exon 8 | |
| Del exon 5 | Fs163 | Exon 5 | |||||
| XP53RO | 35 | France | + | C1066T | Arg-355stop | Exon 9 | |
| Del exon 5; | Fs163 | Exon 5 | |||||
| +T 882 | Fs 294 | Exon 7 | |||||
| XP52RO | 56 | Belgium | ++ | C1066T | Arg-355stop (hom) | Exon 9 | |
| XP62VI | 7 | Algeria | ++ | Del exon 10 | Fs 358 (hom) | Exon 10 | — |
| XP75VI | 17 | Algeria | ++ | Del exon 10 | Fs 358 (hom) | Exon 10 | — |
| XP28VI | 26 | Tunisia | ++ | Del exon 10 | Fs 358 | Exon 10 | — |
| +C 1091 | Fs 364 | Exon 10 | |||||
| XP127VI | 49 | France | +++ | +C 1091 | Fs364 (hom) | Exon 10 | — |
| XP7DU | Scotland | ND | Del 1222–1225 | Fs 407 | Exon 10 | — | |
| ? | ? | ||||||
| XP58RO | 30 | Austria | +++ | Del 224–226 | −Leu-75 (hom) | Exon 3 | |
| XP2DU | 44 | Scotland | ND | Del 224–226 | −Leu-75 | Exon 3 | |
| −G 207 | Fs 69 | Exon 3 | |||||
| XP3DU | 60 | Scotland | ND | Del 224–226 | −Leu-75 | Exon 3 | — |
| −G 207 | Fs 69 | Exon 3 | |||||
| XP6DU | Scotland | +++ | Del 224–226 | −Leu-75 | Exon 3 | — | |
| −G 207 | Fs 69 | Exon 3 | |||||
| XP7BR | 60 | England | ND | Del 224–226 | −Leu-75 | Exon 3 | |
| Ins 764 | Fs 255 | Exon 5 | |||||
| XP36BR | 48 | Scotland | +++ | G332A | Arg-111-His | Exon 4 | |
| Del 1222–1225 | Fs 407 | Exon 10 | |||||
| XP5BI | 7 | England | − | A364C | Thr-122-Pro | Exon 4 | |
| Del 1222–1225 | Fs 407 | Exon 10 | |||||
| XP11BR | 66 | England | +++ | G788T | Gly-263-Val (hom) | Exon 6 | — |
| XP57RO | 57 | Netherlands | + | G1083T | Arg-361-Ser (hom) | Exon 10 | |
| XP86VI | 32 | Algeria | + | C1561T | Gln-521stop (hom) | Exon 11 | + |
| XP1AB | 51 | Scotland | +++ | C1543A | Thr-548stop | Exon 11 | + |
| Del 224–226 | −Leu-75 | Exon 3 | |||||
| XP37BR | 69 | Scotland | + | +C 1668 | Fs 556 (hom) | Exon 11 | + |
−, No tumors; +, less than 10 tumors; ++, 10–50 tumors; +++, more than 50 tumors; ND, no data.
hom, homozygous.
−, defective; +, normal.
Cellular Test for XP-V.
Fibroblasts (1–5 × 104) were plated onto 5-cm dishes and incubated for 2–3 days. The cells were then irradiated with graded doses of UVC light and incubated in medium with or without 1.5 mM caffeine for 2–4 days further. The medium was then replaced with fresh medium, again containing 1.5 mM caffeine, and also 10μCi/ml 3H thymidine (50–80 Ci/mmol), and incubated for 3 h. The cells were scraped off the dishes into 2% SDS or lysed directly in 0.25 M NaOH, and the incorporated radioactivity was measured in a liquid scintillation counter.
Translesion Synthesis.
This assay was carried out essentially as described in Cordonnier et al. (8). In brief, single-stranded plasmids were constructed containing a single AAF adduct at either the first or third G residue in a run of three Gs. 32P end-labeled primers were designed such that they annealed either 91 or 1 nucleotide upstream of the lesion. The annealed template primer was used as substrate in a primer extension assay, with human cell extracts as the source of polymerizing activity. Replication products were digested with EcoRI and PvuII and analyzed on polyacrylamide-urea denaturing gels.
Mutation Analysis.
RNA was extracted from fibroblast cultures using Trizol reagent (Life Technologies, Paisley, U.K.). Total RNA was reverse-transcribed with a first-strand DNA synthesis kit (Amersham Pharmacia), and the polη cDNA was amplified by PCR in one piece, using the primers GATCCCTTCTCGGTTTCTCC (nucleotides −180 to −161) and TCCATGCCTGTGAAGAGATG (nucleotides 2313–2294). The 2.5-kb PCR product was sequenced by using the ThermoSequenase (Amersham Pharmacia) radiolabeled terminator cycle sequencing kit and 33P-labeled dideoxynucleotides.
Results
Identification of XP-V Cell Strains.
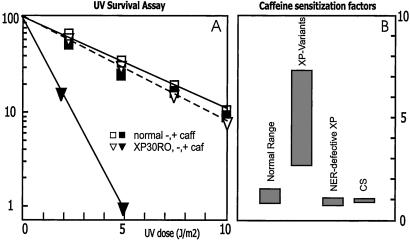
Classically, XP-V cells have been defined by their reduced ability to synthesize high molecular weight daughter-strand DNA after UV irradiation (7, 18). This deficiency is exacerbated by caffeine, which results in a very clear distinction between XP-V cells (daughter strands remain small) and normal cells (daughter strands attain high molecular weight). This analytical test is laborious and does not lend itself to routine screening. The caffeine sensitivity of daughter-strand repair in XP-V cells renders these cells sensitive to killing by UV in the presence of caffeine, whereas in the absence of caffeine this sensitivity is marginal (19). Itoh et al. (20, 21) exploited this UV + caffeine sensitivity and devised a test for measuring the postirradiation recovery of DNA synthesis in the presence of caffeine to distinguish between XP-V and normal cells. We have modified the procedure further by exposing the culture to different doses of UV, incubating in the presence of caffeine and measuring the viability of the culture 2–4 days later. Viability is assessed by the ability of the cells to incorporate [3H]thymidine into DNA, as measured by liquid scintillation counting. Results of a typical experiment are shown in Fig. 1A. The known XP-V strain XP30RO, only marginally sensitive to UV, is sensitized to UV by a factor of 4.5 in the presence of 1.5 mM caffeine, whereas normal cells are hardly affected by the drug. Similar responses were observed in four other known XP variants, showing sensitization factors ranging from 4 to 7 (Table 1). This response is specific to XP-variants, because neither NER-defective XP cell strains nor Cockayne Syndrome cells are sensitized to UV by caffeine (see Table 1 and Fig. 1B). Significantly, patients in the XP-E complementation group with a relatively mild clinical appearance resembling XP variants and a high residual NER activity, which may fall in near-normal ranges in semiquantitative repair assays, can be readily discriminated from XP variants in this assay. Many of the patients with XP-V discussed here were identified by using this simplified assay or slight modifications of it (for examples see Table 1).
Figure 1.
Sensitization to UV by caffeine. (A) Typical thymidine incorporation assay on known XP-V strain (triangles) compared with normal fibroblasts (squares), in absence (open symbols) or presence (closed symbols) of 1.5 mM caffeine. (B) Ranges of caffeine-sensitization factors, calculated from survival curves as indicated in Table 1, specified for various categories of UV-sensitive cell strains.
Table 1.
Sensitization to UV by caffeine
| Fibroblasts | UV-sensitivity factors*
|
Caffeine sensitization† | |
|---|---|---|---|
| − Caffeine | + Caffeine | ||
| Known XP-variants | |||
| XP1RO XP30RO XP37RO XP7TA XP4BE | 1.1–1.5 | 5.1–10.2 | 3.92–7.21 |
| New XP-variants | |||
| XP52RO | 1.5 | 7.5 | 5.0 |
| XP56RO | 1.6 | 6.3 | 4.1 |
| XP53RO | 2.0 | 7.1 | 3.9 |
| XP57RO | 1.4 | 6.9 | 4.9 |
| XP58RO | 1.7 | 6.0 | 3.5 |
| XP59RO | 1.5 | 4.1 | 2.7 |
| Non-XP-V controls | |||
| XP-A: XP25RO | 9.5 | 9.7 | 1.02 |
| XP-B: XPCS1BA | 4.7 | 4.8 | 1.01 |
| XP-C: XP21RO | 5.2 | 5.0 | 0.96 |
| XP-D: n = 3 | 3.9–6.0 | 2.9–5.8 | 0.74–0.91 |
| XP-E: XP2RO | 1.8 | 1.9 | 1.06 |
| XP-F: XP42RO | 2.4 | 2.6 | 1.08 |
| XP-G: n = 3 | 7.7–9.4 | 6.8–9.5 | 0.88–1.03 |
| CS-A: CS3BE | 2.6 | 2.7 | 1.03 |
| CS-B: n = 2 | 2.3–3.8 | 2.3–3.8 | 1.00 |
| Normals: n = 9 | — | — | 0.87–1.32 |
Ratio–D10 of normal control to D10 of XP cell strains.
Ratio–UV-sensitivity factor + caffeine to UV-sensitivity factor − caffeine.
Mutations in the polη Gene.
The 713-aa polη protein can be roughly divided into three regions. The N-terminal 350 amino acids are highly conserved in the polymerase Y family and form the active site (see Discussion). We have shown previously that the C-terminal 120 amino acids are required for nuclear localization and for accumulation into replication foci after irradiation (16). The function of the central 240 amino acids is not yet known.
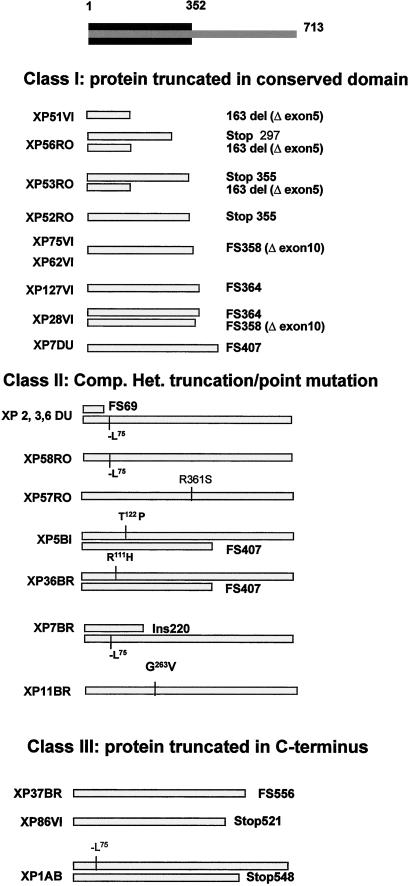
We have analyzed the mutations in polη cDNA from XP-V fibroblasts, using reverse transcription–PCR and direct sequencing. The results are presented in Fig. 2 and Table 2. The mutations can be divided into three categories:
- 1.
Both alleles result in a truncated protein of less than 410 amino acids (severe truncation).
- 2.
At least one allele is a missense mutation.
- 3.
At least one allele results in a truncated protein of more than 520 amino acids (C-terminal truncation).
Figure 2.
Three classes of polη mutations in patients with XP-V. The conserved N-terminal region of the polη protein is indicated in black at the top of the diagram. The gray bars indicate the mutated protein. Two bars are shown for compound heterozygotes, one for homozygotes.
Severe Truncating Mutations.
Nine patients had mutations resulting in severe truncations in both alleles (Fig. 2 Top, Table 2). The mutations in these patients were comprised of one stop codon (homozygous in XP52RO, heterozygous in XP53RO), one +C insertion (homozygous in XP127VI, heterozygous in XP28VI), one +T insertion (heterozygous in XP53RO), and two exon deletions (del exon 5, homozygous in XP51VI, heterozygous in XP56RO and XP53RO; del exon 10, homozygous in XP62VI and XP75VI; heterozygous in XP28VI). In XP51VI, only half of the conserved domains remain. XP53RO had three mutations in the XPV cDNA (stop codon at amino acid 355, +T insertion at nucleotide 882, del exon 5). Because the +T insertion and exon 5 deletion are expressed at lower levels relative to the nonsense mutation, it is likely that they are both on the same allele, and the nonsense mutation is on the other allele.
Three different mutations found in six cell strains curiously all result in truncations within 10 amino acids of each other between amino acids 355 and 364, at the end of the conserved region. Finally, a small deletion resulted in a frame shift at amino acid 407 in three cell strains. XP7DU was heterozygous for this mutation, but we were unable to identify the mutation in the second allele, which was expressed at substantially lower levels.
Missense Mutations.
Seven families had five different missense mutations, as indicated in Fig. 2 Middle and Table 2. An in-frame deletion resulting in loss of Leu-75 was found in one allele of XP6DU, as reported by Johnson et al. (10). This mutation was also found in her two siblings, XP2DU and XP3DU, in two unrelated patients, XP7BR and XP1AB, and in both alleles of XP58RO. Arg-111 was mutated in XP36BR, Thr-122 in one allele of XP5BI, Gly-263 in XP11BR, and Arg-361 in XP57RO.
C-Terminal Truncations.
Mutations in three patients resulted in truncations in the C-terminal region of the protein between amino acids 521 and 556 (Fig. 2 Bottom). XP86VI was homozygous for a nonsense mutation at amino acid 521, XP37BR was homozygous for a +C insertion at amino acid 556, and XP1AB was compound heterozygous for a nonsense mutation at amino acid 548, together with the −Leu-75 mutation in the second allele.
Translesion Synthesis in XP-V Extracts.
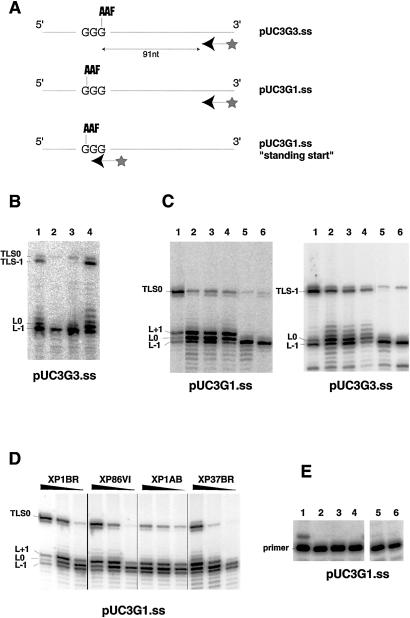
To determine how the different types of mutation affect the ability of polη to carry out TLS, we have measured TLS activity of cell-free extracts by using a single-stranded plasmid with a single AAF adduct at a defined position in the plasmid (8). The ability of the cell-free extracts to extend a primer past this adduct was analyzed. We showed previously that normal cells were able to bypass the lesion, but extracts of five XP-V cell strains, including four analyzed in this study, were blocked at the lesion (8). Here, similar experiments were carried out with a selection of other XP-V extracts, and the results are shown in Fig. 3. In the first set of experiments, the labeled primer was 91 nucleotides away from an AAF lesion located on the most 3′ G residue of a GGG triplet (pUC3G3) (Fig. 3A). With normal cell extracts, there were four major products, resulting from blocking of the polymerase one base before (L-1) or at the lesion (L0), or TLS with (TLS-1) or without (TLS0) one-base slippage. TLS-1 is the major TLS product using normal extracts, but was barely detectable using XP-V extracts in our previous studies (8). In Fig. 3B, XP1BR, an NER-deficient XP cell strain mutated in the XPD gene, is able to form substantial amounts of TLS product (lane 1), similar to amounts detected in wild-type cells (8). The XP-V cell strains, XP28VI and XP127VI, with severe truncating mutations, were blocked at the AAF lesion (lanes 2 and 3), like the XP-V extracts previously examined. In contrast, in extracts of XP86VI, with a truncating mutation close to the C terminus (lane 4), TLS was only slightly reduced when compared with normal extracts. In Fig. 3C, we have used both the pUC3G3 template and pUC3G1, in which the lesion is on the first G of the triplet (see Fig. 3A). We found that not only XP86VI (lanes 4) but also extracts of the other two XP-V cell strains with truncations close to the C terminus, XP37BR (lanes 2) and XP1AB (lanes 3), were able to carry out TLS, in contrast to XP11BR (lanes 5) and CTAg [derived from XP4BE (17)] (lanes 6), which were defective. Fig. 3D shows that TLS activity in XP86VI, XP1AB, and XP37BR and in XP1BR group D extracts was concentration-dependent, with the XP-V cell extracts having somewhat lower activities. Finally, a “standing start” primer, which terminates immediately before the lesion with the pUC3G1 substrate (see Fig. 3A) was not elongated in strains XP75VI (Fig. 3E, lane 2), XP62VI (lane 3), XP51VI (lane 4), XP127VI (lane 5), or XP28VI (lane 6), all of which had severely truncating mutations. In contrast, it was extended in XP86VI (lane 1). We conclude that XP28VI, XP51VI, XP62VI, XP75VI, and XP127VI with severely truncating mutations have defective TLS activity. Three cell strains that have missense mutations, XP3DU, XP6DU, and XP11BR, were shown to be defective in TLS in our previous work (8). In marked contrast, TLS was easily detectable in extracts of XP86VI, XP1AB, and XP37BR, in which the mutations were all close to the C terminus. Quantitation of the levels of TLS by phosphorimagery showed significant variation among extracts. TLS activity using extracts of XP86VI, XP1AB, and XP37BR varied from 10 to 50% of that using normal extracts and was 2–3-fold higher with the pUC3G3 than with the pUC3G1 substrate. TLS in extracts from cells with null mutations was less than 2% of that in normal extracts.
Figure 3.
TLS in XP-V extracts. (A) Different template-primer substrates. (B) Cell extracts were incubated with the pUC3G3.ss template and a 32P end-labeled primer, which annealed 91 nucleotides from the AAF lesion. The products of the reactions were electrophoresed on a sequencing gel. Lane 1, XP1BR; lane 2, XP28VI; lane 3, XP127VI; lane 4, XP86VI. (C) TLS was compared in normal and XP-V extracts, using the pUC3G3.ss and pUC3G1.ss templates. Lane 1, 1BR3; lane 2, XP37BR; lane 3, XP1AB; lane 4, XP86VI; lane 5, XP11BR; lane 6, CTAg. (D) Effect of concentration of various extracts on TLS by using the pUC3G1.ss template. For each strain the amounts of proteins used were 36, 24, and 12 μg. (E) TLS activity of various XP-V extracts using the pUC3G1.ss template and a primer, which anneals immediately upstream of the AAF lesion. Lane 1, XP86VI; lane 2, XP75VI; lane 3, XP62VI; lane 4, XP51VI; lane 5, XP127VI; lane 6, XP28VI.
Discussion
We have identified the inactivating mutations in the polη gene in 21 patients with XP-V. The results are summarized in Fig. 2 and Table 2. A variety of different types and positions of mutations results in the XP-V phenotype. Of the 17 inactivating mutations identified, there were 4 transitions, 4 transversions, 1 in-frame deletion, 5 frame shifts, 1 large insertion, and 2 exon deletions (at the cDNA level). The details of the frame shifts and truncating insertions/deletions are presented in Table 3.
Table 3.
Frame shifts, insertions, and deletions
| Cell strain | Status | Mutation | First mutated codon | Sequence | Size of truncated protein |
|---|---|---|---|---|---|
| XP2DU, XP3DU, XP6DU | Heterozygous | −G 207 | 70 | TAAGAAG | 98 |
| XP51VI | Homozygous | −661–764 | 164 | Exon 5 | 199 |
| XP56RO, XP53RO | Heterozygous | ||||
| XP7BR | Heterozygous | Large insert | 256 | ||
| XP53RO | Heterozygous | +T 882 | 295 | GAATGGG | 305 |
| XP62VI, XP75VI | Homozygous | −1075–1244 | 359 | Exon 10 | 389 |
| XP28VI | Heterozygous | ||||
| XP127VI, XP28VI | Heterozygous | +C 1091 | 365 | CACCCAG | 391 |
| XP7DU, XP5BI, XP36BR | Heterozygous | −ACTT 1222–1225 | 408 | TAATACTTCTGG | 442 |
| XP37BR | Homozygous | +C 1668 | 557 | AACCCAT | 560 |
Of the 16 mutations, 8 resulted in severely truncated products, because of either stop codons, frame shifts, or exon deletions. In several patients, as in those analyzed by others (9, 10), both alleles were affected in this way, and polη in these individuals is unlikely to have any functional ability. This finding confirms that polη is not an essential gene.
Three cell strains have truncations between amino acids 512 and 556, and TLS activity was detectable at a reduced level in extracts from these cells. These data obtained with cell extracts are consistent with findings on purified polη, which had full activity despite containing only the N-terminal 511 amino acids (22). The reduction in activity in extracts of the cells that have C-terminal truncations may result from a deficiency in interactions of the truncated polη with other proteins in the extracts that are important for maximal activity. For example, the extreme C terminus of polη interacts with proliferating cell nuclear antigen (16, 23), and this interaction stimulates the activity of polη in vitro (23).
The polymerase activity of purified polη from Saccharomyces cerevisiae and its ability to bypass cyclobutane pyrimidine dimers unaffected by C-terminal truncations that leave the N-terminal 513 amino acids intact (24). Removal of a further 60 amino acids abolishes these activities. If these results can be extrapolated to human polη, alignment with the yeast protein suggests that the region of human polη that is sufficient for polymerase and bypass activity starts close to the N terminus and ends between amino acids 378 and 437. XP7DU (as well as XP5BI and XP36BR) has a frame shift mutation at amino acid 407, and we have shown previously that extracts of this cell strain are unable to bypass an AAF lesion in our plasmid system (8). This finding suggests that polη is inactive in these extracts and that the boundary for activity probably lies between amino acids 407 and 437.
We showed previously that polη missing the last 120 amino acids (from amino acid 594) is not localized in the nucleus nor relocalized into nuclear foci after UV irradiation, and that constructs lacking this domain failed to correct the UV + caffeine sensitivity of an XP-V cell strain (16). Thus, we expect that polη, although active in the cells containing C-terminal truncating mutations, is not localized in the nucleus. Unfortunately, we have not been able to demonstrate this directly, because none of the available Abs to polη is able to detect endogenous levels of protein. The function of the ≈190-aa region from about amino acid 410 to amino acid 600 is not known.
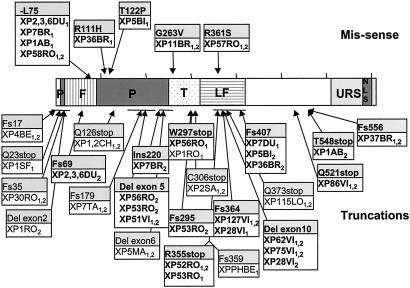
The locations of all of the amino acid changes identified in patients with XP-V in the current and previously reported work are shown in Fig. 4. The mutation spectrum is non-random. The five missense mutations (Fig. 4, upper) are all located in the conserved N-terminal half of the protein and at residues highly conserved in different species. In two of these cell strains that were tested (XP3DU and XP11BR), the mutations abolished polη TLS activity in cell extracts. The N-terminal half of the protein containing the catalytic domains is much more highly conserved than the C-terminal half. Missense mutations in the C-terminal half are therefore less likely to result in a deficient phenotype than those in the N terminus, consistent with the spectrum of missense mutations that we have found.
Figure 4.
Sites of mutations identified in polη. Cell strain designations are indicated in boxes, with those used in this study shown in bold. Others are from Masutani et al. (9), Johnson et al. (10), and Yuasa et al. (26). Subscripts 1 and 2 denote different alleles. Horizontal lines indicate deletions. Single amino acid changes are shown in the upper part of the diagram, and truncations are shown in the lower part. The central part of the diagram shows the different domains in the protein (see Fig. 5), with the finger (F), palm (P), thumb (T), and little finger (LF) domains deduced from ref. 25 delineated by different shading. Also shown are the nuclear localization signal (NLS) and the domain involved in localizing polη into replication foci (URS) (16).
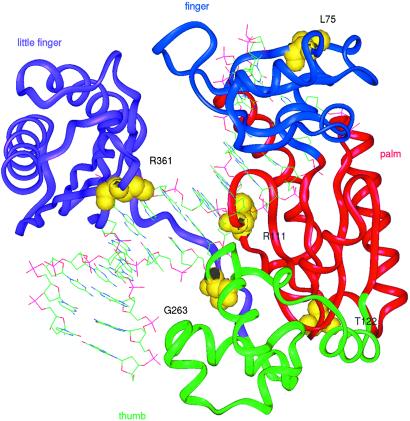
The crystal structure of DNA polymerase IV (Dpo4), a Y-family DNA polymerase from Sulfolobus solfataricus P2, bound to DNA, has been solved recently (25). It shows that the enzyme has finger, palm, and thumb domains, like all known DNA polymerases, despite the lack of sequence similarity. An alignment of polη with Dpo4 enables predictions to be made about the amino acids altered in our patients with XP-V with missense mutations (Fig. 5). Gly-263 and Arg-361 are, respectively, in the thumb and “little finger” domains of the protein that interact with DNA. Gly-263 is close to the primer strand backbone and is conserved in all species of polη. Arg-361 points into the major groove. Arg-111, conserved as arginine or lysine in all species, is likely to interact with Ser-104 and stabilize the palm domain and the catalytic center. Leu-75 points toward the inside of the protein at the beginning of a β-strand in the finger domain. Deletion of this amino acid could disrupt the β-strand. Leucine is found at this position in all orthologs of polη. Although not close to the active site, it probably plays an essential role in keeping finger and palm domains in the correct orientation. Thr-122 is conserved in several species and is important for the structural integrity of the palm domain.
Figure 5.
Sites of missense mutations superimposed on crystal structure of Y-family DNA polymerase. The figure shows the structure of Dpo4 from Sulfolobus solfataricus bound to DNA (25). The predicted sites of the five missense mutations in polη, based on the sequence alignment in ref. 25, are indicated on this structure. The different colors indicate the finger, palm, thumb, and little finger domains.
The truncating mutations are also heavily biased toward the N terminus with 18 different mutations in the N-terminal 407 amino acids, and only 3 in the C-terminal 306 amino acids. This finding is surprising, because the most C-terminal truncations, missing the last 160 amino acids, result in clinical phenotypes very similar to those of patients with truncations in the N-terminal domains. There is, therefore, no expected phenotypic selection against truncations in the C-terminal half of the protein and no obvious explanation for the N-terminal bias. Also unexplained is the apparent “hot region” for truncations between amino acids 355 and 373, with five different mutations identified in this 19-aa region spanning the boundary between exons 10 and 11.
Relationship Between Mutations and Clinical Features.
Assessment of genotype-phenotype relationships in patients with XP is complicated because the clinical features are determined to a large extent by the amount of exposure to sunlight, which is in turn related to the age of the patients. Nevertheless, some general points emerge from our studies. First, even though some patients with XP-V have a high incidence of tumors, many of them seem to survive to a relatively old age compared with NER-defective patients with XP. Almost half of the patients in our study were over 40 years of age. Second, there is no obvious relationship between the site or type of mutation and the incidence of skin tumors. There is a wide variation of tumor incidence in patients with mutations within a particular category (see Table 2). This point is illustrated in the difference in features between patients XP1AB and XP37BR, both of whom live in the north of Scotland and have truncating mutations close to the C terminus (although XP1AB is a compound heterozygote). Patient XP1AB at age 56 is severely affected, having had more than 20 actinic keratoses, more than 100 basal or squamous cell carcinomas, and more than 20 melanomas. Patient XP37BR, in contrast, is a mildly affected patient, who at the age of 62 has had just a few (<5) skin cancers and very few if any actinic keratoses, all the while showing the expected photosensitivity and pigmentation changes characteristic of XP (M. I. White, personal communication).
Acknowledgments
We are indebted to Roger Woodgate and Wei Yang for sharing the information in ref. 25 before publication. This work was supported in part by European Community Contract QLG1-CT1999-0181 (to A.R.L. and R.P.F), European Community Contract FIS5-99-006, and by a Spinoza Award from the Dutch Science Organization (to N.G.J.J.).
Abbreviations
- AAF
acetylaminofluorene
- NER
nucleotide excision repair
- polη
DNA polymerase η
- TLS
translesion synthesis
- XP
xeroderma pigmentosum
- XP-V
XP variant
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kraemer K H, Lee M M, Scotto J. Arch Dermatol. 1987;123:241–250. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- 2.Bootsma D, Kraemer K H, Cleaver J E, Hoeijmakers J H J. In: The Genetic Basis of Human Cancer. Vogelstein B, Kinzler K W, editors. New York: McGraw–Hill; 1998. pp. 245–274. [Google Scholar]
- 3.Berneburg M, Lehmann A R. Advances in Genetics. Vol. 43. San Diego: Academic; 2001. pp. 71–102. [DOI] [PubMed] [Google Scholar]
- 4.Maher V M, Ouellette L M, Curren R D, McCormick J J. Nature (London) 1976;261:593–595. doi: 10.1038/261593a0. [DOI] [PubMed] [Google Scholar]
- 5.Myhr B C, Turnbull D, DiPaolo J A. Mutat Res. 1979;62:341–353. doi: 10.1016/0027-5107(79)90089-7. [DOI] [PubMed] [Google Scholar]
- 6.Arlett C F. In: Progress in Environmental Mutagenesis. Alacevic M, editor. Amsterdam: Elsevier; 1980. pp. 161–174. [Google Scholar]
- 7.Lehmann A R, Kirk-Bell S, Arlett C F, Paterson M C, Lohman P H M, de Weerd-Kastelein E A, Bootsma D. Proc Natl Acad Sci USA. 1975;72:219–223. doi: 10.1073/pnas.72.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordonnier A M, Lehmann A R, Fuchs R P P. Mol Cell Biol. 1999;19:2206–2211. doi: 10.1128/mcb.19.3.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. Nature (London) 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 10.Johnson R E, Kondratick C M, Prakash S, Prakash L. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 11.Ohmori H, Friedberg E C, Fuchs R P P, Goodman M F, Hanaoka F, Hinkle D, Kunkel T A, Lawrence C W, Livneh Z, Nohmi T, et al. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 12.Woodgate R. Genes Dev. 1999;13:2191–2195. doi: 10.1101/gad.13.17.2191. [DOI] [PubMed] [Google Scholar]
- 13.Masutani C, Kusumoto R, Iwai S, Hanaoka F. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng X, Winter D B, Kasmer C, Kraemer K H, Lehmann A R, Gearhart P J. Nat Immunol. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 15.Rogozin I B, Pavlov Y I, Bebenek K, Matsuda T, Kunkel T A. Nat Immunol. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 16.Kannouche P, Broughton B C, Volker M, Hanaoka F, Mullenders L H F, Lehmann A R. Genes Dev. 2001;15:158–172. doi: 10.1101/gad.187501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King S A, Wilson S J, Farber R A, Kaufmann W K, Cordeiro-Stone M. Exp Cell Res. 1995;217:100–108. doi: 10.1006/excr.1995.1068. [DOI] [PubMed] [Google Scholar]
- 18.Lehmann A R, Kirk-Bell S, Arlett C F, Harcourt S A, de Weerd-Kastelein E A, Keijzer W, Hall-Smith P. Cancer Res. 1977;37:904–910. [PubMed] [Google Scholar]
- 19.Arlett C F, Harcourt S A, Broughton B C. Mutat Res. 1975;33:341–346. doi: 10.1016/0027-5107(75)90209-2. [DOI] [PubMed] [Google Scholar]
- 20.Itoh T, Ono T, Yamaizumi M. J Invest Dermatol. 1996;107:349–353. doi: 10.1111/1523-1747.ep12363303. [DOI] [PubMed] [Google Scholar]
- 21.Itoh T, Linn S, Kamide R, Tokushige H, Katori N, Hosaka Y, Yamaizumi M. J Invest Dermatol. 2000;115:981–985. doi: 10.1046/j.1523-1747.2000.00154.x. [DOI] [PubMed] [Google Scholar]
- 22.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haracska L, Johnson R E, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S. Mol Cell Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondratick C M, Washington M T, Prakash S, Prakash L. Mol Cell Biol. 2001;21:2018–2025. doi: 10.1128/MCB.21.6.2018-2025.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling H, Boudsocq F, Woodgate R, Yang W. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 26.Yuasa M, Masutani C, Eki T, Hanaoka F. Oncogene. 2000;19:4721–4728. doi: 10.1038/sj.onc.1203842. [DOI] [PubMed] [Google Scholar]