Abstract
Eleven independent, recessive, N-ethyl-N-nitrosourea-induced mutations that map to a ≈1- to 2-cM region of mouse chromosome (Chr) 7 homologous to human Chr 11p14-p15 were recovered from a screen of 1,218 gametes. These mutations were initially identified in a hemizygous state opposite a large p-locus deletion and subsequently were mapped to finer genomic intervals by crosses to a panel of smaller p deletions. The 11 mutations also were classified into seven complementation groups by pairwise crosses. Four complementation groups were defined by seven prenatally lethal mutations, including a group (l7R3) comprised of two alleles of obvious differing severity. Two allelic mutations (at the psrt locus) result in a severe seizure and runting syndrome, but one mutation (at the fit2 locus) results in a more benign runting phenotype. This experiment has added seven loci, defined by phenotypes of presumed point mutations, to the genetic map of a small (1–2 cM) region of mouse Chr 7 and will facilitate the task of functional annotation of DNA sequence and transcription maps both in the mouse and the corresponding human 11p14-p15 homology region.
Keywords: N-ethyl-N-nitrosourea‖regional mutagenesis‖allelic series‖seizures and runting‖prenatal and juvenile abnormalities
The supermutagenicity of N-ethyl-N-nitrosourea (ENU) (1) has made it possible to recover heritable mutations at high frequency from mouse spermatogonial stem cells. Consequently, it is possible to use phenotype-driven strategies to recover new ENU-induced mutations in specific, preselected loci for the generation of new alleles (1–4), in whole-genome screens for genes comprising components of complex pathways or phenotypes (5–10), or, with the use of chromosomal rearrangements, in a wide variety of genes within specific chromosomal regions (11–18). The phenotyping protocols used in such strategies can range from simple to complex and can be narrowly or broadly focused, with the output of any experiment dependent largely on the discriminating power of the phenotyping used. The extensive homologies between the genomes of mouse and human make it feasible to use the mouse for discovery of new mutations and phenotypes that aid in the functional annotation of the corresponding human DNA sequence and transcription maps.
The region of mouse chromosome (Chr) 7 surrounding the pink-eyed dilution (p) locus that shares homology with human genomic regions 15q11-q13 and 11p14-p15 is being characterized by both genetic and molecular approaches (15, 19–26). We previously reported initial results of a hemizygosity-screen strategy for recovering ENU-induced recessive mutations within the 4- to 5-cM Del(ru2 p)46DFiOD deletion (15); this strategy is similar to a mutation-recovery experiment carried out for a 6- to 11-cM deletion at the more distally mapping Chr 7 tyrosinase [Tyr; previously, albino (c)] locus (14, 16). The initial phase of this “p-region screen” (like the entire Tyr-region screen) used simple phenotyping criteria such as recognizing lethality and externally visible characters or behaviors in mutant lines. We report here initial results of this phase of the p-region screen, in which 1,218 gametes were tested for new mutations mapping to the Del(ru2 p)46DFiOD deletion. Eleven lethal or easily recognized mutations, identifying seven loci, were classified by deletion mapping and complementation analyses into a fine-structure map of the human 11p14-p15 homology region at the proximal end of the Del(ru2 p)46DFiOD deletion in mouse Chr 7.
Materials and Methods
Mice.
All animals were bred at the Mammalian Genetics Research Facility at Oak Ridge National Laboratory (Labcode = “R”). Animals homozygous for p7R75M (abbreviated as px) are darker in color than px/p or px/Del(p) animals. Thus, all lethal p deletions used for fine-structure mapping were maintained by crosses of px/Del(p) heterozygotes to px/px mates. The prenatally lethal Del(ru2 p)46DFiOD deletion (22) was used to detect recessive mutations in the hemizygosity screen (see below) (15). For these crosses, Del(ru2 p)46DFiOD was maintained by alternate crosses of +p/Del(ru2 p)46DFiOD and ru2+/Del(ru2 p)46DFiOD heterozygotes to ru2+/ru2+ and +p/+p mice, respectively. Initial genetic and molecular characterizations of the Oak Ridge p deletions have been reported elsewhere (15, 20–23, 26, 27), and information on each mutation in this article can be found at http://bio.lsd.ornl.gov/mouse.
Mutagenesis.
Four groups of BJR (a/a; ru2 p/ru2 p) males older than 8 weeks old (a total of 225 males) from a closed-colony, noninbred stock were given four weekly i.p. injections (100 mg/kg each) of ENU as described (16). ENU was obtained from Sigma. Males regained fertility on average 12–16 weeks after the last injection and were routinely bred for 5–6 months.
Detection of Mutations.
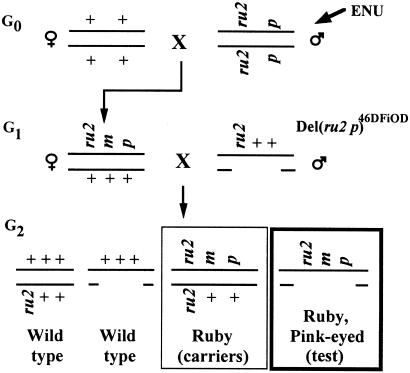
The crosses used to recover new mutations mapping to the Del(ru2 p)46DFiOD deletion have been described (15) and are recapitulated in Fig. 1. The ruby, pink-eyed dilute G2 test-class animals, in which new recessive mutations would become hemizygous, were observed at weaning for visible differences from littermates in body size/weight, skin and hair quality, obvious skeletal abnormalities, ability to swim, balance problems, or abnormal neuromuscular activity. The absence of ruby, pink-eyed dilute animals in any pedigree indicated that that particular ru2 m p/Del(ru2 p)46DFiOD genotype was lethal (m designates a new mutation induced by ENU, not the Chr 4 misty mutation). Whenever possible, we produced at least 30 G2 offspring from each ru2 p/++G1 female so that such lethal mutations could be reliably identified with an acceptably low false-positive rate (15, 16). This was not always possible, however, because some G1 females became sterile or died before producing 30 G2 progeny, and so a number of pedigrees that were potentially segregating an ENU-induced lethal were tested for transmissibility of the new m before 30 G2 progeny had been raised.
Figure 1.
Breeding protocol used to recover ENU-induced recessive mutations mapping within the 4- to 5-cM Del(ru2 p)46DFiOD deletion. The hemizygous test class, in which the phenotype of new recessive mutations can be recognized, is the G2 ruby, pink-eyed-dilute class (heavily outlined box). Carriers of new mutations are the ruby G2 progeny (lightly outlined box), from which detrimental mutations can be propagated. m is a mutation induced by ENU; ru2, ruby-eye-2; p, pink-eyed dilution. Although m is shown here to map between ru2 and p, m can also map to either side of either visible-marker locus.
All new mutations, including lethal or otherwise detrimental or fitness/fertility-reducing ones, were propagated from the ruby-colored (ru2 m p/ru2++) G2 siblings. Once transmissibility of the mutation was demonstrated, the new mutant stock was maintained as described (15) by alternate crosses of ruby carriers [ru2 m p/ru2++] to ++p/Del(ru2 p)46DFiOD mice with selection for the pink-eyed offspring [ru2 m p/++p], followed in the next generation by crossing these ru2 m p/++p carriers to ru2++/Del(ru2 p)46DFiOD mice to regenerate ruby carriers [ru2 m p/ru2++]. This maintenance system provided the opportunity for routine progeny testing in each generation.
Deletion Mapping and Complementation Crosses.
For deletion-mapping crosses, progeny-tested ruby carrier males [ru2 m p/ru2++] were crossed to px/Del(p) females, which were each heterozygous for one of the extensive series of Oak Ridge p deletions. With the exception of the selector deletion Del(ru2 p)46DFiOD, none of these deletions includes ru2. Normal pink-eyed progeny would be expected to comprise ≈25% of the progeny in complementing combinations. A mutation was considered to be included within the deletion if there were no pink-eyed progeny [ru2 m p/+Del(p)] in 30 offspring of this cross (for the lethal mutations), or if the pink-eyed progeny exhibited an abnormal phenotype (for the visible mutations). Before their use in these deletion-mapping crosses, all ruby carrier males [ru2 m p/ru2++] were first proved to carry m by crossing them to female ru2++/Del(ru2 p)46DFiOD or ++p/Del(ru2 p)46DFiOD mice and confirming the lack of ruby, pink-eyed progeny (for lethal mutations) or the presence of abnormal ruby, pink-eyed progeny (for visible mutations). This progeny test was necessary because m might be lost from the original BJR ru2 p chromosome by single crossing-over during gametogenesis in a heterozygous carrier.
For the complementation crosses, ruby [ru2 m1 p/ru2++] or pink-eyed [ru2 m1 p/++p] males proved by progeny test to carry one m (m1) were crossed to female ruby [ru2 m2 p/ru2++] or pink-eyed [ru2 m2 p/++p] mice heterozygous for another m (m2), with observation of the ruby, pink-eyed progeny [ru2 m1 p/ru2 m2 p]. Absence of the ruby, pink-eyed class (for lethal mutations) or presence of an abnormal phenotype in the ruby, pink-eyed class (for visible mutations) indicated noncomplementation, a result that assigned m1 and m2 to the same complementation group (at which time they were considered to be alleles of the same locus until proved otherwise). Initially in these crosses, we used only females proved by progeny test to carry m2; however, this became difficult in many cases because the female often became too old by the end of the progeny test to perform or complete the actual experimental complementation cross with progeny-tested male carriers of m1. Therefore, we instituted a system in which a progeny-tested male obligate carrier of m1 was crossed to three unproved female carriers of an m2, and 20 progeny were raised from each. Conclusions about complementation were then drawn for that combination only if all three females gave similar results.
Results
Recovery of 11 ENU-Induced, Recessive, Lethal or Visible Mutations That Map to the Proximal End of the Del(ru2 p)46DFiOD Deletion.
A total of 2,488 G1 daughters of ENU-treated BJR-ru2 p/ru2 p males were crossed to ru2+/Del(ru2 p)46DFiOD males according to the protocol outlined in Fig. 1. Of these G1 females, 1,218 produced large enough G2 progenies to permit detection of new mutations. Three pedigrees were observed in which the ruby, pink-eyed test-class mice [i.e., ru2 m p/Del(ru2 p)46DFiOD] were externally abnormal, and eight pedigrees were identified in which no or very few test class were found at weaning, thereby suggesting the presence of a lethal mutation (Table 1). All 11 variants proved transmissible to the next generation through the G2 ruby [ru2 m p/ru2++] carrier sibs.
Table 1.
Eleven independent ENU-induced mouse Chr 7 mutations ultimately mapping to the human 11p14-p15 homology region
| Allele | Mutation | Outcome of crosses yielding:*
|
General phenotype† | |
|---|---|---|---|---|
| Homozygotes | Hemizygotes | |||
| l7R21R | 1318SJ | 7 sm/421(9) | 2 sm/829 (0) | PL/NL |
| l7R31R | 735SJ | 0/217 (0) | 0/1047 (8) | PL/NL |
| l7R32R | 951SJ | 50 sm/675 (8) | 7 sm/698 (2) | PL/NL |
| l7R41R | 2033SJ | 7 sm/535 (22) | 5 sm/1085 (10) | PL |
| l7R51R | 403SJ | 5 sm/225 (2) | 14 sm/670 (10) | JLE |
| l7R61R | 88SJ | 0/505 (4) | 0/805 (2) | PL/NL |
| l7R62R | 335SJ | 0/381 (1) | 0/687 (4) | PL/NL |
| l7R63R | 2038SJ | 1 sl/257 (0) | 1 sl/818 (2) | PL/NL |
| psrt1R | 723SJ | 98 sz/631 (7) | 25 sz/729 (2) | JL/R/SZ |
| psrt2R | 1060SJ | 63 sz/497 (3) | 31 sz/696 (4) | JL/R/SZ |
| fit21R | 22SJ | 67 sl 61 sm/834 (5) | 12 sl 91 sm/814 (0) | R |
Shown in each numerator is the number of homozygotes [ru2 m p/ru2 m p] or hemizygotes [ru2 m p/Del(ru2 p)46DFiOD] in the total number of progeny, respectively, from two types of matings that can yield homozygotes: ru2 m p/ru2 + + X ru2 m p/ru2 + + and ru2 m p/ + + p X ru2 m p/+ + p; and from two types of matings that can yield hemizygotes: ru2 m p/ru2 + + X + + p/Del(ru2 p)46DFiOD and ru2 m p/+ + p X ru2 + +/Del(ru2 p)46DFiOD. The homozygous or hemizygous test class in both the intercrosses and the deletion cross, respectively, is ruby, pink-eyed. The genetic distance between ru2 and p is approximately 3 cM. When rare “normal” test-class mice (indicated in the parentheses) were observed, they could be ruby, pink-eyed recombinant chromosomes that lack m as a result of a single crossover between ru2 and m in ru2 m p/+ + p heterozygotes, or between m and p in ru2 m p/ru2 + + heterozygotes. Alternatively, normal test-class mice could potentially still be m hemizygotes at the low end of a broad expressivity scale; distinguishing these outcomes was not attempted in this study. sm, visibly smaller than littermates (at least 70% smaller); sl, slightly smaller than littermates, perceptibly smaller than average, but not as severely growth retarded as the sm class; sz, runted animals under continual seizure.
General phenotype is defined here by that exhibited in hemizygotes and indicates the phenotype that resulted in ascertainment of the mutant pedigree. R, runting; PL/NL, prenatally or neonatally lethal; JLE, juvenile lethal early, associated with death typically between birth and 10 days on this genetic background; JL, juvenile lethal around time of weaning; SZ, almost continual seizures.
Table 1 shows that among the recovered lethal mutations were several (e.g., 951SJ, 403SJ, 1318SJ, and 2033SJ) that demonstrated a “leaky” phenotype, defined by the presence of a small, variable number of variably runted test-class [ru2 m p/ru2 m p or ru2 m p/Del(ru2 p)46DFiOD] animals among large numbers of progeny from intercrosses of heterozygous carriers or from outcrosses of heterozygous carriers to either ru2 +/Del(ru2 p)46DFiOD or +p/Del(ru2 p)46DFiOD mice. We assume that the runted animals, which were anywhere from 50% to 80% of typical littermate size, were either homozygous or hemizygous for m [i.e., ru2 m p/ru2 m p or ru2 m p/Del(ru2 p)46DFiOD, respectively]. However, we did not routinely determine whether apparently normal ruby, pink-eyed animals recovered at low frequency in these lines (parentheses in Table 1) were ru2+ p recombinants that had lost m from the ru2 m p chromosome by a single crossover (in ru2 m p/ru2++ or ru2 m p/++ p meioses; 3-cM distance between ru2 and p), or were m homozygotes or hemizygotes at the low end of a broad phenotype-expressivity scale.
Particularly striking was the leaky nature of the 951SJ mutation (l7R32R) when it was homozygous rather then hemizygous. Fifty runted, presumed ru2 l7R32R p/ru2 l7R32R p homozygotes were detected in 675 classified progeny, but only seven runted, presumed ru2 l7R32R p/Del(p)46DFiOD hemizygotes were observed in a similar number (698) of progeny. Interestingly, no runted homozygotes were observed in 217 progeny of 735SJ heterozygotes, shown to be a probable allele of 951SJ at the l7R3 locus (see below), implying that the 951SJ mutation is hypomorphic relative to wild type but probably is less severe than the 735SJ mutation.
Deletion Mapping and Complementation Analysis of the 11 Mutations.
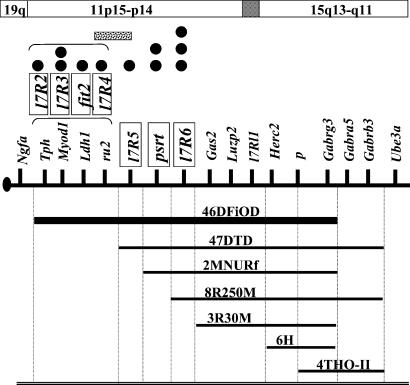
The well-characterized series of Oak Ridge p-locus deletions (22, 23, 26) made it possible to fine-map each mutation to a defined interval within the p-deletion complex (Fig. 2) by a one-cross complementation test based on pseudodominance (see Materials and Methods).
Figure 2.
An ENU-mutation/deletion map of the p region of mouse Chr 7. The lines below the chromosome (identified by the oval centromere at the left) represent a subset of a larger number of Del(p) deletions used to map new ENU-induced mutations; the name of each deletion is indicated above each line. The Del (ru2 p)46DFiOD deletion, used to recognize new mutations initially, is represented by the thick line. The boxes identify the seven loci in this region that were defined as a result of this screen. The black dots above the map denote new ENU-induced mutations, placed into deletion intervals by pseudodominance tests and into specific complementation groups by trans-complementation analyses. Brackets indicate unknown order of loci within the groupings indicated, and the stippled box indicates a possible uncertainty about the map position of l7R4 (see text). Human homology regions are shown in the boxes above the map, with the shaded box representing a break in homology regions and an uncertainty for the human chromosomal position of l7Rl1. No physical distance is implied by placement of any of the markers. A searchable locus list for this and all segments of the mouse genome can be found at http://www.informatics.jax.org.
Lethal Mutations with No Externally Visible Postnatal Phenotype.
A combination of deletion mapping and complementation analyses of the eight lethal mutations (i.e., no ruby, pink-eyed test-class recognized at weaning) provided evidence for four distinct loci, designated l7R2, l7R3, l7R4, and l7R6. Three of the loci (l7R2 through l7R4) were concluded to map in the most proximal segment of the p-deletion complex, because the Del(p)47DTD deletion (and less proximally extending ones as well) complemented mutant alleles at each locus (Fig. 2 and Table 2). The l7R3 locus was found to have two alleles, represented by mutations 735SJ and the more hypomorphic 951SJ (described above), because normal ru2 735SJ p/ru2 951SJ p compound heterozygotes could not be recovered in trans-complementation crosses. l7R6 is defined by three prenatally lethal, apparently fully expressed and nonleaky mutations 88SJ, 335SJ, and 2038SJ. No trans combination between any two of these mutations produced viable ru2 m1 p/ru2 m2 p offspring, showing that all three belonged to the same complementation group. Moreover, 88SJ, 335SJ, and 2038SJ behaved identically in deletion-mapping crosses, all mapping to the a deletion interval distal to the (l7R2, l7R3, l7R4) cluster but still within the human 11p14-p15 homology region (Fig. 2 and Table 2).
Table 2.
Deletion mapping of ENU-induced mutations
| Locus (mutation) | Del(p)
|
||||||
|---|---|---|---|---|---|---|---|
| 46DFiOD | 47DTD | 2MNURf | 8R250M | 3R30M | 6H | 4THO-II | |
| l7R2 (1318SJ) | 2 sm/829 | 5/19 | 5/25 | 5/27 | 7/25 | 8/27 | 5/20 |
| l7R3* (951SJ) | 9/698† | 6/32 | 8/31 | 6/39 | 8/36 | 3/11 | 11/26 |
| l7R4 (2033SJ) | 15/1085‡ | 3/27 | ND | 5/36 | 5/45 | 10/24 | 7/27 |
| l7R4 (2033SJ)§ | 0/40 | 14/97 | 13/66 | 16/82 | ND | ND | ND |
| l7R5 (403SJ) | 24/670¶ | 6/96 | 3/25 | 3/13 | 6/30 | 3/28 | 6/26 |
| l7R5 (403SJ)‖ | 13jv/64** | 23(20jv)/87‡‡ | 9/26 | ND | ND | ND | ND |
| l7R6†† (88SJ) | 2/805 | 0/34 | 0/40 | 1/50 | 10/36 | 10/45 | 3/16 |
| psrt§§ (723SJ) | 25sz/729 | 0/28 | 2sz/18 | 4/12 | 7/23 | 6/27 | 5/17 |
| fit2 (22SJ) | 103sm/917 | 11/40 | 5/23 | 11/49 | 12/43 | 6/38 | 10/49 |
In each case, females heterozygous for the Del(p) deletions shown [px/Del(p)] were crossed to an obligate carrier (progeny-tested) ru2 m p/ru2 + + male, with m representing a mutation at the indicated locus. The ratios represent the number of pink-eyed ru2 m p/+ Del(p) progeny in the total progeny observed at weaning (except for crosses to p
/Del(ru2 p)46DFiOD, in which the test class was ruby, pink-eyed—see ** below). Unless otherwise footnoted or designated, all test-class pink-eyed progeny were normal. Complementation would be indicated by the finding of 25% normal pink-eyed progeny. jv represents pink-eyed progeny that died between birth and weaning, the timing is refined by further footnoting below. sm denotes visibly runted pink-eyed animals, sz represents the typical seizure and extreme runting observed in psrt mutants, and ND denotes that the cross was not done.
Results for the l7R32R (951SJ) mutation are shown; data for the l7R31R (735SJ) mutation show a similar deletion-mapping pattern.
Seven test class were small and two were of normal size.
Five test class were small and 10 were of normal size.
Deletion-mapping crosses with different proved carrier males (ru2 l7R41Rp/ru2 + +) were repeated for the indicated deletions.
Fourteen were small and 10 were of normal size.
Deletion-mapping crosses with different proved carrier males (ru2 l7R51R p/ru2 + +) were repeated for the indicated deletions.
Of 13 ruby, pink-eyed progeny born (the test class for this particular deletion cross), all 13 were missing by 10 days of age and failed to thrive, indicating early selective juvenile loss of the ru2 l7R51R p/Del(ru2 p)46DFiOD mice.
Of 23 pink-eyed progeny born, 19 were missing by weaning and one was small at weaning and failed to thrive, indicating selective juvenile loss of the ru2 l7R51R p/+ Del(p)47DTD mice. We also observed three apparently normal pink-eyed animals at weaning.
Results for the l7R61R (88SJ) mutation are shown; data for the l7R62R (335SJ) and the l7R63R (2038SJ) mutations show a similar deletion-mapping pattern.
Results for the psrt1R (723SJ) mutation are shown, and most hemizygotes were missing by weaning. Data for the psrt2R (1060SJ) mutation indicate a similar deletion-mapping pattern.
Because we were able to recover ru2 l7R41R p/+Del(p)47DTD progeny, we consider the interval for l7R41R (2033SJ) indicated in Fig. 2 [i.e., proximal to the Del(p)47DTD proximal breakpoint] to be the most likely map position for this locus. However, we have observed in the deletion-mapping crosses fewer ru2 l7R41R p/+Del(p)47DTD progeny than might be expected [e.g., we obtained only three such progeny in 27 in the original crosses, and only 14 in 97 progeny in a set of repeat crosses (P < 0.005, by χ2 analysis for the latter crosses)]. Such lower-than-expected numbers could not be correlated with distal extents of deletions, and we saw no evidence of neonatal or juvenile loss of pink-eyed test classes, as was apparent in crosses involving l7R5 (see below). The next smaller proximally extending deletion, Del(p)2MNURf, complements l7R41R, as we observed no statistically significant difference from the expected number of pink-eyed progeny. Consequently, although we believe l7R4 maps proximally to the Del(p)47DTD proximal breakpoint, amid the [l7R2, l7R3, ru2] cluster, we have indicated in Fig. 2 an uncertainty in its map position because of this potential effect of the Del(p)47DTD deletion (possibly caused by a position effect) on the recovery of ru2 l7R41R p/+Del(p)47DTD progeny in mapping crosses.
Visible and Postnatally Lethal Mutations.
The deletion-mapping analyses provided evidence for at least three loci defined by visible or postnatally lethal mutations. First, the one viable runting mutation (22SJ) was complemented by all p deletions tested (Table 2) except Del(ru2 p)46DFiOD, which suggested that this locus, provisionally designated fit2 (fitness-2), mapped between the proximal breakpoints of the Del(ru2 p)46DFiOD and Del(p)47DTD deletions, amid the [l7R2, l7R3, l7R4 ru2] cluster (Fig. 2). However, fit21R (22SJ) was complemented by each of the other 10 mutations identified, indicating that it defines a separate complementation group.
Hemizygosity or homozygosity for either of two mutations (723SJ and 1060SJ) results in mice that remain in almost continual seizure and that invariably die before or around the time of weaning. Complementation crosses demonstrated that ru2 723SJ p/ru2 1060SJ p mice also exhibit the seizure phenotype, indicating that 723SJ and 1060SJ are both likely to be alleles of a single locus. Deletion-mapping results (Table 2) show that this locus, designated psrt (profound seizure and runting; alleles psrt1R and psrt2R, respectively), maps to a deletion interval proximal to l7R6 and Gas2 within the human 11p14-p15 homology region (Fig. 2). This is based on the observation that ru2 psrt1R p/+Del(p)2MNURf pink-eyed mice exhibit the seizure and runting phenotype, whereas ru2 psrt1R p/++Del(p)8R250M pink-eyed mice do not.
Deletion mapping placed the lethal 403SJ mutation (l7R51R) between the [l7R2, l7R3, l7R4, fit2, ru2] cluster and the psrt locus. l7R51R was complemented by Del(p)8R250M and Del(p)3R30M as well as by less proximally extending deletions. However, crosses of ru2 l7R51R p/ru2++ males to ++px/Del(p)47DTD females initially yielded an inconclusive result of six normal pink-eyed progeny in 96 classified progeny (Table 2). We therefore repeated the mapping crosses to the Del(ru2 p)46DFiOD, Del(p)47DTD, and Del(p)2MNURf deletions with different proved ru2 l7R51R p/ru2++ males. With closer examination of progeny before weaning, we found that ru2 l7R51R p/Del(p)47DTD and ru2 l7R51R p/Del(ru2 p)46DFiOD, but not ru2 l7R51R p/Del(p)2MNURf, mice were consistently dying between birth and weaning, typically before 10 days of age, indicating that neither the Del(p)46DFiOD nor Del(p)47DTD deletion complements l7R51R and that hemizygosity for l7R51R was postnatally lethal.
Discussion
We have described the recovery and genetic fine-mapping and complementation analyses of 11 recessive mutations mapping to seven loci within a large p-locus deletion in mouse Chr 7. The seven loci defined here by aberrant phenotypes provide relatively dense functional gene-annotation information for a small segment (perhaps 1–2 cM) of the corresponding human 11p14–15 homology region. Initial glimpses (e.g., http://genome.ucsc.edu) of the DNA sequence and predicted gene content of this human 11p14-p15 surrogately screened in this mouse mutagenesis experiment indicate a minimum of 20 transcription units. Further analysis of these transcription units, in terms of their expression patterns, the proposed biochemical functions of their protein products, and the deletion-map position of the mouse homologues, should inform strategies for mutation identification and gene-phenotype correlation(s).
Because ENU induces primarily single-base pair changes, recessive ENU-induced mutations can range from complete nulls to minor hypomorphs. Having a broad range of phenotypes present in an allelic series at one locus can be particularly useful for additional analyses, because one can often study the effects of minor perturbations on a system, or observe later-acting effects that are brought about by a hypomorphic, rather than a null, allele. A few examples from our own ENU mutagenesis work on mouse Chr 7, plus the experiences of many other groups, illustrate this point. For example, we recently reported (28) two new point-mutation alleles of the fumarylacetoacetate hydrolase (Fah) gene, with one mimicking the neonatal lethality of the acute form of human tyrosinemia, type 1 [much like deletion or knockout alleles (29, 30)]; and another, less-severe mutation mimicking the more chronic form of tyrosinemia in which pups live for several weeks before succumbing to the downstream effects of Fah insufficiency rather than its total absence. Likewise, ENU-induced allelic series with differing severities of effect at, for example, the eed (31–33), fit1 (34), Myo5a (35, 36), and Bmp5 (3) genes provide unique reagents with which to explore gene structure-function relationships.
The two alleles of l7R3 reported here manifest a considerable difference in homozygous phenotype; l7R32R (951SJ) appears to be “leaky” compared with l7R31R (735SJ). A similar hypomorphism or dose relationship may be exhibited by the fit21R (22SJ) mutation, in which homozygotes tend to be less severely affected than hemizygotes; however, we cannot rule out at the present time that this observed difference may be caused solely by the additive detrimental effects of the Del(ru2 p)46DFiOD deletion plus loss of function (either complete or partial) at the fit2 gene in fit2 hemizygotes. A similar situation may exist for both ENU-induced psrt alleles, in which homozygotes are more frequently found than are hemizygotes (Table 1). Another allelic series at l7R6, shows no outward evidence of a range in severity; however, a final conclusion must await closer phenotypic examination (such as a more detailed analysis of the time and/or cause of death of mice hemizygous and homozygous for each of the three alleles of l7R6).
The fine deletion mapping of ENU-induced mutations, such as that performed here and for Tyr-region mutations previously ascertained in a similar type of hemizygosity screen (14, 31, 37), provides a robust way of placing presumed point mutations defined solely by aberrant phenotype into evolving DNA sequence and transcription maps. Indeed, even if recessive mutations are initially recovered by other strategies (such as inversion-based regional homozygosity screens or whole-genome recessive-mutation screens) (8, 9, 13, 17), deletion mapping can be an important tool for correlating point-mutation maps with sequence maps. This is especially true for small genomic regions (such as the human 11p14-p15 homology region discussed here) where mutation ordering with respect to deletion breakpoints greatly facilitates the selection and analysis of candidate genes. New techniques of creating heritable deletions in embryonic stem cells, either by Cre-loxP-mediated chromosome engineering (38–40) or by radiation and negative selection (41–45), will allow the continued development of fine-structure point-mutation maps in other mutagenized regions of the genome that can then be correlated easily with DNA-sequence and transcription maps rapidly emerging from human and mouse genome projects.
Acknowledgments
We thank Drs. L. B. Russell, C. T. Culiat, and M. L. Klebig for comments on the manuscript. This work is currently sponsored by the Office of Biological and Environmental Research, U.S. Department of Energy under Contract DE-AC05–00OR22725 with UT-Battelle and was previously sponsored by the National Human Genome Research Institute (HG 00370).
Abbreviations
- ENU
N-ethyl-N-nitrosourea
- Chr
chromosome
References
- 1.Russell W L, Kelly E M, Hunsicker P R, Bangham J W, Maddux S C, Phipps E L. Proc Natl Acad Sci USA. 1979;76:5818–5819. doi: 10.1073/pnas.76.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson F M, Lewis S E. Proc Natl Acad Sci USA. 1981;78:3138–3141. doi: 10.1073/pnas.78.5.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marker P C, Seung K, Bland A E, Russell L B, Kingsley D M. Genetics. 1997;145:435–443. doi: 10.1093/genetics/145.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox R D, Hugill A, Shedlovsky A, Noveroske J K, Best S, Justice M J, Lehrach H, Dove W F. Genomics. 1999;57:333–341. doi: 10.1006/geno.1999.5804. [DOI] [PubMed] [Google Scholar]
- 5.Shedlovsky A, McDonald J D, Symula D, Dove W F. Genetics. 1993;134:1205–1210. doi: 10.1093/genetics/134.4.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nolan P M, Peters J, Strivens M, Rogers D, Hagan J, Spurr N, Gray I C, Vizor L, Brooker D, Whitehill E, et al. Nat Genet. 2000;25:440–443. doi: 10.1038/78140. [DOI] [PubMed] [Google Scholar]
- 7.Hrabe de Angelis M H, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, Marschall S, Heffner S, Pargent W, Wuensch K, Jung M, et al. Nat Genet. 2000;25:444–447. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- 8.Kasarskis A, Manova K, Anderson K V. Proc Natl Acad Sci USA. 1998;95:7485–7490. doi: 10.1073/pnas.95.13.7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hentges K, Thompson K, Peterson A. Development (Cambridge, UK) 1999;126:1601–1609. doi: 10.1242/dev.126.8.1601. [DOI] [PubMed] [Google Scholar]
- 10.Anderson K V. Trends Genet. 2000;16:99–102. doi: 10.1016/s0168-9525(99)01921-6. [DOI] [PubMed] [Google Scholar]
- 11.Shedlovsky A, Guenet J L, Johnson L L, Dove W F. Genet Res. 1986;47:135–142. doi: 10.1017/s0016672300022977. [DOI] [PubMed] [Google Scholar]
- 12.Shedlovsky A, King T R, Dove W F. Proc Natl Acad Sci USA. 1988;85:180–184. doi: 10.1073/pnas.85.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinchik E M. Mamm Genome. 2000;11:489–499. doi: 10.1007/s003350010095. [DOI] [PubMed] [Google Scholar]
- 14.Rinchik E M, Carpenter D A. Genetics. 1999;152:373–383. doi: 10.1093/genetics/152.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rinchik E M, Carpenter D A, Handel M A. Proc Natl Acad Sci USA. 1995;92:6394–6398. doi: 10.1073/pnas.92.14.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rinchik E M, Carpenter D A, Selby P B. Proc Natl Acad Sci USA. 1990;87:896–900. doi: 10.1073/pnas.87.3.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Justice M J, Noveroske J K, Weber J S, Zheng B, Bradley A. Hum Mol Genet. 1999;8:1955–1963. doi: 10.1093/hmg/8.10.1955. [DOI] [PubMed] [Google Scholar]
- 18.Justice M J, Zheng B, Woychik R P, Bradley A. Methods. 1997;13:423–436. doi: 10.1006/meth.1997.0548. [DOI] [PubMed] [Google Scholar]
- 19.Lyon M F, King T R, Gondo Y, Gardner J M, Nakatsu Y, Eicher E M, Brilliant M H. Proc Natl Acad Sci USA. 1992;89:6968–6972. doi: 10.1073/pnas.89.15.6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholls R D, Gottlieb W, Russell L B, Davda M, Horsthemke B, Rinchik E M. Proc Natl Acad Sci USA. 1993;90:2050–2054. doi: 10.1073/pnas.90.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rinchik E M, Bultman S J, Horsthemke B, Lee S T, Strunk K M, Spritz R A, Avidano K M, Jong M T, Nicholls R D. Nature (London) 1993;361:72–76. doi: 10.1038/361072a0. [DOI] [PubMed] [Google Scholar]
- 22.Russell L B, Montgomery C S, Cacheiro N L, Johnson D K. Genetics. 1995;141:1547–1562. doi: 10.1093/genetics/141.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson D K, Stubbs L J, Culiat C T, Montgomery C S, Russell L B, Rinchik E M. Genetics. 1995;141:1563–1571. doi: 10.1093/genetics/141.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walkowicz M, Ji Y, Ren X, Horsthemke B, Russell L B, Johnson D K, Rinchik E M, Nicholls R D, Stubbs L. Mamm Genome. 1999;10:870–878. doi: 10.1007/s003359901106. [DOI] [PubMed] [Google Scholar]
- 25.Lehman A L, Nakatsu Y, Ching A, Bronson R T, Oakey R J, Keiper-Hrynko N, Finger J N, Durham-Pierre D, Horton D B, Newton J M, et al. Proc Natl Acad Sci USA. 1998;95:9436–9441. doi: 10.1073/pnas.95.16.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhar M S, Johnson D K. Mamm Genome. 1997;8:143–145. doi: 10.1007/s003359900375. [DOI] [PubMed] [Google Scholar]
- 27.Culiat C T, Stubbs L, Nicholls R D, Montgomery C S, Russell L B, Johnson D K, Rinchik E M. Proc Natl Acad Sci USA. 1993;90:5105–5109. doi: 10.1073/pnas.90.11.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aponte J L, Sega G A, Hauser L J, Dhar M S, Withrow C M, Carpenter D A, Rinchik E M, Culiat C T, Johnson D K. Proc Natl Acad Sci USA. 2001;98:641–645. doi: 10.1073/pnas.98.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grompe M, al-Dhalimy M, Finegold M, Ou C N, Burlingame T, Kennaway N G, Soriano P. Genes Dev. 1993;7:2298–2307. doi: 10.1101/gad.7.12a.2298. [DOI] [PubMed] [Google Scholar]
- 30.Kelsey G, Ruppert S, Beermann F, Grund C, Tanguay R M, Schutz G. Genes Dev. 1993;7:2285–2297. doi: 10.1101/gad.7.12a.2285. [DOI] [PubMed] [Google Scholar]
- 31.Rinchik E M, Carpenter D A. Mamm Genome. 1993;4:349–353. doi: 10.1007/BF00360583. [DOI] [PubMed] [Google Scholar]
- 32.Holdener B C, Rinchik E M, Magnuson T. Mamm Genome. 1995;6:474–475. doi: 10.1007/BF00360658. [DOI] [PubMed] [Google Scholar]
- 33.Schumacher A, Faust C, Magnuson T. Nature (London) 1996;383:250–253. doi: 10.1038/383250a0. [DOI] [PubMed] [Google Scholar]
- 34.Potter M D, Shinpock S G, Popp R A, Godfrey V, Carpenter D A, Bernstein A, Johnson D K, Rinchik E M. Blood. 1997;90:1850–1857. [PubMed] [Google Scholar]
- 35.Huang J D, Cope M J, Mermall V, Strobel M C, Kendrick-Jones J, Russell L B, Mooseker M S, Copeland N G, Jenkins N A. Genetics. 1998;148:1951–1961. doi: 10.1093/genetics/148.4.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang J D, Mermall V, Strobel M C, Russell L B, Mooseker M S, Copeland N G, Jenkins N A. Genetics. 1998;148:1963–1972. doi: 10.1093/genetics/148.4.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rinchik E M, Carpenter D A, Long C L. Genetics. 1993;135:1117–1123. doi: 10.1093/genetics/135.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramirez-Solis R, Liu P, Bradley A. Nature (London) 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 39.Liu P, Zhang H, McLellan A, Vogel H, Bradley A. Genetics. 1998;150:1155–1168. doi: 10.1093/genetics/150.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng B, Mills A A, Bradley A. Nucleic Acids Res. 1999;27:2354–2360. doi: 10.1093/nar/27.11.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.You Y, Browning V L, Schimenti J C. Methods. 1997;13:409–421. doi: 10.1006/meth.1997.0547. [DOI] [PubMed] [Google Scholar]
- 42.You Y, Bergstrom R, Klemm M, Lederman B, Nelson H, Ticknor C, Jaenisch R, Schimenti J. Nat Genet. 1997;15:285–288. doi: 10.1038/ng0397-285. [DOI] [PubMed] [Google Scholar]
- 43.Thomas J W, LaMantia C, Magnuson T. Proc Natl Acad Sci USA. 1998;95:1114–1119. doi: 10.1073/pnas.95.3.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kushi A, Edamura K, Noguchi M, Akiyama K, Nishi Y, Sasai H. Mamm Genome. 1998;9:269–273. doi: 10.1007/s003359900747. [DOI] [PubMed] [Google Scholar]
- 45.Schimenti J C, Libby B J, Bergstrom R A, Wilson L A, Naf D, Tanrantino L M, Alavizadeh A, Lengeling A, Bucan M. Genome Res. 2000;10:1043–1050. doi: 10.1101/gr.10.7.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]