Abstract
The human genetic disorder ataxia-telangiectasia (A-T) is characterized by hypersensitivity to ionizing radiation and an elevated risk of malignancy. Epidemiological data support an increased risk for breast and other cancers in A-T heterozygotes. However, screening breast cancer cases for truncating mutations in the ATM (A-T mutated) gene has failed largely to reveal an increased incidence in these patients. It has been hypothesized that ATM missense mutations are implicated in breast cancer, and there is some evidence to support this. The presence of a large variety of rare missense variants in addition to common polymorphisms in ATM makes it difficult to establish such a relationship by association studies. To investigate the functional significance of these changes we have introduced missense substitutions, identified in either A-T or breast cancer patients, into ATM cDNA before establishing stable cell lines to determine their effect on ATM function. Pathogenic missense mutations and neutral missense variants were distinguished initially by their capacity to correct the radiosensitive phenotype in A-T cells. Furthermore missense mutations abolished the radiation-induced kinase activity of ATM in normal control cells, caused chromosome instability, and reduced cell viability in irradiated control cells, whereas neutral variants failed to do so. Mutant ATM was expressed at the same level as endogenous protein, and interference with normal ATM function seemed to be by multimerization. This approach represents a means of identifying genuine ATM mutations and addressing the significance of missense changes in the ATM gene in a variety of cancers including breast cancer.
Ataxia-telangiectasia (A-T) is a rare human genetic disorder characterized by neurodegeneration, cell cycle checkpoint defects, radiosensitivity, and cancer predisposition (1, 2). The gene defective in this syndrome, ATM (A-T mutated), is a member of the phosphatidylinositol 3-kinase family involved in DNA damage recognition and cell cycle control (2, 3). Exposure of cells to ionizing radiation leads to the rapid activation of ATM kinase and in turn the phosphorylation of an array of substrates involved in the recognition and repair of damage in DNA as well as cell cycle checkpoint activation (4–8). Loss of ATM function causes hypersensitivity to ionizing radiation, defective cell cycle control postirradiation, and genomic instability that seems to contribute to the increased incidence of neoplasia (1, 2).
The majority of patients with A-T are compound heterozygotes, and a large majority of the mutations in ATM are truncating (9–11). However, there is evidence for heterogeneity in A-T with some patients having a milder phenotype that seems to be related to the nature of the mutation involved. This less severe phenotype can be explained by the missense mutations or the capacity to produce some normal ATM protein. One such change, an intronic mutation that activates a cryptic splice site resulting in the insertion of 137 nucleotides of intronic sequence (5762 ins137), is leaky and allows the expression of a reduced amount of normal ATM (12). A missense mutation (7271T → G) led to the expression of mutant protein at levels equivalent to that seen in normal cells and had ≈6% of the ATM kinase activity of normal protein (13).
Although A-T is an autosomal recessive disease, there is evidence for some penetrance of the defective gene in heterozygotes. This penetrance is manifested by intermediate sensitivity to ionizing radiation in cells in culture (14–16) and an increased risk of developing cancer, particularly of the female breast (17–23). However, molecular genotyping and mutation analysis have produced mixed results in establishing an association between ATM carrier status and breast cancer (24–31). Gatti et al. (32) in distinguishing between truncating mutations where no ATM protein is detected and missense substitutions where mutant protein of variable stability is observed, have suggested that this mutant protein could produce a dominant negative effect in a heterozygote, resulting in an altered phenotype. Evidence for such interference has been provided by Lim et al. (8), who showed that overexpression of a kinase dead form of ATM inhibited the ATM-dependent phosphorylation of Nbs1 on Ser-343 postirradiation. The presence of a variety of rare variants and polymorphisms in the ATM gene in breast and other cancer patients makes difficult the identification of genuine missense mutations. We describe here the use of in vitro mutagenesis in ATM cDNA to mimic these rare variants followed by transfection into both A-T and control cells to distinguish physiologically important missense mutations from polymorphisms.
Materials and Methods
Cell Culture.
The cells used were Epstein–Barr virus-transformed lymphoblastoid cells. C3ABR is a normal lymphoblastoid cell line, and AT1ABR and AT3ABR are A-T lymphoblastoid cell lines. The cells were cultured in RPMI medium 1640 supplemented with 10% FCS under an atmosphere of 5% CO2.
Site-Directed Mutagenesis.
The QuikChange site-directed mutagenesis kit (Stratagene) was used to introduce the various changes into the ATM cDNA according to the manufacturer's protocol. Because of the combined size of the ATM expression vector (21 kb), it was not possible to introduce changes directly into the full-length construct. Consequently, a 5.1-kb SnaB1–NotI fragment, incorporating the 3′ end of the cDNA, was isolated from pMAT1 (33) and cloned in pSL1180 (Amersham Pharmacia) for the generation of mutants. Mutants were identified by DNA sequencing, and the SnaB1–NotI fragment was subcloned into pMAT1, the full-length ATM cDNA, to generate expression constructs (see Table 1). Presence of the mutation was confirmed further by DNA sequencing of these constructs. All constructs were under control of the inducible metallothionein promoter and contained a FLAG sequence incorporated at the 5′ end.
Table 1.
Missense mutations/alterations introduced into ATM cDNA using in vitro mutagenesis and employed in this investigation
| Amino acid alteration | Nucleotide change | Allelic counts | |
|---|---|---|---|
| A-T mutants | SRI2546del3 | 7636del9 | |
| V2716A | 8147T→C | ||
| R2849P | 8546G→C | ||
| G2867R | 8599G→C | ||
| V2662del | 7987delGTT | ||
| Variants in breast cancer | A2274T | 6820G→A | 1/384 |
| G2287A | 6860G→C | 1/384 | |
| C2464R | 7390T→C | 2/384 | |
| S2592C | 7775C→G | 1/384 | |
| G2772R | 8314G→A | 1/384 |
Nucleotide and the corresponding amino acid alterations are indicated. The allele frequencies of the selected missense substitutions in 192 breast cancer patients are indicated. These missense substitutions were confined to one of both groups and were not observed among 100 control individuals from the general population.
Transfection of Human Lymphoblastoid Cells.
Transfections of Epstein–Barr virus-transformed lymphoblastoid cells were carried out as described (33). Induction of ATM expression in control and A-T lymphoblastoid cells was carried out by using 5 μM CdCl2 for 6 h. To establish cell lines coexpressing different epitope-tagged forms of ATM we transfected stable cell lines expressing the FLAG construct with a full-length ATM cDNA containing a hemagglutinin (HA) sequence (pMAT3) at the 5′ end of the gene. pMAT1 cells were transfected with pMAT3 as described above and plated out as single cells in 96-well plates. Because the selective marker in pMAT3 was also hygromycin, it was necessary to isolate clones expressing both forms of ATM. The presence of both plasmids in a single clone was confirmed by using PCR with primers specific for the HA/FLAG sequences and a sequence within ATM. Four clones in total were isolated from a total of 190 wells (2% efficiency).
Viability of Cells Postirradiation.
Cells were induced or mock-induced with 5 μM CdCl2 for 6 h and irradiated with 1–4 Gy of γ-rays. Cell viability was determined as described (33). The number of viable cells was counted daily up to 4 days postirradiation.
Radiation-Induced Chromosome Aberrations (ICAs).
Cells were induced or mock-induced with 5 μM CdCl2 for 6 h and irradiated with 1 Gy of γ-rays. Fifty metaphases were analyzed for each sample, and ICAs were determined (33).
Kinase Assays.
For ATM kinase assays, control and A-T cells were treated with 6 Gy of ionizing radiation. Kinase assays were performed as described (34). Briefly anti-ATM immunoprecipitation was performed with anti-ATM antibody (ATM-5BA) on 1 mg of total cell extract and protein G-Sepharose for 2 h at 4°C. Immune complex kinase assays were performed by incubating the resulting complexes in kinase buffer and 1 μg of soluble GST fusion protein (p531–40) for 30 min at 30°C and analyzed by SDS/PAGE followed by autoradiography.
Lysate Preparations, Immunoprecipitation, and Western Blotting.
Control and A-T lymphoblastoid cells were exposed to radiation and harvested 1 h later. Cell extracts were prepared as described (7). Immunoprecipitation was carried out by using 1 mg of extract and 1 μg of either anti-ATM (ATM-5BA), anti-FLAG (M2, Sigma), or anti-HA antibodies at 4°C overnight. Whole-cell extract (50 μg) or immune complexes were loaded on 5% SDS/PAGE gels. Samples were transferred to nitrocellulose by using Towbin's buffer at 100 V for 1 h, and membranes were probed with anti-ATM (ATM-2C1, GeneTex, San Antonio, TX) or anti-phospho-p53 (Ser-15; 16G8, Cell Signaling Technology, Beverly, MA) antibodies.
Results
In Vitro Mutagenesis of ATM cDNA.
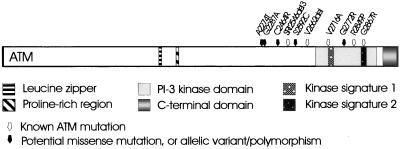
Although several rare missense substitutions have been identified in the ATM gene in early onset breast cancer, their contribution to malignancy cannot be assumed in the absence of a functional assay. We describe here the use of in vitro mutagenesis of full-length ATM cDNA followed by stable transfection of A-T and control cells to distinguish between physiologically important missense mutations and polymorphisms or rare allelic variants. We previously have described the presence of apparent missense mutations in 89 of 192 breast cancer patients (35) and have chosen a representative group of these patients together with known A-T mutations from a study of German A-T patients (ref. 36 and unpublished data) as outlined in Table 1. These missense substitutions were selected because they were found specifically in either the A-T or breast cancer cohort, were rare enough to exclude them as common polymorphisms, and were located in the C-terminal portion of ATM near or within its kinase domain. We used site-directed mutagenesis to incorporate the various nucleotide changes into full-length ATM cDNA. Because of the lack of suitable restriction enzyme sites in the cDNA, changes were created toward the C terminus of ATM, which contains the only well described functional domain (3). Consequently, a 5-kb SnaBI–NotI fragment from the 3′ end of the cDNA (33) was cloned into pSL1180 to generate mutations. Mutants were identified by DNA sequencing, and the SnaBI–NotI fragment was subcloned back into the full-length construct. Presence of the mutation was confirmed further in this construct by DNA sequencing. Because ATM cDNA constructs are very unstable, we also checked the integrity of the constructs with a series of overlapping PCR primers covering the full length of the cDNA (37). In all, 10 mutant cDNAs were made, four of which are previously described A-T mutations (Ataxia-telangiectasia mutation database, www.vmresearch.org./atm.htm) and five are potential missense mutations identified in breast cancer (Table 1). We also included the ATM mutant SRI2546del3, which produces near full-length ATM protein but is less stable than wild-type ATM, as a previously characterized A-T control (38). A schematic representation of the distribution of the amino acid substitutions and deletions produced here appears in Fig. 1.
Figure 1.
Distinguishing between A-T mutations and potential ATM mutations identified in breast cancer patients. Schematic representation of the ATM protein indicating the location of the phosphatidylinositol 3-kinase (PI-3 kinase) domain and other putative domains. The known A-T mutations (white arrows) and breast cancer variants (blank arrows) are those selected for this study.
Distinguishing Missense Mutations from Rare ATM Variants in Stably Transfected A-T Cells.
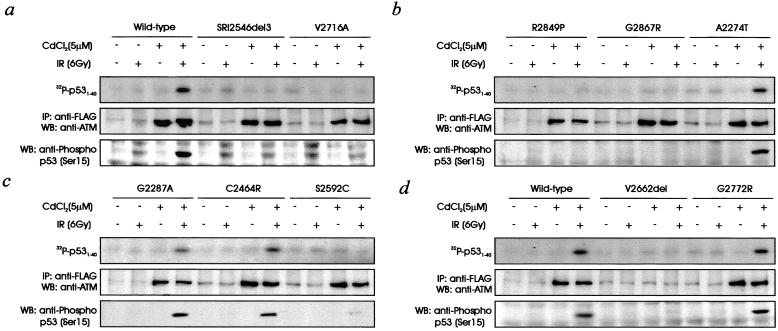
We have shown previously that full-length ATM cDNA (pMAT1, wild type) corrects the radiosensitivity and other aspects of the A-T cellular phenotype (33). When pMAT1 was introduced into A-T cells (AT1ABR) under the control of a metallothionein II-inducible promoter, exposure to ionizing radiation postinduction (CdCl2) caused the activation of ATM kinase as determined in vivo by phosphorylation of p53 Ser-15 at 1 h postirradiation or by immunoprecipitation of ATM from cell extracts followed by phosphorylation of a p53 substrate in vitro (Fig. 2a). No ATM kinase activity was detected in transfected, uninduced AT1ABR cells. Expression of recombinant ATM was demonstrated by using anti-FLAG antibody. The low basal FLAG signal in the uninduced lanes is nonspecific, because this was observed also in untransfected control cells (results not shown). The introduction of an ATM cDNA containing a genuine missense mutation into an A-T cell line would not be expected to correct the phenotype. We used this strategy to distinguish between pathogenic ATM mutants and nonfunctional variants. All three A-T known missense mutants (V2716A, R2849P, and G2867R) together with the deletion mutant SRI2546del3 failed to elicit ATM kinase activity in AT1ABR cells whether ATM kinase activity was determined by phosphorylation of a p53 substrate in vitro or by phosphorylation of p53 Ser-15 in vivo (Fig. 2 a and b). Failure to observe kinase activity was not caused by a lack of expression or instability of the mutant proteins, because immunoprecipitation with anti-FLAG antibodies followed by immunoblotting with ATM antibodies revealed the presence of approximately equal amounts of ATM in all cases (Fig. 2 a and b). In a lymphoblastoid cell line, established from a patient with A-T who was compound heterozygous for V2716A, and a truncating ATM mutation on the second allele, ATM kinase activity was reduced markedly, and mutant ATM protein levels were low (results not shown). No kinase activity was observed in AT1ABR transfected with cDNA for the A-T deletion V2662 del (Fig. 2d). In this case the amount of expression of ATM was undetectable, which would explain the failure to see induction of kinase activity. To test ATM expression, we analyzed a lymphoblastoid cell line from a patient with A-T who was homozygous for V2662del. Low levels of ATM protein and an absence of induced ATM kinase activity also were characteristic of this cell line (results not shown), indicating that this substitution destabilized the protein. This phenomenon has been described previously for some missense ATM mutations (39).
Figure 2.
Effect of stable transfection of A-T lymphoblastoid cells (AT1ABR) with full-length or mutant forms of ATM on radiation-induced ATM kinase activity and radiosensitivity. (a) AT1ABR cells were transfected with wild-type, SRI2546del3, or V2716A, and stable lines were established by selection with hygromycin (200 μg/ml). Expression of ATM protein was induced with CdCl2 for 6 h before irradiation (6 Gy; IR) and incubation for 1 h. ATM kinase was determined in vivo by immunoblotting extracts with anti-p53 Ser-15 phosphorylation-specific antibody (Lower). In vitro ATM kinase activity was determined by immunoprecipitating (IP) ATM with anti-ATM antibodies and assaying for phosphorylation of p531–40 substrate (Upper). Expression of ATM cDNA was determined by immunoprecipitation with anti-FLAG antibodies followed by immunoblotting with anti-ATM antibodies (Middle). WB, Western blot. (b) AT1ABR cells transfected with R2849P, G2867R, or A2274T. Cell lines were established, and ATM expression and ATM kinase activity were determined as described for a. (c) AT1ABR cells transfected with G2287A, C2464R, or S2592C. Conditions for ATM kinase and ATM expression were as described for a. (d) AT1ABR cells transfected with wild-type, V2662del, or G2772R and ATM expression and kinase activity were determined as described for a.
We used the same approach for the five ATM variants described in patients with breast cancer as outlined in Table 1. Four of these (A2274T, G2287A, C2464R, and G2772R) had normal induction of kinase activity in vitro and in vivo when assayed in AT1ABR (Fig. 2 b–d). Again, the level of ATM expression was confirmed in these transfected lines with anti-FLAG antibodies. When Ser-2592 was mutated to Cys (S2592C), no kinase activity was observed when the cDNA was introduced into AT1ABR cells, indicating that this was a genuine missense mutation (Fig. 2c). ATM protein expression was detected also in these cells with anti-FLAG antibody.
Because A-T cells are hypersensitive to ionizing radiation, the introduction of ATM cDNA containing a genuine missense mutation also should fail to correct this cellular phenotype in A-T cells. All five known A-T mutations (SRI2546del3, V2716A, R2849P, G2867R, and V2662del) failed to alter the extent of radiosensitivity when introduced into AT1ABR cells (Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). Elevated levels of radiation ICAs also are characteristic of A-T (33). As observed for survival postirradiation, the known A-T cDNA mutants did not reduce the level of aberrations (2.98–3.20 ICAs per metaphase) in A-T cells (Table 2). On the other hand A2274T, G2287A, C2464R, and G2772R cDNAs corrected the radiosensitive phenotype of the A-T cell line, AT1ABR (Fig. 5). These cDNAs also reduced the levels of radiation ICAs from ≈3 ICAs per metaphase to ≈1 ICA per metaphase (Table 2). Both sets of data provide further evidence that A2274T, G2287A, C2464R, and G2772R are missense variants not interfering with the function of ATM. However, S2592C failed to correct the radiosensitivity and elevated chromosome aberrations in AT1ABR cells, confirming that it was a genuine mutation (Fig. 5 and Table 2).
Table 2.
γ radiation-induced G2 phase chromosome aberrations in A-T cells (AT1ABR) transfected with wild-type or mutant ATM constructs
| Cell line | Aberrations, no.
|
No. ICAs per metaphase | ||
|---|---|---|---|---|
| sb | cb | Int | ||
| C3ABR | 54 | 2 | 0 | 1.12 |
| AT1ABR | 168 | 1 | 0 | 3.38 |
| AT1ABR + Wild type | 58 | 1 | 0 | 1.18 |
| AT1ABR + SRI2546del3 | 153 | 2 | 2 | 3.14 |
| AT1ABR + V2716A | 148 | 1 | 0 | 2.98 |
| AT1ABR + R2849P | 161 | 1 | 0 | 3.20 |
| AT1ABR + G2867R | 156 | 1 | 0 | 3.14 |
| AT1ABR + A2274T | 54 | 0 | 0 | 1.08 |
| AT1ABR + G2287A | 57 | 1 | 0 | 1.16 |
| AT1ABR + C2464R | 55 | 2 | 0 | 1.14 |
| AT1ABR + S2592C | 161 | 0 | 1 | 3.24 |
| AT1ABR + V2662del | 155 | 2 | 0 | 3.14 |
| AT1ABR + G2772R | 59 | 1 | 0 | 1.20 |
sb, chromatid breaks; cb, chromosome breaks; Int, interchanges. Fifty metaphases were analyzed for each sample after exposure to 1 Gy of radiation and in the presence of CdCl2. Data for ICAs are the mean.
Missense Mutations in ATM cDNA Interfere with Normal ATM Function.
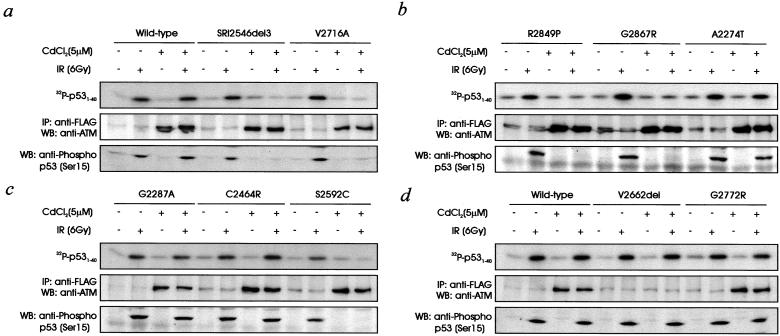
Lim et al. (8) have shown recently that radiation-induced phosphorylation of nibrin on Ser-343 is inhibited in a dominant negative manner by overexpression of ATM containing a double mutation (D2870A,N2875K) in the kinase domain, which renders the molecule inactive. We predicted that this also might be the case for the known ATM mutants used in the present study and would be another means of distinguishing neutral variants from pathogenic mutations. As expected, introduction of wild-type cDNA into a control cell line (C3ABR) did not interfere with normal ATM kinase activity (Fig. 3a). When ATM kinase activity was determined in normal control cells transfected with V2716A, R2849P, and G2867R, there was no evidence for radiation-induced phosphorylation of p53 on Ser-15 in any case (Fig. 3 a and b). Interference with radiation-induced kinase activity was observed also in in vitro immunoprecipitation of ATM by using p531–40 as substrate (Fig. 3 a and b). ATM kinase activity was abolished also in cell lines expressing cDNA with a 3-aa deletion (SRI 2546del 3; Fig. 3a). This inhibition was also the case when in vitro kinase activity was determined. In all these examples, ATM protein was detected by using anti-FLAG antibodies suggesting that the observed effects were caused by mutant protein expression. The inhibition observed with immunoprecipitated ATM suggests that the mutant forms of ATM interfere with the activation of ATM kinase after irradiation. The V2662del mutant did not interfere with activation, again most likely because of the low expression of mutant ATM protein in vitro. When four of the rare variant forms of ATM cDNA (A2274T, G2287A, C2464R, and G2772R) were transfected into control cells, there was no interference with ATM kinase activity measured in vivo or in vitro (Fig. 3 b–d). However, S2592C, which we have shown to express mutant ATM protein, behaved similar to the known A-T mutants in that it prevented the activation of endogenous ATM kinase in vivo and in vitro (Fig. 3c).
Figure 3.
Effect of stable transfection of control lymphoblastoid cells (C3ABR) with full-length and mutant forms of ATM on ATM kinase activity and radiosensitivity. (a) C3ABR cells were stably transfected with wild type, SRI 2546del3, or V2716A. ATM protein was induced, and cells were irradiated (IR) as described in the Fig. 2a legend. (Lower) ATM kinase in vivo; (Upper) In vitro ATM kinase using p531–40 as substrate; (Middle) expression of FLAG-tagged ATM determined by immunoprecipitation (IP) with anti-FLAG antibodies followed by immunoblotting with anti-ATM antibodies. WB, Western blot. (b) C3ABR cells stably transfected with R2849P, G2867R, or A2274T and ATM kinase activity and protein expression were determined as described in Fig. 2a the legend. The panels are as described for a. (c) C3ABR cells stably transfected with G2287A, C2464R, or S2592C. ATM protein and kinase were induced and assayed as described for a. (d) C3ABR cells stably transfected with wild-type, V2662 del, or G2772R and ATM kinase activity and expression were determined as described for a.
Because the mutant forms of ATM interfered with endogenous ATM kinase, it seemed likely that they also would sensitize these cells to ionizing radiation. After exposure to radiation, cells transfected with the known A-T mutant cDNAs (SRI 2546del3, V2716A, R2849P, and G2867R) as well as the initially identified variant S2592C showed enhanced sensitivity to radiation similar to that observed in an A-T cell line (Fig. 6, which is published as supporting information on the PNAS web site). This sensitization was supported by radiation ICAs, which were of the same order as those seen in irradiated A-T cells (Table 3; 2.92–3.42 ICAs per metaphase). None of the other variant cDNAs (A2274T, G2287A, C2464R, and G2772R) or the V2662del mutant had an effect on the cellular radiation response, whether determined by cell survival or level of ICAs (Fig. 6 and Table 3).
Table 3.
γ radiation-induced G2 phase chromosome aberrations in control cells (C3ABR) transfected with wild-type or mutant ATM constructs
| Cell line | Aberrations, no.
|
No. ICAs per metaphase | ||
|---|---|---|---|---|
| sb | cb | Int | ||
| C3ABR | 54 | 2 | 0 | 1.12 |
| AT1ABR | 168 | 1 | 0 | 3.38 |
| C3ABR + Wild-type | 57 | 0 | 0 | 1.14 |
| C3ABR + SRI2546del3 | 145 | 0 | 1 | 2.92 |
| C3ABR + V2716A | 147 | 2 | 0 | 2.98 |
| C3ABR + R2849P | 150 | 0 | 0 | 3.00 |
| C3ABR + G2867R | 149 | 1 | 1 | 3.02 |
| C3ABR + A2274T | 58 | 1 | 0 | 1.18 |
| C3ABR + G2287A | 61 | 1 | 0 | 1.24 |
| C3ABR + C2464R | 59 | 0 | 2 | 1.22 |
| C3ABR + S2592C | 143 | 2 | 1 | 2.90 |
| C3ABR + V2662del | 58 | 0 | 1 | 1.18 |
| C3ABR + G2772R | 63 | 1 | 0 | 1.28 |
sb, chromatid breaks; cb, chromosome breaks; Int, interchanges. Fifty metaphases were analyzed for each sample after exposure to 1 Gy of radiation and in the presence of CdCl2. Data for ICAs are the mean.
Mode of Interference of Mutant ATM Protein.
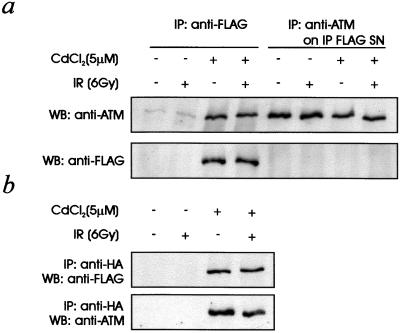
We have described here that mutant ATM protein interferes with normal ATM kinase activity. However, in A-T heterozygotes, although intermediate sensitivity to radiation has been reported (14–16), radiation-induced kinase activity is normal (results not shown). Normal kinase activity can be explained by instability of the mutant ATM protein, which in reduced amounts does not discernibly interfere with ATM. To explain the dominant interference observed in these studies we determined the relative amounts of mutant (S2592C) and endogenous protein in transfected cells. Mutant protein was immunoprecipitated with anti-FLAG antibody from transfected, induced control (C3ABR) cells followed by immunoblotting with anti-ATM and anti-FLAG antibodies (Fig. 4a). Immunoprecipitation of the resulting supernatant with anti-ATM antibodies revealed approximately equal amounts of endogenous ATM regardless of whether the mutant ATM protein was being expressed (Fig. 4a). Absence of a FLAG signal in these immunocomplexes (ATM-IP) demonstrated that mutant protein was not contributing to the ATM measured under these conditions.
Figure 4.
Expression levels of mutant protein in stably transfected cells and determination of ATM-ATM association. (a) C3ABR cells transfected with the S2952C mutant cDNA were induced and irradiated (IR) as indicated (see Fig. 2a legend). Cells extracts were immunoprecipitated (IP) initially with anti-FLAG antibodies followed by immunoblotting with either anti-ATM or anti-FLAG antibodies. The supernatant remaining after immunoprecipitation with anti-FLAG antibodies was subsequently immunoprecipitated with antibodies against ATM protein. This immunocomplex then was resolved on SDS/PAGE followed by immunoblotting with anti-ATM and anti-FLAG antibodies. WB, Western blot. (b) AT3ABR cells stably transfected with pMAT1 (FLAG-ATM) and pMAT3 (HA-ATM) were induced and irradiated as described in the Fig. 2a legend. Cell extracts were immunoprecipitated with anti-HA antibodies followed by immunoblotting with anti-FLAG antibodies. These membranes were immunoblotted also with an antibody against ATM protein.
We also determined whether the dominant interference might be due to ATM–ATM association. To check for this we used an A-T cell line (AT3ABR) not expressing detectable levels of ATM protein (38). These cells were stably transfected with ATM-FLAG (pMAT1) and ATM-HA (pMAT3) sequentially, and clones were isolated as described under Materials and Methods. Coimmunoprecipitation was demonstrated by using anti-HA antibodies followed by immunoblotting with anti-FLAG antibodies (Fig. 4b). Immunoblotting with anti-ATM antibodies confirmed that this protein was ATM.
Discussion
Intermediate sensitivity to ionizing radiation is well documented for cell lines from A-T heterozygotes employing a variety of methods based on cell survival and chromosome damage (14–16). Haploinsufficiency of the gene is evident also in mice where Atm+/− carriers display increased sensitivity to radiation, manifested by decreased survival and premature graying after exposure to sublethal doses of radiation (40). Adverse clinical manifestation of the abnormal gene in human carriers is supported also by higher mortality rates and earlier age of death from cancer and ischemic heart disease (41). Nevertheless the relationship between mutations in the ATM gene and cancer predisposition has remained controversial (42). Added to this controversy is the inability to distinguish between physiologically important missense mutations in the ATM gene and polymorphic variants, which is complicated further by the identification of only one functional domain in this protein thus far (3). The assay system we described here provides an approach to identifying functionally relevant missense mutations in coding regions of the ATM gene and distinguishing them from neutral missense variants. However, some missense changes may not interfere with ATM kinase activity and radiosensitivity but have more subtle effects on the interaction of ATM with other proteins important on other aspects of radiation signaling. This assay also can be adapted readily to include intronic sequences in the cDNA construct to detect nucleotide changes giving rise to splicing defects. Variants in the ATM gene have been reported widely, and a recent study described 34 intragenic polymorphisms or rare variants in the gene for which it was not possible to rule out functional effects on ATM (43). Of the five breast cancer variants investigated here, only one, S2592C, was shown to be a genuine missense mutation. The S2592C mutation is in the same exon (exon 54) as SRI2546del3, and both produce mutant protein lacking ATM kinase activity, which has a dominant interfering effect. This substitution could alter the conformation of ATM or disrupt a potential phosphorylation site in the SSQL sequence involved and thus render the kinase inactive.
We have demonstrated that missense mutations in regions of the ATM protein, away from the kinase domain, lead to a dominant interfering effect on the activation of normal ATM kinase. Because we observe interference with normal kinase activity under the conditions of the in vitro assay, both mutant and wild type would be expected to be in the same immune complex. Indeed, there is evidence that ATM is present in a complex termed Brca1-associated genome surveillance complex (BASC; ref. 44). We have shown here that different epitope-tagged forms of ATM coimmunoprecipitate, which is suggestive of multimerization. Smith et al. (45) have described a higher order of assemblies (tetramers) of ATM using atomic force microscopy also supporting ATM–ATM association. In addition, it has been reported previously that overexpression of a kinase dead ATM cDNA has a dominant interfering effect on ATM kinase activity (8). However, with the system used here, mutant ATM protein expression is approximately equal to that found for the wild-type allele and, if ATM is present in a multimeric form, could explain how equal expression gives rise to a dominant interfering effect.
ATM protein is not detectable in the presence of truncating mutations, and instability is observed also with some missense mutations. No ATM protein is detectable in a patient homozygous for a double missense mutation, and 9 of 10 A-T heterozygotes have been reported to have reduced expression of ATM protein (39). However, mutant ATM protein was detected in two families carrying the presumed missense mutation V2424G, which is strongly associated with breast cancer (12). In addition, a stably expressed exon 24 polymorphism of ATM, substituting arginine for proline, was also associated with breast cancer (46). We have described here reduced but detectable levels of ATM protein containing rare missense mutations. Thus even low levels of mutant ATM protein have the potential to interfere with ATM function and might in this way contribute to cancer susceptibility, as envisaged in the model outlined by Gatti et al. (32). It seems likely that a reduced amount of ATM protein in heterozygotes is responsible for the widely described intermediate sensitivity to radiation. This hypothesis is supported by transfection experiments with antisense ATM cDNA where reduced ATM protein sensitizes cells to radiation (47). A dominant interfering effect of mutant ATM protein together with the reduced amount of wild-type protein in A-T heterozygotes may tip the balance further in favor of genome instability and cancer predisposition. The case of S2592C in our study shows that functionally and clinically relevant missense mutations exist beyond those that were identified within A-T families. The approach we have described here offers the possibility of extending mutagenesis of ATM cDNA to include any missense mutation/rare variant observed in patients with breast and other cancers to test for interference with normal ATM function. This approach would be a forerunner to generating an animal model of such a missense mutation to study cancer susceptibility.
Supplementary Material
Acknowledgments
Thanks to Aine Farrell for technical support and Tracey Laing for typing the manuscript. We thank the Australian National Health and Medical Research Council, the A-T Research Foundation (Los Angeles), and the A-T Children's Project for support.
Abbreviations
- A-T
ataxia-telangiectasia
- HA
hemagglutinin
- ATM
A-T mutated
- ICA
induced chromosome aberration
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
To whom reprint requests should be addressed at: Queensland Institute of Medical Research, 300 Herston Road, Herston, Brisbane 4029, Australia. Email: martinL@qimr.edu.au.
References
- 1.Boder E, Sedgwick R P. Psychiatr Neurol Med Psychol Beih. 1970;13–14:8–16. [PubMed] [Google Scholar]
- 2.Lavin M F, Shiloh Y. Annu Rev Immunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- 3.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 4.Banin S, Moyal L, Shieh S, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 5.Kim S T, Lim D S, Canman C E, Kastan M B. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 6.Cortez D, Wang Y, Qin J, Elledge S J. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 7.Gatei M, Scott S P, Filippovich I, Sorokina N, Lavin M F, Weber B, Khanna K K. Cancer Res. 2000;60:3299–3304. [PubMed] [Google Scholar]
- 8.Lim D S, Kim S T, Xu B, Maser R S, Lin J, Petrini J H, Kastan M B. Nature (London) 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 9.Byrd P J, Cooper P R, Stankovic T, Kullar H S, Watts G D, Robinson P J, Taylor M R. Hum Mol Genet. 1996;5:1785–1791. doi: 10.1093/hmg/5.11.1785. [DOI] [PubMed] [Google Scholar]
- 10.Gilad S, Bar-Shira A, Harnik R, Shkedy D, Ziv Y, Khosravi R, Brown K, Vanagaite L, Xu G, Frydman M, et al. Hum Mol Genet. 1996;5:2033–2037. doi: 10.1093/hmg/5.12.2033. [DOI] [PubMed] [Google Scholar]
- 11.Telatar M, Wang Z, Udar N, Liang T, Bernatowska-Matusziewicz E, Lavin M, Shiloh Y, Concannon P, Good R A, Gatti R A. Am J Hum Genet. 1996;59:40–44. [PMC free article] [PubMed] [Google Scholar]
- 12.Stankovic T, Kidd A M, Sutcliffe A, McGuire G M, Robinson P, Weber P B, Bedenham T, Bradwell A R, Easton D F, Lennox G G, et al. Am J Hum Genet. 1998;62:334–345. doi: 10.1086/301706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart G S, Last J I K, Stankovic T, Haites N, Kidd A M J, Byrd P J, Taylor A M R. J Biol Chem. 2001;276:30133–30141. doi: 10.1074/jbc.M103160200. [DOI] [PubMed] [Google Scholar]
- 14.Chen P C, Lavin M F, Kidson C, Moss D. Nature (London) 1978;274:484–486. doi: 10.1038/274484a0. [DOI] [PubMed] [Google Scholar]
- 15.Paterson M C, MacFarlane S J, Gentner N, Smith B P. In: Ataxia-telangiectasia: Genetics, Neuropathology and Immunology of a Degenerative Disease of Childhood. Gatti R A, Swift M, editors. New York: Liss; 1985. pp. 73–87. [Google Scholar]
- 16.Shiloh Y, Parshad R, Sanford K K, Jones G M. Lancet. 1986;1:689–690. doi: 10.1016/s0140-6736(86)91773-3. [DOI] [PubMed] [Google Scholar]
- 17.Swift M, Morrell D, Massey R B, Chase C L. N Engl J Med. 1991;325:1831–1835. doi: 10.1056/NEJM199112263252602. [DOI] [PubMed] [Google Scholar]
- 18.Easton D F. Int J Radiat Biol. 1994;66:S177–S182. doi: 10.1080/09553009414552011. [DOI] [PubMed] [Google Scholar]
- 19.Pippard E C, Hall A J, Barker D J, Bridges B A. Cancer Res. 1988;15:2929–2932. [PubMed] [Google Scholar]
- 20.Borresen A L, Andersen T I, Tretli S, Heiberg A, Moller P. Genes Chromosomes Cancer. 1990;2:339–340. doi: 10.1002/gcc.2870020412. [DOI] [PubMed] [Google Scholar]
- 21.Janin N, Andrieu N, Ossian K, Lauge A, Croquette M F, Griscelli C, Debre M, Bressac-de-Paillerets B, Aurias A, Stoppa-Lyonnet D. Br J Cancer. 1999;80:1042–1045. doi: 10.1038/sj.bjc.6690460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inskip H M, Kinlen L J, Taylor A M, Woods C G, Arlett C F. Br J Cancer. 1999;79:1304–1307. doi: 10.1038/sj.bjc.6690209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsen J H, Hahnemann J M, Borresen-Dale A L, Brondum-Nielsen K, Hammarstrom L, Kleinerman R, Kaariainen H, Lonnqvist T, Sankila R, Seersholm N, et al. J Natl Cancer Inst. 2001;93:121–127. doi: 10.1093/jnci/93.2.121. [DOI] [PubMed] [Google Scholar]
- 24.Athma P, Rappaport R, Swift M. Cancer Genet Cytogenet. 1996;92:130–134. doi: 10.1016/s0165-4608(96)00328-7. [DOI] [PubMed] [Google Scholar]
- 25.Vorechovsky I, Luo L, Lindblom A, Negrini M, Webster A D, Croce C M, Hammarstrom L. Cancer Res. 1996;56:4130–4133. [PubMed] [Google Scholar]
- 26.Fitzgerald M G, Bean J M, Hedge SR, Unsal H, MacDonald D J, Harkin D P, Finkelstein D M, Isselbacher K J, Haber D A. Nat Genet. 1997;15:307–310. doi: 10.1038/ng0397-307. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Birkholtz G G, Lindblom P, Rubio C, Lindblom A. Cancer Res. 1998;58:1376–1379. [PubMed] [Google Scholar]
- 28.Bay J O, Grancho M, Pernin D, Presneau N, Rio P, Tchirkov A, Uhrhammer N, Verrelle P, Gatti R A, Bignon Y J. Int J Oncol. 1998;12:1385–1390. doi: 10.3892/ijo.12.6.1385. [DOI] [PubMed] [Google Scholar]
- 29.Bebb D G, Yu Z, Chen J, Telatar M, Gelmon K, Phillips N, Gatti R A, Glickman B W. Br J Cancer. 1999;80:1979–1981. doi: 10.1038/sj.bjc.6690630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Broeks A, Urbanus J H, Floore A N, Dahler E C, Klijn J G, Rutgers E J, Devilee P, Russell N S, van Leeuwen F E, van't Veer L J. Am J Hum Genet. 2000;66:494–500. doi: 10.1086/302746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izatt L, Greenman J, Hodgson S, Ellis D, Watts S, Scott G, Jacobs C, Liebmann R, Zvelebil M J, Mathew C, Solomon E. Genes Chromosomes Cancer. 1999;26:286–294. [PubMed] [Google Scholar]
- 32.Gatti R A, Tward A, Concannon P. Mol Genet Metab. 1999;68:419–423. doi: 10.1006/mgme.1999.2942. [DOI] [PubMed] [Google Scholar]
- 33.Zhang N, Chen P, Khanna K K, Scott S, Gatei M, Kozlov S, Watters D, Spring K, Yen T, Lavin M F. Proc Natl Acad Sci USA. 1997;94:8021–8026. doi: 10.1073/pnas.94.15.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canman C E, Lim D S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 35.Dörk T, Bendix R, Bremer M, Rades D, Klöpper K, Nicke M, Skawran B, Hector A, Yamini P, Steinmann D, et al. Cancer Res. 2001;61:7608–7615. [PubMed] [Google Scholar]
- 36.Sandoval N, Platzer M, Rosenthal A, Dörk T, Bendix R, Skawran B S M, Wegner R D, Sperling K, Banin S, Shiloh Y, et al. Hum Mol Genet. 1999;8:69–79. doi: 10.1093/hmg/8.1.69. [DOI] [PubMed] [Google Scholar]
- 37.Khanna K K, Keating K E, Kozlov S, Scott S, Gatei M, Hobson K, Taya Y, Gabrielli B, Chan D, Lees-Miller S P, Lavin MF. Nat Genet. 1998;20:398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- 38.Watters D, Khanna K K, Beamish H, Birrell G, Spring K, Kedar P, Gatei M, Stenzel D, Hobson K, Kozlov S, et al. Oncogene. 1997;14:1911–1921. doi: 10.1038/sj.onc.1201037. [DOI] [PubMed] [Google Scholar]
- 39.Becker-Catania S G, Chen G, Hwang M J, Wang Z, Sun X, Sanal O, Bernatowska-Matuszkiewicz E, Chessa L, Lee E Y, Gatti R A. Mol Genet Metab. 2000;70:122–133. doi: 10.1006/mgme.2000.2998. [DOI] [PubMed] [Google Scholar]
- 40.Barlow C, Eckhaus M A, Schäffer A A, Wynshaw-Boris A. Nat Genet. 1999;21:359–360. doi: 10.1038/7684. [DOI] [PubMed] [Google Scholar]
- 41.Su Y, Swift M. Ann Intern Med. 2000;133:770–778. doi: 10.7326/0003-4819-133-10-200011210-00009. [DOI] [PubMed] [Google Scholar]
- 42.Swift M. J Natl Cancer Inst. 2001;93:84–85. doi: 10.1093/jnci/93.2.84. [DOI] [PubMed] [Google Scholar]
- 43.Castellvi-Bel S, Sheikhavandi S, Telatar M, Tai L Q, Hwang M, Wang Z, Yang Z, Cheng R, Gatti R A. Hum Mutat. 1999;14:156–162. doi: 10.1002/(SICI)1098-1004(1999)14:2<156::AID-HUMU7>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Cortez D, Yazdi P, Neff N, Elledge S J, Qin J. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 45.Smith G C, Cary R B, Lakin N D, Hann B C, Teo S H, Chen D J, Jackson S P. Proc Natl Acad Sci USA. 1999;96:11134–11139. doi: 10.1073/pnas.96.20.11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larson P S, de las Morenas A, Cupples L A, Huang K, Rosenberg C L. Am J Pathol. 1998;152:1591–1598. [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang N, Chen P, Gatei M, Scott S, Khanna K K, Lavin M F. Oncogene. 1998;17:811–818. doi: 10.1038/sj.onc.1202007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.