Abstract
We report that feeding Drosophila throughout adulthood with 4-phenylbutyrate (PBA) can significantly increase lifespan, without diminution of locomotor vigor, resistance to stress, or reproductive ability. Treatment for a limited period, either early or late in adult life, is also effective. Flies fed PBA show a global increase in histone acetylation as well as a dramatically altered pattern of gene expression, including induction or repression of numerous genes. The delay in aging may result from the altered physiological state.
Aging is a common biological phenomenon shared by animals and plants (1), but our understanding of why and how we age remains limited. It would be of great biological interest and practical importance if we could gain insight into the molecular basis of aging, learn to delay the aging process, and maintain the vigor of youth.
Recent studies show that longevity can be altered by genetic manipulations in various organisms. The mutations age-1 (2), daf-2 (3, 4), and clk-1 (5) in Caenorhabditis elegans increase longevity of the worm. In yeast, Sir genes extend the budding lifespan of the mother cell (6). Drosophila lifespan is increased by mutations in a G protein-coupled receptor in methuselah (7), a sodium dicarboxylate cotransporter in Indy (8), an insulin receptor substrate in chico (9), or an insulin-like receptor in InR (10). Studies with Drosophila by transgenic methods recently have been reviewed (11). It is obviously of great interest to discover drugs that can extend lifespan by simple feeding. Mimetics of superoxide dismutase and catalase have been shown to extend lifespan in C. elegans (12). While studying the mechanism of drug action on neurodegenerative mutants (13), we discovered that sodium 4-phenylbutyrate (PBA) can increase both the median and maximum lifespan of Drosophila.
PBA is known to inhibit the activity of histone deacetylase, thus inducing hyperacetylation of histones, hence tending to release histones from their binding to chromatin, with consequent effects on gene transcription (14). Originally approved by the FDA as a drug to treat urea cycle disorder (15), PBA has also been tested in treatment of several diseases. In sickle cell anemia, PBA stimulates transcription of the normal fetal hemoglobulin gene, which is ordinarily silent in the adult, thus substituting for the mutated adult form (16–18). In adrenoleukodystrophy, PBA increases production of ALDRP, a protein that can replace the mutated ABC transporter, thus preventing the accumulation of very long chain fatty acids (19). In cystic fibrosis, PBA prevents degradation of the mutated cystic fibrosis transmembrane conductance regulator (ΔF508-CFTR) (20). PBA is considered to favor differentiation of tumors, as observed in acute promyelocytic leukemia (21) and prostate cancer cells (22), and is therefore being used in various clinical cancer trials. In Fragile X syndrome, due to expanded CGG sequences in the FMR1 gene, production of the FMR1 protein can be restored by DNA demethylation with 5-azadeoxycytidine, combined with hyperacetylation of histones by PBA (23). Beneficial effects on polyglutamine toxicity have also been reported (24). Recent genome-wide studies in yeast of trichostatin-A sensitive histone deacetylase function indicated its effects on transcription of various genes (25). The relation of histone deacetylation to gene silencing and associated changes in budding lifespan has been documented in yeast (26, 27).
We present below the finding of extension of lifespan in Drosophila by feeding PBA, along with data on changes in histone acetylation and the spectrum of genes induced and suppressed by PBA.
Materials and Methods
Fly Strains and Phenylbutyrate.
The Drosophila strains w1118 and wild-type Canton-S were used. PBA acid, sodium salt, was a generous gift from Joseph Cooper of Medicis, Scottsdale, AZ. The purity of the chemical was reported to be 99.6%.
Lifespan Determination.
Newly eclosed flies were collected and raised in standard corn meal agar medium (28). For each experiment, 10 vials, each containing 20 flies, were maintained at either 29°C or 25°C and were transferred to fresh vials every 3 days. The number of dead flies was counted every day.
Membrane Hybridization.
The smart PCR cDNA synthesis kit (CLONTECH) was used to synthesize probes for hybridization. Total RNA was prepared from flies fed for 10 days at 29°C with medium containing 10 mM PBA, or with plain medium. After first strand synthesis of cDNAs by Maloney murine leukemia virus (MMLV)-reverse transcriptase, the cDNA was amplified by PCR, the reaction consisting of 95°C for 1 min, followed by 24 cycles of 15 seconds at 95°C, 30 seconds at 65°C, and 6 min at 68°C. Probes were prepared by a random primed DNA labeling method with [α-32P]dCTP. Filters containing 7,500 Drosophila unique EST clones (Research Genetics, Huntsville, AL) were prehybridized for 4 h, then probes were added to hybridize for 16 h at 58°C in a buffer containing 1 M NaCl, 0.05 M Tris (pH 8.0), 5 mM EDTA, 1% SDS, and 10% dextran sulfate. After hybridization, filters were washed several times and exposed to x-ray film to identify genes that were induced or repressed by PBA.
Reverse-Northern Blots.
To quantitate changes in abundance of individual transcripts, ≈500 base pairs of each candidate gene was synthesized by PCR, and 100 ng was mounted on a nylon transfer membrane (hybond-N+, Amersham Pharmacia). Newly emerged adult flies were fed standard fly food containing 10 mM PBA, or plain food, for 10 days at 29°C. Messenger RNA was extracted and used to synthesize cDNA by MMLV-reverse transcriptase, and probes were prepared by a random priming and [α-32P]dCTP. The nylon membrane containing candidate genes was hybridized with the probe prepared from control flies, and exposed to a PhosphorImager screen (Molecular Dynamics). After the exposure, the probe was stripped off in 10 mM EDTA/0.1% SDS at 80°C, and exposed to a PhosphorImager screen for 18 h, to confirm that no residual probe remained. The membrane was then hybridized again with the second probe, prepared from PBA-fed flies. The expression level of each gene was quantified by using the IMAGE QUANT program (Molecular Dynamics).
Western Blotting of Histones.
Batches of 100 flies were used to prepare histones (29). Homogenates of whole flies were centrifuged at 2,500 × g for 10 min in a medium containing 0.05 M glycine, 10 mM Tris potassium maleate, 5 mM MgCl2, and 10 mM mercaptoethanol (pH 7.3) to get rid of cell debris. HCl was added to the supernatant to a final concentration of 0.25 M, which was kept on ice for 1 h. Acid-soluble proteins were recovered by centrifugation at 13,000 rpm for 10 min at 4°C, then trichloroacetic acid (TCA) was added to precipitate proteins. Ten-microgram samples of proteins were loaded on a 16.5% polyacrylamide gel for electrophoresis, then transferred to a poly(vinylidene difluoride) (PVDF) membrane, followed by hybridization with antibodies to acetylated and nonacetylated H3 and H4 (Upstate Biotechnology, Lake Placid, NY). To verify that any residual PBA in fly bodies would not affect acetylation of histones during their purification, we added 30 mM PBA to the homogenate buffer before isolation of nuclei and histones, and found no difference.
Results and Discussion
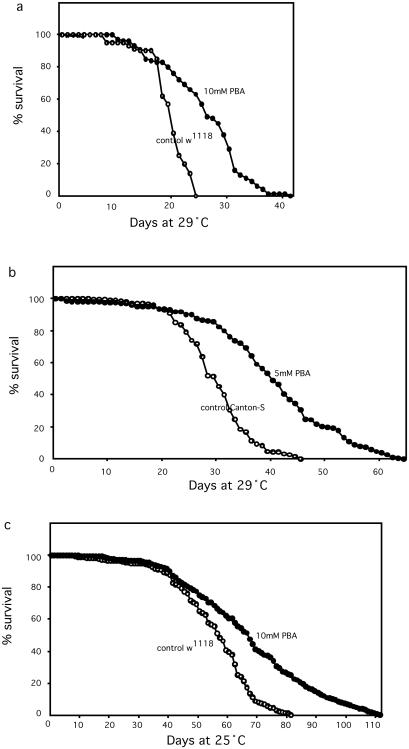
To test the action of PBA in vivo, we fed newly eclosed flies of the w1118 strain, throughout their lifetime, with standard fly medium (cornmeal, agar, dextrose, yeast; ref. 28) containing various concentrations of the drug (0, 0.1, 1, 2.5, 5, 10, 20, and 40 mM). Lifespan was measured both at 29°C and 25°C. Ten millimolar PBA showed a striking extension of both median and maximum lifespan (Fig. 1 a and c). At the higher concentrations, 20 mM and 40 mM, PBA was apparently toxic, reducing survival; there is a narrow range of PBA concentration that is effective for lifespan extension. To determine whether the drug is also effective on a different Drosophila strain, we also measured the effect of PBA on lifespan of the wild-type strain Canton-S (C-S), using 0, 2, 5, and 10 mM PBA. Five millimolar increased lifespan as much as did 10 mM with the w1118 strain (Fig. 1b), whereas 2 mM had no effect and 10 mM was toxic. Thus, the optimal PBA concentration appears to vary with genetic background. As shown below, the degree of inhibition of histone deacetylase also differed in the same way between these two strains.
Figure 1.
Lifespan extension by feeding PBA. Newly emerged adult flies were fed continuously on standard cornmeal/yeast/agar medium, with or without added PBA, and survival curves were measured. Each initial population was 400 flies. Data were combined from two separately run groups of 100 males plus two separately run groups of 100 virgin females. (a) w1118 strain at 29°C. Ten millimolar PBA extends median survival by 33% (mean survival by 36%, maximum survival by 52%). (b) Canton-S wild type strain at 29°C. Five millimolar PBA extends median survival by 36% (mean survival by 40%, maximum by 41%). (c) w1118 strain at 25°C. PBA is also effective at the lower temperature.
It has been suggested that early events in life can delay the onset of aging, thus extending longevity (1). We were therefore curious to know whether a limited period of PBA treatment, early or late, can extend longevity. Newly emerged adult w1118 flies were fed with PBA, at the optimal concentration of 10 mM, from emergence to 12 days (before survival begins any rapid decline), then transferred to plain medium for the rest of their lifetime. Alternatively, flies were fed plain medium for their first 12 days, then medium containing the drug for the remainder of life. In both cases, the PBA-treated flies showed increased lifespan compared with untreated flies (Fig. 2 a and b). In females, treatment initiated at a later age was even more effective than the same duration of treatment at young age. The results suggest that establishment of an altered cellular environment by PBA, albeit for a limited period, can extend the lifespan of flies, possibly by inhibiting the accumulation of damage, and/or stimulating repair mechanisms.
Figure 2.
PBA extends lifespan whether fed early or late in life. Newly emerged w1118 flies were fed on medium plus 10 mM PBA for 12 days, then transferred to normal medium for the rest of their lifespan. Converse group was fed on normal medium for the first 12 days, then with PBA for the remainder of life. Arrows indicate day 12. Each initial population was 200 flies. (a) Virgin females; (b) males.
To test the issue of possible caloric restriction effects, we compared the flies for weight and size after 10 days of feeding at 25°C, with or without PBA, and no difference was observed. We measured the number of eggs produced, the percentage of eggs yielding adult progeny, and the weight and size of the progeny. In all those measurements, there was no detriment due to feeding PBA (Table 1). Qualitative observations on ingestion of food dye were consistent with that conclusion.
Table 1.
Comparison of weight, size, and reproductive ability of w1118 flies after being maintained on food with or without 10 mM PBA for 10 days at 25°C
| Control | PBA | |
|---|---|---|
| Parent | ||
| Weight of 5 flies, mg (n = 10) | ||
| Females | 5.1 ± 0.2 | 5.1 ± 0.2 |
| Males | 3.3 ± 0.1 | 3.3 ± 0.4 |
| Size of a fly, mm (n = 40) | ||
| Female | 5.5 ± 0.1 | 5.5 ± 0.1 |
| Male | 4.5 ± 0.1 | 4.6 ± 0.1 |
| Egg laying (3 males + 3 females a vial for 16 h; n = 20 vials) | 66 ± 16 | 67 ± 15 |
| % of eggs yielding adults (n = 20 vials) | 66 ± 15 | 70 ± 16 |
| Progeny | ||
| Weight of 5 flies, mg (n = 10) | ||
| Females | 5.3 ± 0.4 | 5.2 ± 0.2 |
| Males | 3.4 ± 0.6 | 3.5 ± 0.3 |
| Size of a fly, mm (n = 40) | ||
| Female | 5.4 ± 0.2 | 5.5 ± 0.1 |
| Male | 4.5 ± 0.2 | 4.6 ± 0.2 |
Flies were weighed in batches of five. Size is the length of the fly from front of head to tip of abdomen as measured with an eyepiece micrometer. Eggs laid were counted after 16 h with three males and three females per vial. Values represent mean ± SD. None of these parameters showed any significant differences due to feeding PBA.
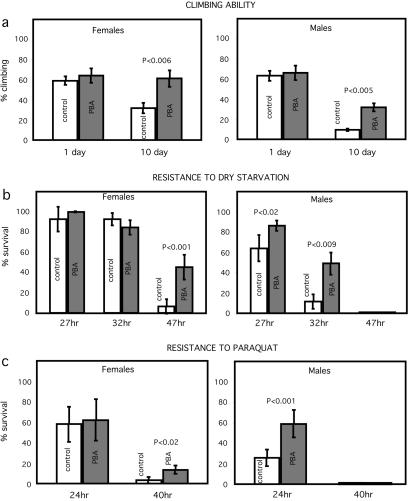
We tested other parameters to determine whether, associated with the extension in lifespan, there was also extended maintenance of vigor. Locomotor ability, as measured by climbing in the negative geotactic response (Fig. 3a), resistance to starvation (Fig. 3b), and resistance to the free radical generator paraquat (Fig. 3c) all showed extension of healthy lifespan, as compared with controls not fed PBA.
Figure 3.
PBA inspires maintenance of locomotor ability and resistance to stress. For each experiment, n = 80 w1118 flies, repeated four times. (Left) Virgin females; (Right) males. In all these paradigms, both male and female PBA-treated flies showed superior performance. (a) Locomotor ability assayed by climbing test for three transfers in a “counter current distribution” apparatus (41) held vertically. Flies that climbed to the upper tube three times out of three trials were counted as positive. Flies were maintained at 29°C for 10 days with or without PBA before testing. (b) Resistance to dry starvation. Flies raised for 10 days at 25°C with or without 10 mM PBA were put into empty vials to test their survival measured vs. time. (c) Resistance to paraquat, a free-radical generator. Flies raised for 10 days at 25°C with or without 10 mM PBA were first deprived of water in an empty vial for 3 h, then transferred to a vial containing filter paper wetted with 350 μl of 20 mM paraquat in 5% sucrose. Values represent mean ± SD for the four runs. Student's t test was used to analyze the significant difference between controls and PBA-treated flies.
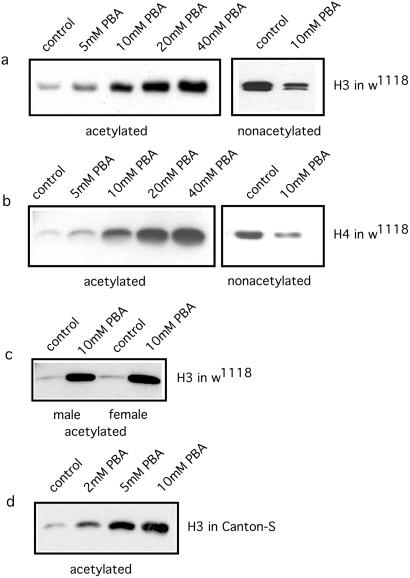
Because PBA, an inhibitor of histone deacetylase, is known to enhance acetylation of the tails of histones H3 and H4, we investigated the level of histone acetylation in flies with and without PBA feeding, using specific antibodies on Western blots. As shown in Fig. 4 a and b, the nonacetylated forms of these histones were predominant in untreated flies, but PBA feeding increased the proportions of their acetylated forms. Similar results were obtained for both males and females (Fig. 4c). These changes in histone acetylation suggested possible modifications of chromatin structure, which might change the regulation of transcription.
Figure 4.
Effect of feeding PBA on acetylation of histones H3 and H4. w1118 flies were raised for 10 days at 29°C on medium containing various concentrations of PBA. Histones prepared from whole flies were probed on Western blots with antibodies specific for acetylated or nonacetylated forms of the tails of histones H3 and H4. Ten micrograms of histone were used per lane. In flies treated with PBA, the acetylated forms of both H3 and H4 were increased, whereas the nonacetylated forms decreased. (a) Histone H3 in w1118 flies. Abundance of the acetylated form increases with concentration of PBA added to the food. The nonacetylated form decreases (mixed males and virgin females). (b) Similar result for histone H4 in w1118 flies (mixed males and virgin females). (c) w1118 males and virgin females assayed separately. The effect of PBA is similar in both sexes. (d) Increase of acetylation of histone H3 in wild-type strain Canton-S (mixed males and virgin females).
To investigate the differential patterns of gene activity resulting from PBA treatment, high density membrane arrays containing an estimated 7,500 unique Drosophila EST clones (Research Genetics, Huntsville, AL) were used. Messenger RNA was prepared from flies maintained at 29°C on food containing 10 mM PBA from emergence to 10 days of age, and a similar preparation was made from untreated flies. These were labeled with 32P and used as probes on the membranes.
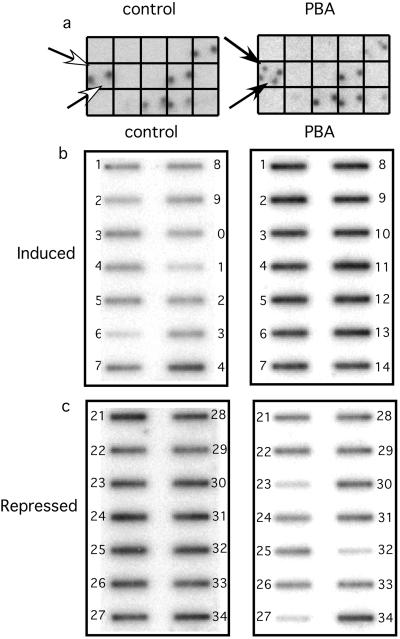
Fig. 5a shows a sample portion of the array hybridized with each of the two different probes, to illustrate the choosing of 100 genes that were induced by PBA, based on increased intensity of hybridization. Conversely, we chose 48 genes that were suppressed by the treatment, as judged by disappearance of a visible spot. The patterns were reproducible with two independent preparations of mRNA each from treated and untreated flies, and with two independent pairs of membranes for each preparation.
Figure 5.
Induction or repression of transcription of various genes by PBA, verified by reverse-Northern blots. (a) Sample area of membrane microarray of cDNA clones hybridized with mRNA probes prepared from w1118 flies raised with or without 10 mM PBA for 10 days at 29°C. The area shown contained 180 clones of the array. Arrows at right illustrate typical spots selected as candidates for increased abundance of mRNA in PBA-treated flies. (b) Reverse-Northern blots for genes induced by PBA: 1, heat shock protein cognate 70–4; 2, daughterless; 3, elongation factor 1α 48D; 4, heat shock protein 60; 5, mitochondrial phosphate carrier protein; 6, inebriated; 7, dnaJ-like protein2; 8, imaginal disc growth factor1; 9, cytochrome P450–4d1; 10, transportin; 11, superoxide dismutase; 12, epididymal secretory protein; 13, glutathione S-transferase; 14, Control = nina E, which showed similar expression with or without PBA treatment. (c) Genes repressed by PBA: 21, glyceraldehyde-3-phosphate dehydrogenase1; 22, porin; 23, NADH: ubiquinone reductase 75-kDa subunit precursor; 24, cytochrome c oxidase; 25, osa; 26, hexokinase; 27, dnaJ-like protein 1; 28, cytochrome c oxidase subunit VIb; 29, peptidylglycine-α-hydroxylating monooxygenase; 30, peroxysomal farnesylated protein; 31, fatty acid synthetase; 32, calreticulin; 33, cyclin-dependent kinase 9; 34, nina E control.
The 100 induced genes included 3 that previously have been implicated in detoxification, 3 chaperones, 2 involved in translation machinery, 3 transcription factors, 7 involved in signal transduction pathways, 25 involved in metabolism, 11 ribosomal proteins, 11 proteases, 4 kinases, 13 genes that function as transporters or carriers, and 18 others involved in other miscellaneous functions. Among the 48 repressed genes, there were 21 involved in metabolism, 4 proteases, 2 ribosomal proteins, and 21 others having other miscellaneous functions.
To verify the changes in transcription levels indicated on the microarrays, we used reverse-Northern blots on a subset of the candidate genes. Seven of the induced genes were chosen because of their putative involvement in enhancing longevity: superoxide dismutase, elongation factor1α, glutathione S-transferase, cytochrome P450, and three chaperones. In Drosophila, transgenic flies with superoxide dismutase genes have been reported to show extended mean lifespan (30–32). Elongation factor 1α plays a critical role in maintaining the level of protein synthesis, which normally declines with age (33). Glutathione S-transferase and cytochrome P450 are involved in detoxification, one of the determinants of aging (1, 34). Heat shock proteins enhance resistance to stress and extend lifespan (35, 36). All of these genes were induced by PBA, as confirmed in Fig. 5b. Note that superoxide dismutase (SOD) expression, in particular, was dramatically increased.
Nineteen additional genes were chosen at random, 6 of the induced group and 13 of the repressed group. All were confirmed by reverse-Northern blot, supporting the membrane screening method used in identifying the genes. The genes are listed in Table 2, grouped according to their putative functions.
Table 2.
Partial list of genes induced or repressed in w1118 flies by 10 days of feeding with PBA at 29°C
| Function | Clone ID | Gene | Fold change |
|---|---|---|---|
| Genes induced in PBA-treated flies (confirmed by Northern blot) | |||
| Detoxification | CG8905 | Superoxide dismutase | 51.9 |
| CG3656 | Cytochrome P450-4d1 | 7.2 | |
| CG10045 | Glutathione S-transferase | 4.6 | |
| Chaperon | CG4264 | hsc70 | 4.5 |
| CG12101 | hsp60 | 6.7 | |
| CG3061 | dnaJ like2 | 2.9 | |
| Translation | CG8280 | Translational elongation factor1 α | 4.1 |
| Neurotransmitter | CG15444 | inebriated | 26.8 |
| Transcription factor | CG5102 | daughterless | 8.0 |
| Ligand binding | CG7398 | transportin | 8.0 |
| Signal transduction | CG7291 | Epididymal secretory protein | 7.5 |
| Transporter | CG4994 | Mitochondrial phophate carrier protein | 5.1 |
| Growth factor | CG4472 | Imaginal disc growth factor1 | 5.3 |
| Genes repressed in PBA-treated flies (confirmed by Northern blot) | |||
| Metabolism | CG12055 | Glyceraldehyde-3-phosphate dehydrogenase1 | 3.7 |
| CG2286 | NADH:ubiquinone reductase 75-kD subunit precursor | 25.3 | |
| FBgn0013674 | Cytochrome c oxidase | 6.6 | |
| CG3832 | Peptidyl gycine α hydroxylating monooxygenase | 1.4 | |
| CG3523 | Fatty acid synthetase | 2.4 | |
| CG14235 | Cytochrome c oxidase subunitVIb | 2.2 | |
| CG3001 | Hexokinase | 5.0 | |
| Chaperon | CG10578 | dnaJ like1 | 13.2 |
| DNA binding | GH03026 | osa | 5.0 |
| Ligand binding | GH25160 | calreticulin | 26.5 |
| Signal transduction | GH03076 | Peroxisomal farnesylated protein | 1.8 |
| Kinase | GH21935 | Cyclin-dependent kinase9 | 2.7 |
| Ion channel | GH08586 | porin | 1.4 |
All were confirmed by reverse-Northern blots and fold change is indicated for each. The first seven of the induced genes were chosen for confirmation because of previous literature indicating their involvement in aging; six others were chosen at random from the remaining set of induced clones. The 13 genes in the repressed group were chosen at random. Data are based on two separately prepared batches of mRNA, using the same membrane before and after stripping.
Whether the observed changes in gene transcription, or a subset of them, are responsible for the increased longevity will require testing each candidate gene individually by overexpression in transgenic flies, or by specific inhibition. Also, which are primary and which are downstream effects of PBA remains to be determined. It will be a challenge to define constellations of genes and their networks that can be responsible for increasing lifespan.
There have been various reports of global molecular changes associated with aging, by comparing tissues from young and old animals (37–39), but it is difficult to determine which events are directly involved in the aging process. The extension of lifespan of Drosophila by treatment with PBA may serve as a useful model. In Drosophila, transgenic constructs can readily be made to test for the effects of overexpression or silencing of individual genes. This system may also be useful in understanding basic mechanisms in histone acetylation and transcriptional regulation of gene expression, by identifying common features of control regions for the different gene responses. We observed both induction and repression of transcription by PBA. It is interesting to note that overexpression of Sir2 protein in both yeast (26) and C. elegans (40), which has NAD-dependent histone deacetylase activity, is implicated in silencing of gene transcription, associated with extension of the lifespan, whereas deletion of a different histone deacetylase (RPD3) also extends lifespan (27). This conundrum suggests that the underlying mechanism for the extension of lifespan may be an optimal balance of expression and repression of various genes to regulate an optimal physiological and cellular environment for longevity. That finding would be consistent with our observation on the concentration dependence of PBA fed to flies, which shows counterproductive effects at excess levels as is typical of drug-dosage effects. The genes induced or suppressed by PBA therefore warrant equal attention in further study.
It is intriguing that PBA treatment late in the life of the adult fly can still be effective. Whatever the mechanisms, our observation that simple feeding of a drug can enhance lifespan, along with improved maintenance of vigor, strongly suggests the feasibility of high throughput screening, for which Drosophila represents a convenient model organism. It will also be of interest to determine whether PBA treatment of other animals can affect their lifespan.
Acknowledgments
We especially thank Joseph Cooper at Medicis (Scottsdale, AZ) for his generous gift of PBA. This research, initiated at Caltech, was supported by an intramural grant to K.-T.M. from the National Institute of Neurological Disorders and Stroke and the Ellison Foundation, and grants to S.B. from the National Science Foundation, the National Institutes of Health, the Ellison Foundation, and the James G. Boswell Foundation.
Abbreviation
- PBA
4-phenylbutyrate
References
- 1.Arking R. Biology of Aging: Observations and Principles. 2nd Ed. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 2.Friedman D B, Johnson T E. Genetics. 1998;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R A. Nature (London) 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 4.Kimura K D, Tissenbaum H A, Liu Y, Ruvkun G. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 5.Wong A, Boutis P, Hekimi S. Genetics. 1995;139:1247–1259. doi: 10.1093/genetics/139.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson F B, Sinclair D A, Guarente L. Cell. 1999;96:291–302. doi: 10.1016/s0092-8674(00)80567-x. [DOI] [PubMed] [Google Scholar]
- 7.Lin Y J, Seroude L, Benzer S. Science. 1998;282:943–946. doi: 10.1126/science.282.5390.943. [DOI] [PubMed] [Google Scholar]
- 8.Rogina B, Reenan R A, Nilsen S P, Helfand S L. Science. 2000;290:2137–2140. doi: 10.1126/science.290.5499.2137. [DOI] [PubMed] [Google Scholar]
- 9.Clancy D J, Gems D, Harshman L G, Oldham S, Stocker H, Hafen E, Leevers S J, Partridge L. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 10.Tatar M, Kopelman A, Epstein D, Tu M P, Yin C M, Garofalo R S. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 11.Tower J. Mech Ageing Dev. 2000;118:1–14. doi: 10.1016/s0047-6374(00)00152-4. [DOI] [PubMed] [Google Scholar]
- 12.Melov S, Ravenscroft J, Malik S, Gill M S, Walker D W, Clayton P E, Wallace D C, Malfroy B, Doctrow S R, Lithgow G J. Science. 2000;289:1567–1569. doi: 10.1126/science.289.5484.1567. [DOI] [PubMed] [Google Scholar]
- 13.Min K-T, Benzer S. Science. 1999;284:1985–1988. doi: 10.1126/science.284.5422.1985. [DOI] [PubMed] [Google Scholar]
- 14.Lea M A, Randolph V M. Anticancer Res. 1998;18:2717–2722. [PubMed] [Google Scholar]
- 15.Brusilow S W, Danney M, Waber L J, Batshaw M, Burton B, Levitsky L, Roth K, McKeethren C, Ward J. N Engl J Med. 1984;310:1630–1634. doi: 10.1056/NEJM198406213102503. [DOI] [PubMed] [Google Scholar]
- 16.Perrine S P, Ginder G D, Faller D V, Dover G H, Ikuta T, Witkowska H E, Cai S P, Vichinsky E P, Olivieri N F. N Engl J Med. 1993;328:81–86. doi: 10.1056/NEJM199301143280202. [DOI] [PubMed] [Google Scholar]
- 17.Dover G J, Brusilow S, Charache S. Blood. 1994;84:339–343. [PubMed] [Google Scholar]
- 18.Collins A F, Pearson H A, Giardina P, McDonagh K T, Brusilow S W, Dover G J. Blood. 1995;85:43–49. [PubMed] [Google Scholar]
- 19.Kemp S, Wei H M, Lu J F, Braiterman L T, McGuinness M C, Moser A B, Watkins P A, Smith K D. Nat Med. 1998;4:1261–1268. doi: 10.1038/3242. [DOI] [PubMed] [Google Scholar]
- 20.Rubenstein R C, Zeitlin P L. Am J Respir Crit Care Med. 1998;157:484–490. doi: 10.1164/ajrccm.157.2.9706088. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Saunthararahah Y, Render R L, Liu J M. Cancer Res. 1999;59:2766–2769. [PubMed] [Google Scholar]
- 22.Melchior S W, Brown L G, Figg W D, Quinn J E, Santucci R A, Brunner J, Thuroff J W, Lange P H, Vessella R L. Int J Oncol. 1999;14:501–508. doi: 10.3892/ijo.14.3.501. [DOI] [PubMed] [Google Scholar]
- 23.Chiurazzi P, Pomponi M G, Pietrobono R, Bakker C E, Neri G, Oostra B A. Hum Mol Genet. 1999;8:2317–2323. doi: 10.1093/hmg/8.12.2317. [DOI] [PubMed] [Google Scholar]
- 24.Steffan J S, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol B L, Kazantsev A, Schmidt E, Zhu Y Z, Greenwald M, et al. Nature (London) 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 25.Bernstein B E, Tong J K, Schreiber S T. Proc Natl Acad Sci USA. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. . (First Published November 28, 2000; 10.1073/pnas.250477697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imai S, Amstrong C M, Kaeberlein M, Guarente L. Nature (London) 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 27.Kim S, Benguria A, Lai C, Jazwinski S M. Mol Biol Cell. 1999;10:3125–3136. doi: 10.1091/mbc.10.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis E B. Drosophila Inf Serv. 1960;34:117–118. [Google Scholar]
- 29.Alfageme C R, Zweidler A, Mahowald A, Cohen L H. J Biol Chem. 1974;12:3729–3736. [PubMed] [Google Scholar]
- 30.Orr W C, Sohal R S. Science. 1994;263:1128–1130. doi: 10.1126/science.8108730. [DOI] [PubMed] [Google Scholar]
- 31.Parkes T L, Elia A J, Dickinson D, Hilliker A J, Phillips J P, Boulianne G L. Nat Genet. 1998;19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- 32.Orr W C, Sohal R C. Arch Biochem Biophys. 1993;301:34–40. doi: 10.1006/abbi.1993.1111. [DOI] [PubMed] [Google Scholar]
- 33.Webster G C, Webster S L. Mech Ageing Dev. 1984;24:335–342. doi: 10.1016/0047-6374(84)90118-0. [DOI] [PubMed] [Google Scholar]
- 34.Mannervik B. Adv Enzymol Relat Areas Mol Biol. 1985;57:357–417. doi: 10.1002/9780470123034.ch5. [DOI] [PubMed] [Google Scholar]
- 35.Tatar M, Khazaeli A A, Curtsinger J W. Nature (London) 1997;390:30. doi: 10.1038/36237. [DOI] [PubMed] [Google Scholar]
- 36.Lithgow G J. Rev Clin Gerontol. 1996;6:119–127. [Google Scholar]
- 37.Lee C, Klopp R G, Weindruch R, Prolla T A. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 38.Shelton D N, Chang E, Whittier P S, Choi D, Funk W D. Curr Biol. 1999;9:939–945. doi: 10.1016/s0960-9822(99)80420-5. [DOI] [PubMed] [Google Scholar]
- 39.Zou S, Meadows S, Sharp L, Jan L Y, Jan Y N. Proc Natl Acad Sci USA. 2000;97:13726–13731. doi: 10.1073/pnas.260496697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tissenbaum H A, Guarente L. Nature (London) 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 41.Benzer S. Proc Natl Acad Sci USA. 1967;58:1112–1119. doi: 10.1073/pnas.58.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]