Abstract
In present studies, six Schiff bases were prepared, characterized and evaluated for their anti-tumor activity against the colorectal cancer cell line SW-480. The test compounds were characterized by various physico-chemical techniques such as M. P., TLC, UV, FT-IR, elemental analysis, 1H-NMR spectroscopy etc. and investigated for their non-covalent DNA binding potential. The electronic absorption and hydrodynamic studies expressed strong complementary evidence that the Schiff bases are binding between the narrow walls of the helical DNA grooves and were stabilized via electrostatic interactions through groove binding as the dominant binding mode. Moreover, these studies also revealed that the tested compound had significant non-covalent binding to chicken (ck) blood ds-DNA at blood pH (7.4) and body temperature 310 K: the calculated values of standard Gibbs free energy changes (ΔG = −RT ln Kf) for all compounds were negative which manifested the spontaneity of binding for all compounds. The cytotoxicity of the compounds was found through triplicate testing and the O. D. values were compared to find the percentage viability of the cells. The IC50 values of the compounds were estimated through dose-dependent curves. HSB3, HSB4 and HSB1 showed relatively potent anti-cancer activity with IC50 values of 7.0913 μg mL−1, 17.1469 μg mL−1 and 17.5254 μg mL−1, respectively. The same compounds had also exhibited relatively better ds-DNA binding efficacy with binding constant values (9.1 × 105, 3.5 × 105 and 5.13 × 104 respectively).
Synthesis, characterization, ds-DNA binding and anticancer studies of synthesized Schiff base.
1. Introduction
The perpetual battle to discover and introduce significant non-toxic anti-tumor drugs continues to establish sustainable health standards as per the World Health Organization (WHO)'s charter. With an ever-increasing annual number of deaths due to cancer, this crucial issue has attained the unique focus of the entire research and development industry throughout the entire world. As per the WHO's estimates, cancer is the second most common cause of death, accounting for 9.6 million deaths in 2018, causing around one in six deaths worldwide.1 Cancer is a complex disease that develops in multiple stages, due to uncontrolled replication of cells by the mutation caused in the genetic structure of normal cells.2–4 Normal cells become malignant and stop apoptosis, form larger populations and begin invasion into neighboring organs and also circumvent cell checkpoints.5,6 While early diagnosis and treatment can help manage cancer, delayed detection and advanced stages can be life-threatening.7 Malignant cells may develop multidrug resistance (MDR), a condition in which they become resistant to medicine, affects more than 270 different forms of cancer, causing a relapse after initial treatment.8–10 Apoptosis, a natural death mechanism of injured, or abnormal cells which inhibits the growth of cancer, is suppressed in malignant cells can be reactivated by anticancer medications. To cure cancer with minimal side effects, new anticancer drugs are still of utmost relevance to the global scientific community.11
It is anticipated that there would be over 35 million new cases of cancer in 2050, a 77% increase over the projected 20 million cases in 2022.12 Therefore, development of therapeutic drugs that target MDR cancer cells and disease relapse are major challenges in anticancer drug research and development. Schiff bases are considered as one of the most significant compounds extensively studied in this field since these are diverse in structures, cost-effective and easy to synthesize.8
Schiff bases are organic compounds synthesized from condensation of an aldehyde or a ketone with primary amine.13–15 The biologically active Schiff bases have diverse medicinal applications including anticancer and antiviral activities.16–18 Presence of imine group and other donor functional groups like –Cl, –OH, –CH3, play a key role in tuning their biological activities.19–21 Due to their adaptability and simplicity of production, these are common ligand precursors for various applications.22 The various reported medications such as thioacetazone (antituberculosis), nifuroxazide (antibiotic) and dantrolene (muscle relaxant) contain Schiff base functionality.23 Significant and targeted anticancer activity against BT549 and A549 cells was reportedly demonstrated by a novel Schiff base that was synthesized from 1,10-phenanthroline-2,9-dicarboxaldehyde and from 2,2′-bipyridyl-4,4-dialdehyde.24
According to reports, Schiff bases derived from benzaldehyde have exhibited cancer inhibition against various human cell lines. Anti-cancer activity of new promising class of Schiff bases and azo dyes of pyranquinolinone derivatives against MCF-7 breast cancer, HepG2 liver cancer and HCT-116 colon carcinoma in comparison with a standard drug known as 5-fluorouracil was used as a reference drug.25 Due to its extensive biological implications, including its potential in combating cancer, advancements in bio-inorganic chemistry have garnered significant attention to the azomethine compound and its corresponding complexes. Researchers are captivated by the antitumor properties exhibited by Schiff base and its complexes against a range of tumor cell lines and are striving to develop novel, side-effect-free anticancer medications as a result.
In present studies, an attempt is made to synthesize, characterize, and investigate the interaction of six Schiff bases with chicken blood double stranded (ds)-DNA using UV-visible spectroscopy and hydrodynamic technique at blood (7.4) pH under body temperature (37 °C). These compounds were also investigated for their antitumor potential against the SW-480 colorectal cell line. The viability of cells was evaluated using the MTT assay and the optimal cell density was determined after assessing cytotoxicity in triplicate. To calculate the IC50 values, a dose-dependent curve was employed.
2. Materials and methods
Chemicals used during the reactions are: 4-(diethylamino)-2-hydroxybenzaldehyde (98%), benzaldehyde (97%), 4-nitro-1,2-phenylenediamine (97%), ortho-tolidine (98%), 2,4,6-trimethylaniline (99%),1-naphthylamine (97%) and cinnamaldehyde (99%) have been purchased from Sigma Aldrich (USA). Solvents of analytical grade were used without any purification include ethanol, methanol, ethyl acetate, n-hexane, dichloromethane, acetone, and chloroform. The chicken blood was obtained from chickens during slaughter via neck cut under “perfectly bled method” to have maximum blood discharge and collection for ds-DNA extraction. The Cell Lines were purchased from ATCC (American Type Culture Collection) SW480 (CCL-228). Chemicals were weighed using electronic analytical balance ATY 224, Japan. Progress of reaction was checked by taking TLC at regular intervals and seen under UV lamp. The Bauchi Rotavapor R-210 was used to evaporate different solvents from reaction mixtures. The melting point apparatus SMP10 was employed to find the melting points of the final synthesized products. The functional groups present in the end products were investigated using FT-IR equipped with an ATR model ALPHA 20488. The 1H-NMR was conducted using a 500 MHz BRUKER spectrometer in DMSO as a solvent. The microanalysis was carried out using CHNS/O Analyzer 2400 Series PerkinElmer Company prepared by Soung Ryu & Gwen Tenney 2005.
2.1. General procedure for synthesis of compounds (HSB1–HSB6)
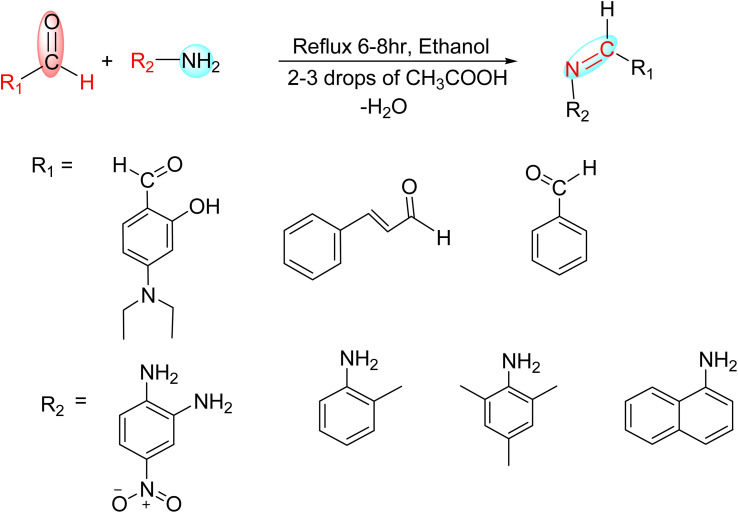
All the targeted Schiff bases were synthesized by condensing stoichiometric ratios of corresponding aldehyde and amine in dry ethanol under reflux at slight acidic pH (6–6.5) (Scheme 1) in the light of reported methods.26,27 In case of HSB1 to HSB4, 4-(diethylamino)-2-hydroxybenzaldehyde (10 mmole, 1.93 g) was treated with equivalent masses of corresponding amines (4-nitro-1,2-phenylenediamine (5.0 mmole, 0.76 g, 2 : 1 mole ratio); 2,4,6-trimethylaniline (10 mmole, 1.35 g); ortho-tolidine (10 mmole, 1.7 g); and 1-naphthylamine(10 mmole, 1.43 g) respectively) whereas for HSB5 and HSB6, 4-nitro-1,2-phenylenediamine (5.0 mmole, 0.76 g) was condensed with 2 equivalent of benzaldehyde (10 mmole, 1.06 mL)and cinnamaldehyde (10 mmole, 1.32 g). Both aldehydes and amines were dissolved in 10 mL dry ethanol and inter-mixed in dry 25 mL two neck round bottom flask duly fitted with reflux condenser. After mixing, the pH of final reaction mixture was slightly acidified by adding 2–3 drops of glacial acetic acid to have final pH (6–6.5). The reaction mixture was refluxed for about 6–7 hours depending upon the progress of reaction, which was monitored by TLC (10% methanol : chloroform) after regular intervals till completion of reaction. Upon completion of reaction as indicated by TLC, the reaction mixture was filtered and before drying, the solid products were washed by cold ethanol (three time) and then with ethyl acetate. These synthesized compounds were then characterized by elemental analysis, FTIR, and NMR spectroscopy.
Scheme 1. General synthesis of Schiff bases.
2.2. Biological evaluation
The antitumor assay was evaluated on SW-480 colorectal cell line using MTT assay.28 Ninety-six well plates containing DMEM media supplemented with 10% fetal bovine serum and 1% antibiotics (penicillin G and streptomycin) were used to culture the cells. The plates were then incubated for twenty-four hours at 37 °C with 5% CO2 and humidity. Trypsinization was performed once the actively dividing cells formed a confluent monolayer. A cell suspension of 105 cells per milliliter was then seeded in the wells having culture media and varying concentrations of HSB1 to HSB6 (200 μg mL−1, 100 μg mL−1, 50 μg mL−1, 25 μg mL−1, 12.5 μg mL−1, 6.25 μg mL−1). After that, the plate was incubated in a 5% CO2 atmosphere for 48 hours at 37 °C. Following incubation, the cellular viability of each derivative concentration was assessed. Next, 20 μL of MTT (5 mg mL−1 PBS) was added to each well and the plates were incubated once more for three hours at 37 °C in a 5% CO2 atmosphere. Following the incubation period, 100 μL of DMSO was added to dissolve the formazan crystals that were formed by metabolically active cells. The medium was then carefully removed. Using a micro plate reader, the optical density (O. D.) of the wells was determined at 570 nm, with a reference wavelength of 655 nm. An IC50 was calculated using a dose-dependent curve. The growth inhibition rate for each dilution was calculated by the following formula:
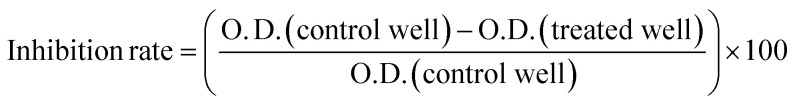 |
1 |
2.3. DNA binding studies
2.3.1. UV-visible absorption spectroscopic measurements
The normal electronic absorption spectra of different test compounds were recorded on Shimadzu 1800 UV-visible spectrophotometer using matched quartz cuvettes of 3 mL capacity. The extinction coefficients were calculated using Lambert Beer law to optimize the concentrations of all pro-drug candidates for DNA non-covalent binding studies. The ds-DNA extracted from chicken blood was dissolved in the double distilled autoclaved water for making ds-DNA stock solution of 5.4 × 10−5 M strength while concentration of DNA was spectrophotometrically calculated by using its optical density at 260 nm and molar extinction coefficient ε260 = 6600 cm−1 M−1. The purity of DNA was checked by taking a ratio of optical density (O. D.) at 260 nm to that at 280 nm, O. D260/O. D280 > 1.8 and O. D260/O. D280 < 2.1. The optimized constant concentrations of the test compounds were subjected to UV-visible spectroscopic titrations at body temperature (310 K) and stomach pH (7.4) by varying concentrations of ds-DNA (0–5 μM) under standard addition method using 0.1 M sodium phosphate buffer. In measuring each absorption spectrum, the mixture was allowed to incubate for 10 minutes within the cell cavity to assure the formation of compound–DNA adduct at the required temperature before recording the related spectrum.
2.3.2. Hydrodynamic measurements
To further confirm the binding mode of the test Schiff bases, viscosity measurements were conducted by using an Ubbelodhe Viscometer. The temperature was kept at 310.0 ± 0.1 K by immersing viscometer in a thermostat water bath. Titrations were performed by keeping DNA concentration constant at 4 μM and steadily increasing concentration of test compounds from 0.4 μM to 4 μM with an increment of 0.4 μM.
3. Results and discussion
3.1. Physical data
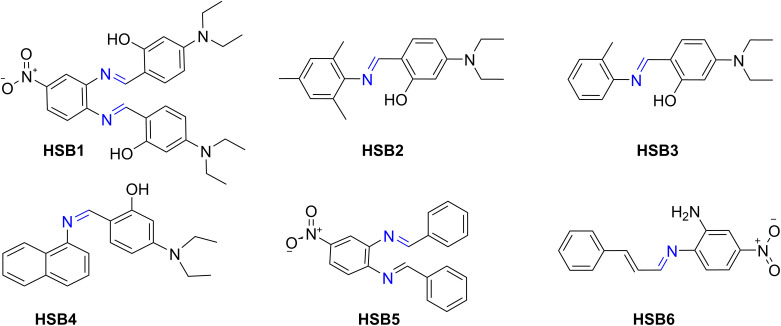
The physical data of all the compounds (HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6) regarding atom economy, yield, color, melting point, and solubility are given in Table 1. The structures of all the compounds are given in Fig. 1.
Table 1. Physical data of synthesized compounds (HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6).
| Sr. no. | Percentage atom economy | Percentage yield | Color | M. P.a (°C) | Solubility |
|---|---|---|---|---|---|
| HSB1 | 93.24 | 84 | Orange | 205 | Acetone, ethanol (hot) and DMSO |
| HSB2 | 94.14 | 84 | Yellow | 80 | Acetone, ethanol (hot) and DMSO |
| HSB3 | 93.54 | 85 | Yellow | 98 | Acetone, ethyl acetate and DMSO |
| HSB4 | 94.58 | 76 | Green | 125 | Acetone, ethanol (hot) and DMSO |
| HSB5 | 90.14 | 84 | Yellow | 199 | Acetone, ethanol (hot) and DMSO |
| HSB6 | 93.68 | 87 | Yellow | 200 | Acetone, ethanol (hot) and DMSO |
Melting point (M.P).
Fig. 1. Structures of synthesized compounds (HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6) where as HSB1: 6,6'-((1E,1'E)-((4-nitro-1,2-phenylene)bis(azanylylidene))bis(methanylylidene))bis(3-(diethylamino)phenol), HSB2: (E)-5-(diethylamino)-2-((mesitylimino)methyl)phenol, HSB3: (E)-5-(diethylamino)-2-((o-tolylimino)methyl)phenol, HSB4: (Z)-5-(diethylamino)-2-((naphthalen-1-ylimino)methyl)phenol, HSB5: (N1E,N2E)-N1,N2-dibenzylidene-4-nitrobenzene-1,2-diamine, HSB6: (E)-4-nitro-N1-((E)-3-phenylallylidene)benzene-1,2-diamine.
3.2. Microanalysis
Elemental analysis of all synthesized Schiff bases was carried on PerkinElmer CHNS Analyzer 2400 Series. Properly weighed and encapsulated samples were subjected to combustion in a pure oxygen environment. The evolutions of resultant combustion gases were measured in an automated fashion to assess the weight% composition of CHN. The same weight% composition was used to set up chemical formula. So, the results of net calculations for molecular formula, molecular weight and yield of synthesized compounds are given in Table 2.
Table 2. Microanalysis data of synthesized Schiff bases (HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6)a.
| Compound | Calculated (%) | Found (%) | Molecular formula | Molecular weight (g mol−1) | ||||
|---|---|---|---|---|---|---|---|---|
| C | H | N | C | H | N | |||
| HSB1 | 61.64 | 6.90 | 16.04 | 61.20 | 5.80 | 16.4 | C28H33N5O4 | 503 |
| HSB2 | 77.38 | 8.44 | 9.02 | 77.10 | 8.20 | 8.94 | C20H26N2O | 309 |
| HSB3 | 76.56 | 7.85 | 9.92 | 76.20 | 7.70 | 9.60 | C18H22N2O | 281 |
| HSB4 | 79.21 | 6.96 | 8.80 | 79.01 | 6.70 | 8.60 | C21H22N2O | 318 |
| HSB5 | 64.28 | 4.20 | 17.45 | 64.10 | 4.10 | 17.20 | C20H15N3O2 | 329 |
| HSB6 | 72.82 | 5.65 | 15.79 | 72.60 | 5.20 | 15.20 | C15H13N3O2 | 267 |
Elemental analysis for C, H, N are within ±0.4% of the theoretical values.
3.3. Spectroscopic analysis
The FTIR spectrum of all the synthesized Schiff bases showed common characteristic absorption band between 1570 to 1620 cm−1 for C N– which exclusively indicate the formation of Schiff base. Similarly, 1H-NMR of all the bases showed characteristic singlet peak between 7.5 to 8.7 ppm which confirmed the presence of azomethine proton and formation of Schiff bases.29 The complete FTIR and 1H-NMR spectra of all the compounds are given in the ESI† whereas the overall NMR spectral data of the synthesized compounds is summarized as follows:
3.3.1. HSB1
FT-IR, (cm−1), O–H (3229), C–H (3000), C N (1577), NO2 (1518, 1390), C–N (1327), C–O (1130). 1H-NMR, (DMSO, Hz, ppm): 1.15 (6H, t, J = 7.0), 3.5 (4H, q, J = 7), 6.1 (1H, d, J = 2), 6.3 (1H, dd, J = 9, 2.5), 6.5 (2H, s), 6.8 (1H, d, J = 9), 7.5 (1H, J = 9), 7.85 (1H, d, J = 2.5), 7.9 (1H, dd, J = 8.5, 2.5), 8.7(1H, s).
3.3.2. HSB2
FT-IR, (cm−1), O–H (3510–3280), C–H (2968), C N (1596), C C (1520), C–N (1339), C–O (1128). 1H-NMR, (DMSO, Hz, ppm): 1.1 (6H, t, J = 6.5), 2.1 (6H, s), 2.2 (3H, s), 3.5 (4H, q, J = 6.5), 6.1 (1H, s), 6.3 (1H, d, J = 6.5), 6.8 (2H, s), 7.3 (1H, d, J = 6.5), 8.3 (1H, s).
3.3.3. HSB3
FT-IR, O–H (3093), C–H (2968), C N (1604), C C (1517), C–N (1346), C–O (1211). 1H-NMR (DMSO, Hz, ppm): 1.15 (6H, t, J = 7), 2.3 (3H, s), 3.4 (4H, q, J = 7), 6.05(1H, d, J = 2.5), 6.3(1H, dd, J = 8.5, 2.5), 7.15 (1H, m), 7.25(3H, m), 7.35 (1H, d, J = 9), 8.7 (1H, s).
3.3.4. HSB4
FT-IR, O–H (3055), C–H (2964), C N (1619), C C (1514), C–N (1343), C–O (1132). 1H-NMR (DMSO, Hz, ppm):1.3 (6H, t, J = 6.9), 3.5 (4H, q, J = 6.9), 6.34 (2H, m), 7.2 (1H, dd, J = 7.2, 0.6), 7.3 (1H, m), 7.5 (3H, m), 7.75 (1H, d, J = 9, J = 9), 7.9 (1H, m), 8.38 (1H, m), 8.5 (1H, s).
3.3.5. HSB5
FT-IR, C N (1612), C C (1569), NO2 (1486, 1361), C–N (1281). 1H-NMR (CDCl3, Hz, ppm), 6.8 (3H, m), 7.6 (2H, m), 7.9 (1H, dd, J = 7.2, 1.6), 8.1 (2H, m), 8.85 (1H, s).
3.3.6. HSB6
FTIR data (cm−1) R, O–H (3055), C–H (2964), C N (1619), C C (1514), C–N (1343), C–O (1132) 1H-NMR (CDCl3, Hz, ppm),5.1 (2H, s), 6.7 (1H, d, J = 12), 7.2 (1H, q, J = 11.6), 7.3 (2H, m), 7.5 (3H, m), 7.6 (2H, m), 8.1 (1H, m), 8.5 (1H, d, J = 12).
3.4. In vitro anticancer screening
Discovery of the anti-cancer activity of different Schiff bases and their metal complexes against various cancer cell lines intrigues the researchers to develop new anticancer drugs with reduced side effects. To evaluate the anti-cancer potential of synthetic compounds (HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6), we conducted 3-(4,5-dimethyl2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay on colorectal cancer cell line (SW-480). More than 1.85 million instances of colorectal cancer (CRC) are reported each year, making up 9.8% of all cancer cases and there will be approximately 3.2 million cases of colorectal cancer in 2040.30 In this study, doxorubicin was used as a standard anti-cancer drug. Table 3 depicts the IC50 values of all synthetic Schiff base compounds and their comparison with standard drug. It was found that 5 out of a total of six synthetic compounds (HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6) with IC50 values of 17.1469 μg mL−1, 34.0821 μg mL−1, 7.0913 μg mL−1, 17.5254 μg mL−1 and 34.0335 μg mL−1, respectively, demonstrated potent anticancer activity against the SW-480 cell line. Among these 5 compounds, HSB3 exhibited promising anticancer activity when compared to the standard drug Doxorubicin. The findings of this study reveal the promising anti-cancer activity of imine Schiff-bases, which were synthesized through the condensation of 4-(diethylamino)-2-hydroxybenzaldehyde, cinnamaldehyde and 4-(dimethyl amino) benzaldehyde with four distinct primary amines using the simple and facile method. The results of this research accentuate the need for extensive screening of these newly developed Schiff-bases derivatives against cancer, particularly colorectal carcinoma. Further mechanistic studies have shown that the synthesized Schiff base compounds produced anticancer activity by inducing apoptosis or cell cycle arrest in SW-480 cancer cells. These substances regulate various tumor cells through a variety of mechanisms, such as tumor-induced angiogenesis, apoptosis, mitochondrial permeabilization, blocking of signal transduction pathways by blocking important enzymes, cytomorphological changes brought on by disruptions in cellular differentiation, compounds intercalating with duplex DNA, suppression of survival signaling, pore formation that causes cancer cell membrane leakage, substantial rise in the sub-G1 population, G2/M phase cell cycle arrest, inhibition of mitotic spindle formation, disruption of the membrane fatty acids, cation carrier, ion channel formation and survival signaling suppression.31 The structure activity relationship (SAR) analysis revealed that the ligand structure and different functional groups pose great variation in making any Schiff base a potent anticancer drug. The mechanism being involved in the cytotoxic activity of these compounds is not fully understood, but it is likely that they induce cell death by inhibiting critical cellular processes such as DNA synthesis, cell proliferation and cell cycle progression.32–35
Table 3. IC50 data of (HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6) against SW-480 (colorectal cancer) cancerous cell line.
| IC50 (μg mL−1) against cell lines | |
|---|---|
| Compounds | SW 480 |
| HSB1 | 17.2414 |
| HSB2 | 34.3335 |
| HSB3 | 7.0913 |
| HSB4 | 17.1454 |
| HSB5 | 34.6754 |
| HSB6 | 42.422 |
| Standard drug (doxorubicin) | 2.3159 |
The synthesized Schiff bases in recent study proved promising anticancer activity against the colorectal cancer cell line (SW-480). This suggests that these compounds could potentially serve as ligands for the development of new and effective anticancer agents. However, further studies are necessary to elucidate the underlying mechanism of their cytotoxic activity and to improve their structure for improved potency and selectivity.
3.5. DNA binding studies
3.5.1. Absorption spectra of synthesized compounds
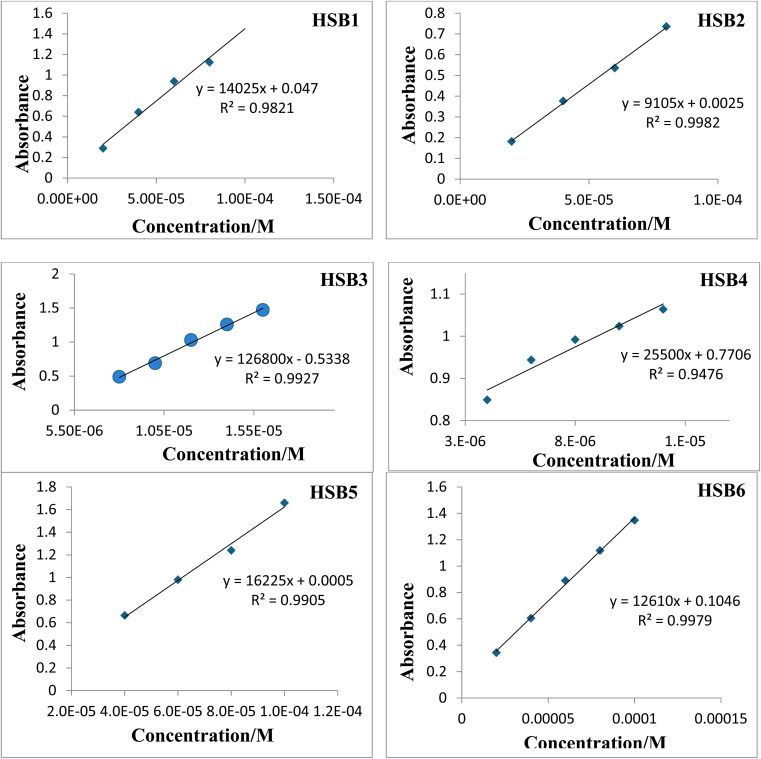
Initially full range (200 nm to 800 nm) UV-visible spectra of all Schiff bases (HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6) were carried out in 4% aqueous CH3OH and internal transitions (π–π*, n–π*) of the test compounds were observed in UV-region as the most appropriate and intense peaks at 258 nm, 370 nm, 259 nm, 258 nm, 265 nm and 261 nm respectively. The absorption spectra of each compound were separately recorded with gradually increasing concentration of compound as a function of absorbance. The plot of absorbance at λmax for each concentration versus concentration of each compound showed that absorbance increases linearly with the increasing concentrations as reflected by the values of linear regression: R2 = 0.978, 0.998, 0.993, 0.948, 0.990, 0.998 for HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6, respectively. The molar extinction coefficients in units of cm−1 M−1 as found from the slope values of test compounds (HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6) are 148 083, 9146, 84 658, 11 122, 16 231, 14 036, respectively. Therefore, in terms of the Burawoy classification, absorption bands in all synthesized compounds can be assigned one of the following possible empirical designations:
(1) B (fine structure benzenoid) and E (ethylenic) bands (local excitation or LE bands) for π–π* transitions.
(2) K band for π–π* transitions involving a conjugation group (electron transfer or ET-band).
(3) R band for n–π* transitions when a group on aromatic ring has non-bonded electron pairs.
Experimentally determined values of λmax in the pure UV range from 258 nm to 370 nm and molar extinction values predicted that both K and R bands for respective transitions are operative for all bases.36
3.5.2. DNA binding study by UV-spectroscopy
Both the qualitative and quantitative investigation of binding small molecules to DNA is classically carried out by UV-absorption spectroscopy as a unique complement to other techniques such as cyclic voltammetry, FT-IR, fluorescence spectroscopy etc.37 In UV-spectroscopic titrations, the physical inspection of electronic absorption spectra of the free compound and its DNA adduct usually show difference either in magnitude of absorption intensity called chromic effect or in position of maximum absorption (λmax) called chromic shift (Δλ). Additionally, one or more additional bands might also emerge, possibly corresponding to compound–DNA adduct intermolecular charge transfer transitions.38 Upon interaction with DNA, either blue or red chromic shift (hypos chromic or bathochromic shift) of the maximum absorption or chromic effect in the form of change in magnitude of absorption intensity may be observed separately or in combination.39 The apparent change in the magnitude of absorption intensity without any noticeable wavelength shift of absorption maxima in absorption spectra of small molecules is an illustration of groove binding or electrostatic interaction40 whereas shift of the absorption maxima point out the presence of π-stacking interaction binding mode called hydrophobic interaction or intercalation. Peculiarly π-stacking interaction, the most common binding mode between the compound and DNA base pairs results either in hypos chromic shift plus hyperchromic effect or in bathochromic shift plus hypochromic effect.41
Spectrophotometric responses of HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6 were investigated at physiological pH (7.4) and at 310 K by using solution of phosphate buffers, showed no variation in the wavelengths, which could indicate non-interactive behavior of these compounds with buffer solution.
UV-spectra of each synthesized Schiff base were separately recorded with gradual addition of DNA (1–5 μM) in the constant and optimized concentration of the test compound (HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6) (concentration of compounds as per Benesi–Hildebrand) under standard addition method. During all experiments, the concentration of the test compounds was kept constant at their optimized level in the sample cell, while the concentrations of DNA were varied (1–5 μM) both in the sample as well as in the reference cell. The changes in the absorption spectra with the addition of DNA indicate the formation of Schiff base-DNA adduct. Thus, the dynamic equilibrium so established, can be generally represented as:
| Schiff base + DNA ⇌ Schiff base − DNA | 2 |
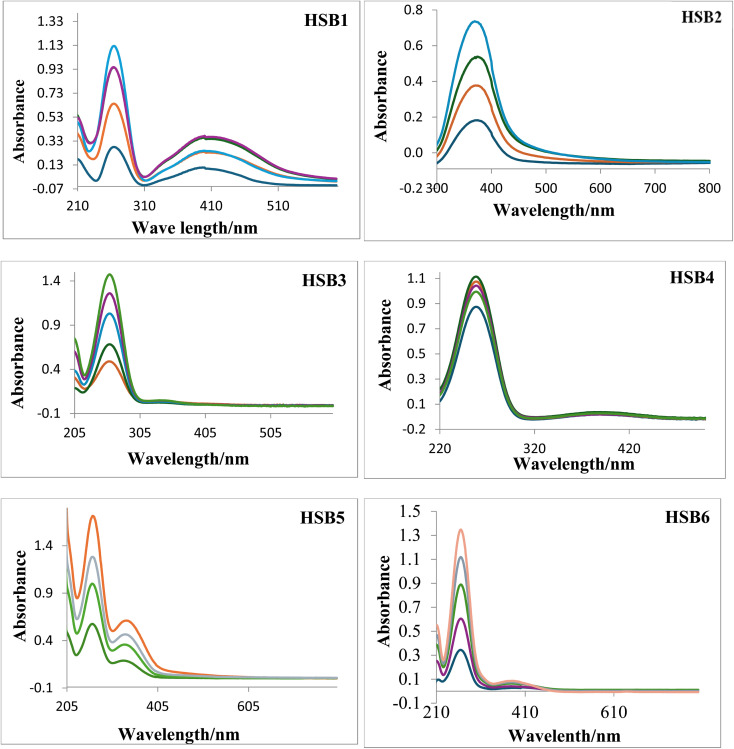
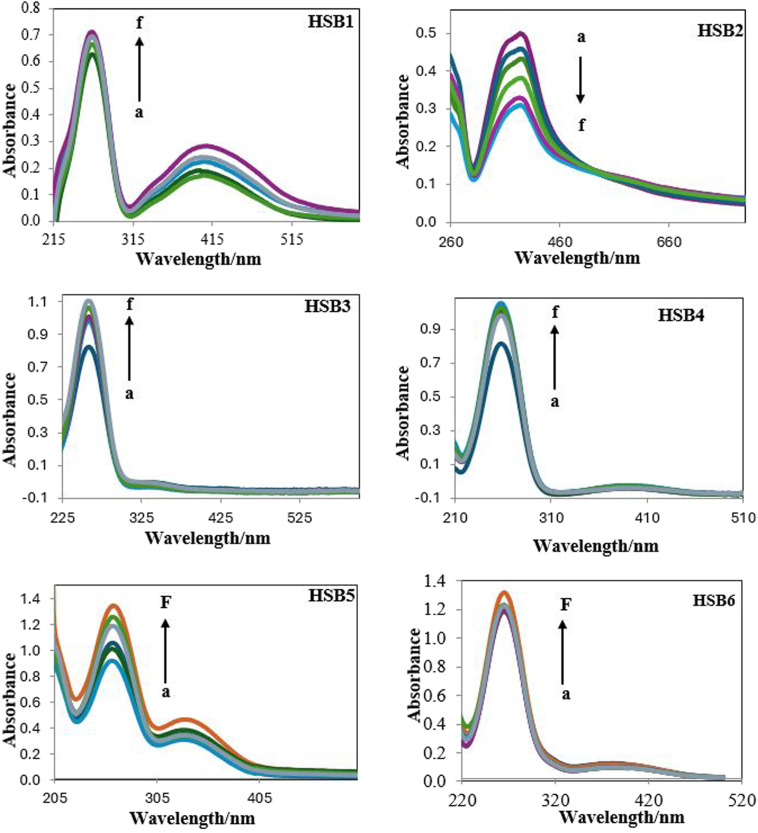
The effect of the varying concentration of DNA on the electronic absorption spectra of the test compounds at pH 7.4 (0.1 M phosphate buffer) and at 310 K is shown in Fig. 2, respectively. Overall, an increase of DNA concentration for all Schiff bases resulted in the net increase of absorbance (hyperchromic) as well as a negligible or no chromic shift except for HSB2 where steady increase of DNA concentration has resulted in decrease in absorbance intensity (hypochromic) with no hypos/or bathochromic shift (Fig. 3). These pronounced hyperchromic effects (in case of HSB1, HSB3, HSB4, HSB5 and HSB6) show the interaction of the electronic states of the non-classically intercalating chromophores of the investigated compounds and those of the stacked base pairs of ds-DNA. In the same way, a decrease in face-to-face base stacking (exposed electrons) results in an increase in absorption intensity (hyperchromic).40,42 Whereas in case of HSB2 decrease in absorbance intensity (hypochromic) is observed as the coupling of π-orbital is partially filled with electrons, thus decreasing transition probabilities and concomitantly resulting in hypochromic.43–45 Percentage of chromicity for compounds (HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6) were calculated by using following relation:
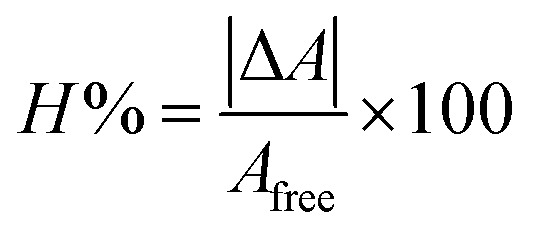 |
3 |
where ΔA = Afree − Abound
Fig. 2. UV-spectra of the investigated compounds at different concentrations.
Fig. 3. Concentration profiles of HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6 in terms of their linear regression values and extinction coefficient.
The binding constant Kf of each Schiff base-DNA adduct was evaluated spectrophotometrically by applying double reciprocal Benesi–Hildebrand equation on the optimized band of each drug [HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6]:
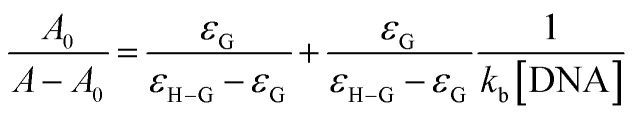 |
4 |
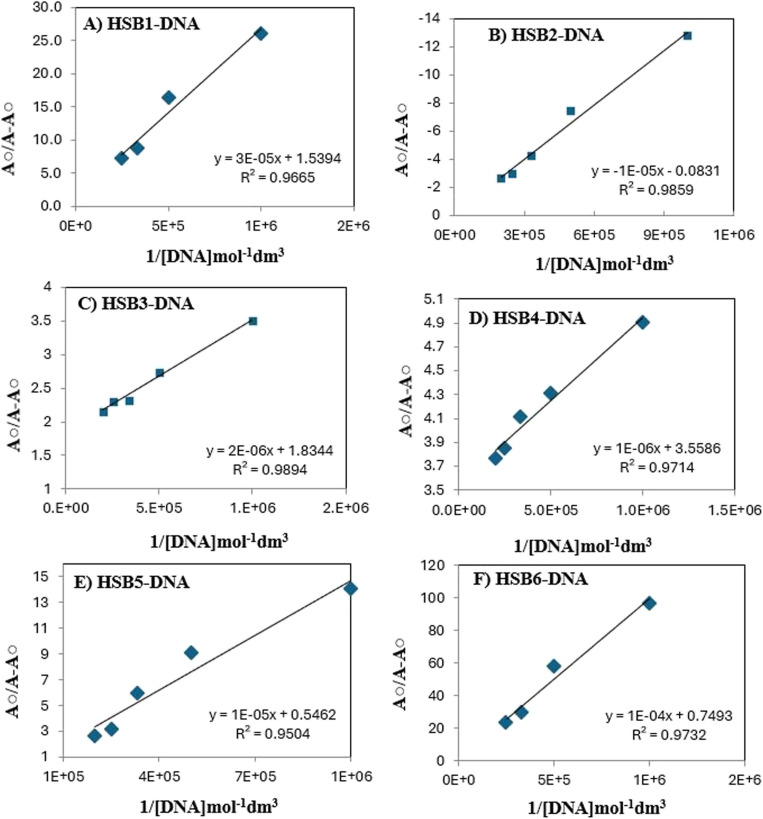
where the molar extinction coefficients of the free drug and complex are denoted by EG and EH–G, respectively, while the absorbances of the free Schiff base and complex are represented by A0 and A. Based on linear regression and almost equivalent size, it is presumed that the adduct of pro-drug candidate (small molecule) with DNA (per nucleotide phosphate) on the DNA polynucleotide chain is to be a kind of 1 : 1 complex. From the plot of A0/(A − A0) to 1/[DNA], the ratio of the intercept to the slope gives the binding constant “Kf”.
The binding constant values, calculated at pH values of 7.4 and 310 K temperature are given in Fig. 4, 5 and Table 4. The Kf values indicate that binding affinity of HSB3 is greater than that of other synthesized compounds. The orders of formation constants at pH 7.4 were found as follow:
Fig. 4. UV-spectra of HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6 (10 × 10−6 mol dm−3) in the absence and presence of different concentrations of DNA at pH 7.4. Arrows show status of relative absorption intensity on incremental addition of different concentrations (1 × 10−6 mol dm−3) of DNA (a) 0, (b) 1, (c) 2, (d) and (e) 4 (f) 5 into the constant concentration of each Schiff base under standard addition method.
Fig. 5. Plot of A0/A − A0vs. 1/[DNA] mol−1 dm3 for the application of Benesi–Hildebrand equation for calculation of (A) HSB1–DNA, (B) HSB2–DNA, (C) HSB3–DNA, (D) HSB4–DNA, (E)HSB5–DNA and (F)HSB6–DNA formation constants at pH 7.4/temperature 310 K.
Table 4. Binding constant values for HSB1–DNA, HSB2–DNA, HSB3–DNA, HSB4–DNA, HSB5–DNA and HSB6–DNA complexes from UV-spectrophotometric data at 7.4 and at body temperature (310 K)a.
| Compound | Photophysical properties Schiff base-DNA adducts | Binding constant Kf/M−1 | Free energy (−ΔG) kJ mol−1 | ||||
|---|---|---|---|---|---|---|---|
| Absorption maxima (λmax) | Chromic effect/change in absorption intensity | ||||||
| Free | Bound | Free | Bound | H% (hypo/or hyperchromicity) | |||
| HSB1 | 263 | 263 | 0.626 | 0.712 | 13.74% (hyperchromicity) | 5.13 × 104 | 27.95 |
| HSB2 | 387 | 387 | 0.499 | 0.310 | 37.33% (hypochromicity) | 8.31 × 103 | 23.26 |
| HSB3 | 258 | 258 | 0.834 | 1.073 | 28.74% (hyperchromicity) | 9.1 × 105 | 34.02 |
| HSB4 | 259 | 259 | 0.824 | 1.043 | 26.58% (hyperchromicity) | 3.5 × 105 | 32.94 |
| HSB5 | 261 | 261 | 0.91 | 1.33 | 37.78% (hyperchromicity) | 4.09 × 104 | 27.37 |
| HSB6 | 265 | 265 | 1.163 | 1.213 | 4.29% (hyperchromicity) | 7.49 × 103 | 23.05 |
K f – binding constant; ΔG – Gibbs free energy.
K HSB3 > KHSB4 > KHSB1 > KHSB5 > KHSB2 > KHSB6 (pH 7.4).
3.5.3. Calculation of free energy
From the data of bind affinity (binding constant “Kf”), free energy “−ΔG/kJ mol−1” of compound–DNA complex were calculated (Table 4). Formation constants are measure of stability of compound–DNA complexes while free energy “ΔG” indicates the spontaneity of binding of compounds with DNA. The formation constant and free energy are affected by the change of pH.
| ΔG = −RT ln K (kJ mol−1) | 5 |
The order for the values of free energy ΔG of the compounds at pH 7.4 are as follows:
ΔGHSB3 > ΔGHSB4 > ΔGHSB1 > ΔGHSB5 > ΔGHSB2 > ΔGHSB6
These results show that HSB3 binds to the DNA more spontaneously as compared to the others at pH 7.4 and 310 K.
3.5.4. Hydrodynamic approach
Internal resistance to the flow of liquid is called viscosity. Liquids flow in the form of layers. Layer in contact with wall of pipe is almost stationary due to adherence. Next layer is moving with velocity until the central layer has maximum velocity. Each layer exerts drag or pull on the next layer due to the velocity difference or energy exchange. Five factors affect the magnitude of viscosity: molecular shape, intermolecular forces, molecular size, temperature, and density of the liquid. Keeping in view the interdependency of molecular size intermolecular forces and molecular shape in the measurement of viscosity at constant temperature and density (almost) hydrodynamic measurements can be used to manipulate the interactional binding between the investigated compound and DNA. In solution double strands of polyanion DNA macromolecules are more extended to counteract and minimize the repulsion of negative charges at the phosphate backbone. The regular shape of the DNA macromolecule is further changed on interaction with drug or pro-drug candidate therefore relative specific viscosity of DNA solution will also be changed. When a cationic entity binds to the negatively charged phosphate group of the DNA helix through electrostatic interaction, the net negative charge on the polyanionic chain is reduced. Hence repulsion will be reduced leading to DNA contraction/shortening and decrease in the viscosity of DNA. Furthermore, during intercalation imbibed ds-DNA helix is enlarged linearly. Larger DNA macromolecules with the higher molecular weights (due to drug invasion) are difficult to slide over or slip over one another. Hence relative specific viscosity will be increased. Thus, viscosity measurement technique is an important hydrodynamic technique to verify the mode of interaction of small molecules with DNA.
3.5.4.1. Viscosity measurements for evaluation of binding modes
The viscosity measurements are directly dependent on the macromolecular length and therefore sensitive to length variations. Hence these are unambiguously employed as the most critical investigation tool to assess the binding model in the solution phase. In general intercalation causes an increase in viscosity of DNA solution due to lengthening of DNA helix as the base pair pockets are widened to accommodate the binding molecule45 or in other words the binding molecules invade the helix to imbibe into it irregularly and to increase its length. These shape and size deformations lead to the increase in viscosity. The partial or non-classical intercalator which bends the DNA helix due to the presence of uneven opening of strands by intercalation to exposed base pairs reduces the viscosity concomitantly. Conversely, the compounds that exclusively bind in the DNA grooves by partial intercalation and/or non-classical intercalation, typically produce less pronounced (positive or negative) or no change in viscosity of DNA solution under the same conditions. Such surface/or groove binders may invoke either regularity in the helix by filling surface gaps or contraction of double strands by neutralizing the polyanionic negative charge, concomitantly resulting in slight increase or even decrease in relative viscosity of DNA. This compactness and aggregation of DNA helix with binding molecule reduces the number of independently moving macromolecules and resultantly lowering of the solution viscosity. Moreover the compounds which interact with DNA via electrostatic binding mode have either no influence on the DNA viscosity10 or decreased degree of relative DNA viscosity.46
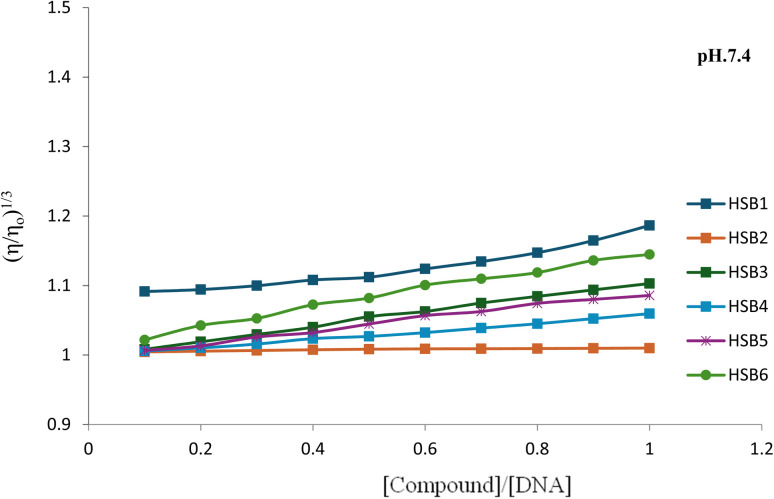
Initially viscosity of DNA solution (η0) in buffer was determined at 310 K at human blood pH i.e., 7.4 and body temperature (310 K). Then specific viscosity contribution (η) of different aliquots of each investigated compound into the constant concentration of DNA solution were determined by the standard addition method. The results were presented as (η/η0)1/3versus (r).
(i) ‘r’ is the ratio of the concentration of test compound and DNA.
(ii) ‘η’ is the viscosity of DNA in presence of compound and ‘η0’ is the viscosity of free DNA.
The relative kinematic viscosity of DNA both in the absence and presence of the test compounds were separately calculated from the following equation:
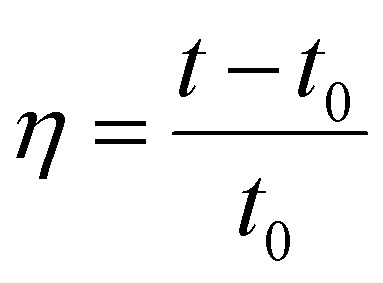 |
6 |
where t is the observed flow time of the DNA containing solution and t0 is the flow time of 0.1 molar phosphate buffer.
On increasing the amount of test compound, the relative viscosity of DNA increased steadily which indicated that the minor groove binding mode is the dominant binding mode. This indication was consistent with the results of photophysical titrations/UV spectral finings. Thus, hydrodynamic studies of all compounds also supported the minor groove binding mode because the effect of an increasing Schiff base concentration on the DNA viscosity was less pronounced as shown in the Fig. 6. This minor increase of DNA viscosity on addition of test compounds is the confirmatory aptitude of groove binders as the test compounds were either invoking regularity in the DNA helix by filling surface gaps and result in small increase in relative viscosity of DNA or engaging in the DNA surface binding with their sterically bulky size affecting an increase in DNA viscosity. This compactness and aggregation of DNA helix with binding molecule reduces the number of independently moving macromolecules and resultantly lowering of the relative viscosity.43 Here several steric factors are tempting to inhibit classical intercalation into the base pairs. Overall the compounds bind to DNA by groove binding through non-classical intercalation with minor widening and no net increase in the counter length of the ds-DNA strands. The order of increasing degree of viscosity which solely depends the binding affinity of compound to DNA is as follows:
Fig. 6. Plot of relative specific viscosity vs. [compound]/[DNA] for HSB1, HSB2, HSB3, HSB4, HSB5 and HSB6 at pH 7.4 and 310 K.
HSB2 < HSB4 < HSB5 < HSB3 < HSB6 < HSB1.
The viscometric studies provided a compelling argument for groove binding as observed through the results of photophysical studies.
Very interestingly the slight increase or no change in viscosities measurements is not only the indication of groove binding mode but also foreshadows the pronounced hyperchromism and hypochromism with no bathochromism or hypsochromism of the compounds in the presence of DNA.47
4. Conclusion
Based on net results of spectroscopic and hydrodynamic measurements for ds-DNA interaction studies with the investigated Schiff bases and the overall structural outlook of the compounds it is concluded that all the compounds interact with DNA through minor groove binding predominantly. The binding constants of all the Schiff bases were in the order of 103–105. The photophysical as well as hydrodynamic findings of the ds-DNA binding studies remained not only comparable and consistent to each other but also in good agreement with the results of in vitro anti-cancer studies. Among these compounds, three (HSB3, HSB4 and HSB1) displayed significant potency against the SW-480 cell line than the others. Whereas HSB3 demonstrated exceptional anticancer activity as compared to the standard drug Doxorubicin. Additionally, the same Schiff base had the highest binding affinity in terms of binding constant values.
Ethical statement
This research exclusively utilized in vitro experimental systems, with no involvement of human participants or animal subjects. Therefore, ethical approval and informed consent were not applicable. All procedures were conducted in strict accordance with institutional protocols and regulatory standards governing the use of in vitro methodologies.
Data availability
All data generated or used for characterization of our synthetic compounds during this research work are included in this submitted article.
Author contributions
Hammad Nasir carried out the synthetic work. Muhammad Arfan provided oversight and supervision throughout the research process. Naeem Abbas and Hamna Hafeez conducted the dsDNA binding assays. Usman Aftab carried out the anticancer activity assays. Muhammad Latif provided critical revisions and write up of manuscript. All authors reviewed and approved the final manuscript prior to submission for publication.
Conflicts of interest
The authors of the submitted manuscript declare that they have no conflicts of interest to disclose.
Supplementary Material
Acknowledgments
We are thankful for financial assistance provided, by National University of Sciences and Technology, Islamabad, 44000 Pakistan.
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra06962e
References
- Siegel R. L. Miller K. Jemal A. Cancer Statistics, 2018. Ca—Cancer J. Clin. 2018, 68, 7– 30, Cancer statistics, 2018Siegel Rebecca L; Miller Kimberly D; Jemal AhmedinCA. Ca-Cancer J. Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. https://dx.doi.org/10.3322/caac.214421 [DOI] [PubMed] [Google Scholar]
- Wild C. P. Espina C. Bauld L. Bonanni B. Brenner H. Brown K. et al., Cancer Prevention Europe. Mol. Oncol. 2019;13:528–534. doi: 10.1002/1878-0261.12455. https://dx.doi.org/10.1002/1878-0261.12455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alyar S. Şen T. Özmen Ü. Ö. Alyar H. AdemandC Ş. Şen J. Synthesis, Spectroscopic Characterizations, Enzyme Inhibition, Molecular Docking Study and DFT Calculations of New Schiff Bases of Sulfa Drugs. J. Mol. Struct. 2019;1185:416–424. doi: 10.1016/j.molstruc.2019.03.002. https://dx.doi.org/10.1016/j.molstruc.2019.03.002 [DOI] [Google Scholar]
- Amiri M. Ajloo D. Fazli M. Mokhtarieh A. Grivani G. Saboury A. A. J. et al., Spectroscopic, Electrochemical, Docking and Molecular Dynamics Studies on the Interaction of Three Oxovanadium (IV) Schiff Base Complexes with Bovine Serum Albumin and their Cytotoxicity Against Cancer. J. Biomol. Struct. Dyn. 2018;36:3753–3772. doi: 10.1080/07391102.2017.1400467. https://dx.doi.org/10.1080/07391102.2017.1400467 [DOI] [PubMed] [Google Scholar]
- Hyndman I. J. The contribution of both nature and nurture to carcinogenesis and progression in solid tumours. Cancer Microenviron. 2016;9:63–69. doi: 10.1007/s12307-016-0183-4. https://dx.doi.org/10.1007/s12307-016-0183-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. New and Emerging Factors in Tumorigenesis: An Overview. Cancer Manage. Res. 2015;7:225. doi: 10.2147/CMAR.S47797. https://dx.doi.org/10.2147/CMAR.S47797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virnig B. A. Baxter N. N. Habermann E. B. Feldman R. D. Bradley C. J. A Matter of Race: Early-versus Late-stage Cancer Diagnosis. Health Aff. 2009;28:160–168. doi: 10.1377/hlthaff.28.1.160. https://dx.doi.org/10.1377/hlthaff.28.1.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolski-Renić A. Čipak Gašparović A. Valente A. Lopez O. Schiff bases and their metal complexes to target and overcome (multidrug) resistance in cancer. Eur. J. Med. Chem. 2024:116363. doi: 10.1016/j.ejmech.2024.116363. https://dx.doi.org/10.1016/j.ejmech.2024.116363 [DOI] [PubMed] [Google Scholar]
- Cree I. A. Charlton P. Molecular Chess? Hallmarks of Anti-cancer Drug Resistance. BMC Cancer. 2017;17:1–8. doi: 10.1186/s12885-016-3022-6. https://dx.doi.org/10.1186/s12885-016-2999-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudry P. Slaby O. Neradil J. Soukalova J. Melicharkova K. Rohleder O. et al., Case Report: Rapid and Durable Response to PDGFR Targeted Therapy in a Child with Refractory Multiple infantile myofibromatosis and a heterozygous germline mutation of the PDGFRB gene. BMC Cancer. 2017;17:1–7. doi: 10.1186/s12885-017-3115-x. https://dx.doi.org/10.1186/s12885-017-3115-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M. Zhivotovsky B. Caspases and Cancer. Cell Death Differ. 2011;18:1441–1449. doi: 10.1038/cdd.2011.30. https://dx.doi.org/10.1038/cdd.2011.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, Global cancer burden growing, amidst mounting need for services, 2024-02-01, [2024-02-25], https://www.who.int/news/item/01-02-2024-global-cancer-burden-growing--amidst-mountingneed-for-services, 2024 [PMC free article] [PubMed]
- Crissman H. A. Tobey R. A. J. S. Cell-cycle Analysis in 20 Minutes. Methods Cell Biol. 1974;184:1297–1298. doi: 10.1126/science.184.4143.1297. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz Z., Halicka H. D. and Zhao H. J. P., Analysis of Cellular DNA Content by Flow and Laser Scanning Cytometry, Polyploidization and Cancer, Springer Link, 2010, pp. 137–147, 10.1007/978-1-4419-6199-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehkhodaei M. Sahihi M. Amiri Rudbari H. J. Spectroscopic and Molecular Docking Studies on the Interaction of Pd (II) & Co (II) Schiff base Complexes with β-lactoglobulin as a Carrier Protein. J. Biomol. Struct. Dyn. 2018;36:3130–3136. doi: 10.1080/07391102.2017.1380537. https://dx.doi.org/10.1080/07391102.2017.1380537 [DOI] [PubMed] [Google Scholar]
- Deshmukh P. Soni P. K. Kankoriya A. Halve A. K. Dixit R. J. I. 4-Aminoantipyrine: A Significant Tool for the Synthesis of Biologically Active Schiff bases and metal Complexes. Int. J. Pharm. Sci. Rev. Res. 2015;34:162–170. [Google Scholar]; .http://www.globalresearchonline.net/
- Sinha D. Tiwari A. K. Singh S. Shukla G. Mishra P. Chandra H. et al., Synthesis, Characterization and Biological Activity of Schiff base Analogues of Indole-3-carboxaldehyde. Eur. J. Med. Chem. 2008;43:160–165. doi: 10.1016/j.ejmech.2007.03.022. https://dx.doi.org/10.1016/j.ejmech.2007.03.022 [DOI] [PubMed] [Google Scholar]
- Doonan F. Cotter T. G. Morphological Assessment of Apoptosis. Methods. 2008;44:200–204. doi: 10.1016/j.ymeth.2007.11.006. https://dx.doi.org/10.1016/j.ymeth.2007.11.006 [DOI] [PubMed] [Google Scholar]
- Elmore S. Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. https://dx.doi.org/10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muiva-Mutisya L. M. Atilaw Y. Heydenreich M. Koch A. M. Akala H. Cheruiyot A. C. Antiplasmodial prenylated flavanonols from Tephrosia subtriflora. Nat. Prod. Res. 2017;32:1407–1414. doi: 10.1080/14786419.2017.1353510. https://dx.doi.org/10.1080/14786419.2017.1353510 [DOI] [PubMed] [Google Scholar]
- Hyndman I. J. The Contribution of Both Nature and Nurture to Carcinogenesis and Progression in Solid Tumours. Cancer Microenviron. 2016;9:63–69. doi: 10.1007/s12307-016-0183-4. https://dx.doi.org/10.1007/s12307-016-0183-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel P. Kumar D. Chandra S. Schiff's Base Ligands and Their Transition Metal Complexes as Antimicrobial Agents. J. Chem., Biol. Phys. Sci. 2014;4(3):1946–1964. [Google Scholar]
- Hassan A. S. Hafez T. S. Osman S. A. M. Ali M. M. Synthesis and in vitro cytotoxic activity of novel pyrazolo [1, 5-$ a $] pyrimidines and related Schiff bases. Turk. J. Chem. 2015;39(5):1102–1113. doi: 10.3906/kim-1504-12. https://dx.doi.org/10.3906/kim-1504-12 [DOI] [Google Scholar]
- Majeed R. H. A. Hussein H. A. Mohd A. A. Preparation and Characterization of Novel Schiff Base Derived From 4-Nitro Benzaldehyde and Its Cytotoxic Activities. Int. J. Mol. Cell. Med. 2022;11:4–285. doi: 10.22088/IJMCM.BUMS.11.4.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abd-Elzaher M. M. L. Mousa A. A. Moustafa H. A. Ali S. A. El-Rashedy M. M. Synthesis, Anticancer Activity and Molecular Docking Study of Schiff base Complexes Containing Thiazole Moiety. Beni-Suef Univ. J. Basic Appl. Sci. 2016;5(1):85–96. doi: 10.1016/j.bjbas.2016.01.001. [DOI] [Google Scholar]
- Venkatesh G. Vennila P. Kaya S. Ahmed S. B. Sumathi P. Siva V. Rajendran P. Kamal C. Synthesis and Spectroscopic Characterization of Schiff Base Metal Complexes, Biological Activity, and Molecular Docking Studies. ACS Omega. 2024;9(7):8123–8138. doi: 10.1021/acsomega.3c08526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval J. P. Desai K. R. A Comparative Study Of Microwave-Assisted and Conventional Synthesis of Novel 2-(4-Diethylamino-2-Hydroxyphenyl)-3-Substituted-Thiazolidin-4-One Derivatives. Chemija. 2009;20(2):101–108. [Google Scholar]
- Uddin M. N. Ahmed S. S. Alam S. R. J. Biomedical Applications of Schiff base Metal Complexes. J. Coord. Chem. 2020;73:3109–3149. doi: 10.1080/00958972.2020.1854745. https://dx.doi.org/10.1080/00958972.2020.1854745 [DOI] [Google Scholar]
- Aftab U. Sajid I. J. I. Antitumor Peptides From Streptomyces sp. SSA 13, Isolated From Arabian Sea. Int. J. Pept. Res. Ther. 2017;23:199–211. doi: 10.1007/s10989-016-9552-6. https://dx.doi.org/10.1007/s10989-016-9552-6 [DOI] [Google Scholar]
- Uddin N. Rashid F. Ali S. Tirmizi S. A. Ahmad I. Zaib S. Synthesis, Characterization, and Anticancer Activity of Schiff bases. J. Biomol. Struct. Dyn. 2020;38:3246–3259. doi: 10.1080/07391102.2019.1654924. https://dx.doi.org/10.1080/07391102.2019.1654924 [DOI] [PubMed] [Google Scholar]
- Morgan E. A. Gini M. Lorenzoni A. Cabasag V. Laversanne C. J. Vignat M. Ferlay J. Murphy J. Bray N. Global Burden of Colorectal Cancer in 2020 and 2040: Incidence and Mortality Estimate from GLOBOCAN. GutBMG Journal. 2023;72(2):338–344. doi: 10.1136/gutjnl-2022-327736. [DOI] [PubMed] [Google Scholar]
- Abd-Elzaher M. M. L. Mousa A. A. Moustafa H. A. Ali S. A. El-Rashedy M. M. Synthesis, Anticancer Activity and Molecular Docking Study of Schiff base Complexes Containing Thiazole Moiety. Beni-Suef Univ. J. Basic Appl. Sci. 2016;5(1):85–96. doi: 10.1016/j.bjbas.2016.01.001. [DOI] [Google Scholar]
- Abd-Elzaher M. M. L. Mousa A. A. Moustafa H. A. Ali S. A. El-Rashedy M. M. Synthesis, Anticancer Activity and Molecular Docking Study of Schiff base Complexes Containing Thiazole Moiety. Beni-Suef Univ. J. Basic Appl. Sci. 2016;5(1):85–96. doi: 10.1016/j.bjbas.2016.01.001. [DOI] [Google Scholar]
- Saeed A. M. AlNeyadi S. S. Abdou I. M. Anticancer Activity of Novel Schiff bases and Azo Dyes Derived From 3-amino-4-hydroxy-2H-pyrano [3, 2-c] quinoline-2, 5 (6H)-dione. Heterocycl. Commun. 2020;26(1):192–205. doi: 10.1515/hc-2020-0116. https://dx.doi.org/10.1515/hc-2020-0116 [DOI] [Google Scholar]
- Gupta S. Pandey S. K. Kumar S. Ram A. K. Gautam P. N. Bharty M. K. Kushwaha D. Acharya A. Butcher R. J. Experimental, Spectroscopic, and theoretical investigation on structural and anticancer activities of Schiff bases derived from isonicotinohydrazide. J. Mol. Struct. 2023;1293:136212. doi: 10.1016/j.molstruc.2023.136212. https://dx.doi.org/10.1016/j.molstruc.2023.136212 [DOI] [Google Scholar]
- Shukla S. Srivastava R. S. Shrivastava R. K. Sodhi A. Kumar P. Synthesis, Characterization, In vitro Anticancer Activity, and Docking of Schiff bases of 4-Amino-1,2-naphthoquinone. Med. Chem. Res. 2012:1604–1617. doi: 10.1007/s00044-012-0150-7. [DOI] [Google Scholar]
- Saygıdeğer Demir B. Gönül İ. Gümüşgöz Çelik G. İpekbayrak S. Synthesis and Anticancer Activities of Water-Soluble Schiff Base Metal Complexes. Adıyaman Univ. J. Sci. Tech. 2020;10(2):441–456. doi: 10.37094/adyujsci.738743. [DOI] [Google Scholar]
- Rehman S. U. Sarwar T. Husain M. A. Ishqi H. M. Tabish M. Arch. Biochem. Biophys. 2015;576:49–60. doi: 10.1016/j.abb.2015.03.024. https://dx.doi.org/10.1016/j.saa.2008.10.049 [DOI] [PubMed] [Google Scholar]
- Ronconi L. Marzano C. Zanello P. Corsini M. Miolo G. Macca C. Trevisan A. Fregona D. J. Med. Chem. 2006;49(5):1648–1657. doi: 10.1021/jm0509288. https://dx.doi.org/10.1016/j.saa.2008.10.049 [DOI] [PubMed] [Google Scholar]
- Arshad N. Abbas N. Perveen F. Mirza B. Almuhaini A. M. Alkahtani S. J. Saudi Chem. Soc. 2021;25(9):101323. doi: 10.1016/j.jscs.2021.101323. https://dx.doi.org/10.1016/j.saa.2008.10.049 [DOI] [Google Scholar]
- Shahabadi N. Kashanian S. Darabi F. Eur. J. Med. Chem. 2010;45(9):4239–4245. doi: 10.1016/j.ejmech.2010.06.020. https://dx.doi.org/10.1016/j.saa.2008.10.049 [DOI] [PubMed] [Google Scholar]
- Arshad N. Abbas N. Bhatti M. H. Rashid N. Tahir M. N. Saleem S. Mirza B. J. Photochem. Photobiol., B. 2012;117:228–239. doi: 10.1016/j.jphotobiol.2012.10.003. https://dx.doi.org/10.1016/j.saa.2008.10.049 [DOI] [PubMed] [Google Scholar]
- Hobro A. J. Rouhi M. Blanch E. W. Conn G. L. Nucleic Acids Res. 2007;35(4):1169–1177. doi: 10.1093/nar/gkm012. https://dx.doi.org/10.1016/j.saa.2008.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. Bai G. Dong C. Bioorg. Med. Chem. 2005;13(20):5694–5699. doi: 10.1016/j.bmc.2005.06.023. https://dx.doi.org/10.1016/j.saa.2008.10.049 [DOI] [PubMed] [Google Scholar]
- Yang X. Shen G.-L. Yu R.-Q. Microchem. J. 1999;62(3):394–404. doi: 10.1006/mchj.1999.1747. https://dx.doi.org/10.1016/j.saa.2008.10.049 [DOI] [Google Scholar]
- Li Y. Yang Z.-Y. Wu J.-C. Eur. J. Med. Chem. 2010;45(12):5692–5701. doi: 10.1016/j.ejmech.2010.09.025. https://dx.doi.org/10.1016/j.saa.2008.10.049 [DOI] [PubMed] [Google Scholar]
- Zhou C.-Y. Xi X.-L. Yang P. Biochemistry. 2007;72:37–43. doi: 10.1016/j.saa.2008.10.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or used for characterization of our synthetic compounds during this research work are included in this submitted article.