Abstract
Efficient budding of HIV-1 from the plasma membrane of infected cells requires the function of a 6-kDa protein known as p6. A highly conserved Pro-Thr-Ala-Pro (PTAP) motif (the “late” or “L” domain), is critical for the virus-budding activity of p6. Recently, it was demonstrated that the product of tumor susceptibility gene 101 (TSG101), which contains at its N terminus a domain highly related to ubiquitin-conjugating (E2) enzymes, binds HIV-1 Gag in a p6-dependent fashion. We examined the impact of overexpressing the N-terminal region of TSG101 on HIV-1 particle assembly and release. We observed that this domain (referred to as TSG-5′) potently inhibits virus production. Examination of cells coexpressing HIV-1 Gag and TSG-5′ by electron microscopy reveals a defect in virus budding reminiscent of that observed with p6 L domain mutants. In addition, the effect of TSG-5′ depends on an intact p6 L domain; the assembly and release of virus-like particles produced by Gag mutants lacking a functional p6 PTAP motif is not significantly affected by TSG-5′. Furthermore, assembly and release of murine leukemia virus and Mason–Pfizer monkey virus are insensitive to TSG-5′. TSG-5′ is incorporated into virions, confirming the Gag/TSG101 interaction in virus-producing cells. Mutations that inactivate the p6 L domain block TSG-5′ incorporation. These data demonstrate a link between the E2-like domain of TSG101 and HIV-1 L domain function, and indicate that TSG101 derivatives can act as potent and specific inhibitors of HIV-1 replication by blocking virus budding.
Retroviral Gag proteins are necessary and sufficient for the formation and release of virus-like particles from Gag-expressing cells. The Gag proteins of type C retroviruses and lentiviruses are synthesized as polyprotein precursors that are expressed in the cytosol and transported to the plasma membrane. At the membrane, Gag–Gag interactions direct the assembly of virus particles that eventually bud off from the plasma membrane. Distinct domains in Gag provide the functions required for binding of Gag to membrane, Gag–Gag multimerization, and virus release. In HIV-1, membrane-binding determinants map to the N-terminal region of the matrix (MA or p17) domain; Gag interaction is mediated largely by a region spanning the C terminus of the capsid (CA or p24) domain, the p2 spacer peptide, and the N terminus of the nucleocapsid (NC or p7) domain; and the budding-off of particles from the plasma membrane is promoted by a PTAP motif located near the N terminus of p6 (1, 2).
Domains have been defined in the Gag proteins of several retroviruses (2), and in the structural proteins of the filoviruses (3) and rhabdoviruses (4, 5) that encode a virus release function analogous to that provided by p6 of HIV-1. These domains are collectively referred to as “late” or “L” domains to reflect their role late in the budding process (6). One of the intriguing characteristics of these L domains is that they all contain highly conserved motifs known to play a role in mediating protein–protein interactions among cellular proteins. For example, the L domain of HIV-1 p6 contains a PXXP motif (7, 8); L domain function of EIAV p9 is mediated by a YXXL motif (9); and PPXY motifs are responsible for the L domain activity of MLV p12 (10), Rous sarcoma virus p2b (11), pp16 of Mason–Pfizer monkey virus (M-PMV) (12), and the rhabdovirus M protein (4, 5). The filovirus matrix protein, VP40, contains overlapping PTAP and PPXY motifs (3). PXXP, YXXL, and PPXY motifs present in cellular proteins mediate interactions with Src-homology region 3 (SH3) domains, clathrin-associated adapter protein complexes, and WW domains, respectively (13–15). In several instances, interactions or subcellular colocalization between L domain proteins and cellular proteins have been reported (3, 5, 16, 17). These observations suggest that retroviral L domains function by interacting with host factors.
Although the physiological relevance of potential interactions between L domains and host factors remains to be established, a variety of lines of evidence suggest that L domains interact with the host ubiquitination machinery (reviewed in ref. 18): (i) The L domain protein of MLV (p12) and HIV-1 (p6) are themselves ubiquitinated (19). (ii) Depletion of free ubiquitin (Ub) in virus-producing cells with proteasome inhibitors impairs virus budding, producing a defect reminiscent of that observed with L domain mutations (20–22). (iii) The Ub ligase Nedd4 has been shown to interact with Rous sarcoma virus Gag, and the WW domain region from Nedd4 inhibits Rous sarcoma virus particle production (23). Finally, (iv) the cellular protein TSG101, which contains at its N terminus a domain related to Ub conjugating (E2) enzymes, binds Gag in a p6-dependent manner (24). The latter observation raises the possibility that TSG101 may play a physiologically relevant role in HIV-1 budding.
TSG101 was originally identified in a random RNA knockout screen for genes whose homozygous disruption led to neoplasia (25). Subsequently, the TSG101 protein, and its yeast homologue Vps23, have been implicated in a number of cellular functions, including mitotic spindle formation (26), genome stability (26), transcriptional transactivation (27, 28), regulation of MDM2 and p53 levels (29); and endosomal sorting (30–33). TSG101/Vps23 contains several domains (Fig. 1A); as mentioned above, at its N terminus is a region that bears sequence and predicted structural similarity with E2 Ub-conjugating enzymes (34). E2 (also known as UBC) enzymes play a critical role, in conjunction with E3 Ub ligases, in attaching Ub to target proteins. E2s contain at their active sites a Cys residue to which Ub is directly conjugated via a high-energy thioester bond (35). The E2-like domain of TSG101 and other E2-like cellular proteins lacks the catalytic site Cys and is therefore incapable of directly conjugating Ub. TSG101/Vps23 has recently been shown to assemble into a high molecular weight (350 kDa) complex (termed ESCRT-1) that functions in endosomal sorting (31, 33). Despite the fact that the E2-like domain of TSG101/Vps23 cannot directly conjugate Ub, it seems to interact directly with ubiquitinated proteins and sort them into the multivesicular body pathway (33).
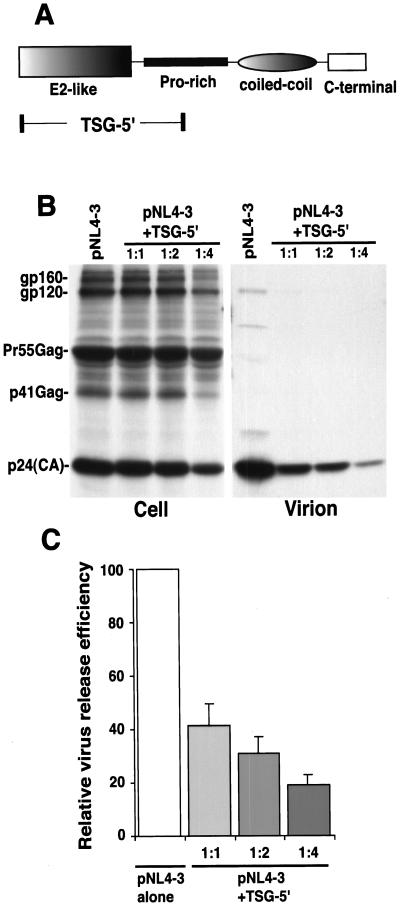
Figure 1.
TSG-5′ inhibits HIV-1 particle production. (A) Schematic organization of TSG101, depicting the major domains of the protein. The region of TSG101 expressed by the TSG-5′ vector is indicated. (B) Radioimmunoprecipitation analysis of cell and viral lysates obtained from HeLa cells transfected with pNL4–3 alone or cotransfected with pNL4–3 + TSG-5′ at 1:1, 1:2, or 1:4 DNA ratios. The positions of the Env glycoproteins gp160 and gp120; the Gag precursor, Pr55Gag; the p41Gag processing intermediate, and the mature CA protein are shown on the left. (C) Quantitative analysis of the effect of TSG-5′ on the production of virion-associated Gag. Relative virus release efficiency was calculated as the amount of virion-associated p24(CA) as a fraction of total Gag [(cell-associated Pr55Gag + p41Gag + p24(CA) + virion p24(CA)]. Data are averages from PhosphorImager analysis of at least four experiments, ± SE.
To determine whether TSG101 plays a role in HIV-1 budding, we overexpressed the N-terminal region of the protein, containing the E2-like domain (TSG-5′), with the full-length HIV-1 molecular clone pNL4–3. We observed that TSG-5′ expression markedly inhibited HIV-1 budding from virus-producing cells. This effect was specific for HIV-1 and depended on an intact p6 L domain. Furthermore, we demonstrated that TSG-5′ is incorporated into wild-type (WT) but not L-domain-deficient virus particles. We believe that these results have significant implications for our understanding of HIV-1 budding.
Materials and Methods
Cells and Plasmids.
HeLa and 293T cells were passaged as reported previously (36). The full-length HIV-1 molecular clone pNL4–3 (37) and the p6-mutant derivatives p6/L1Term, p6/PTAP−, and p6/S14Term have been described previously (8, 38), as have the protease (PR)-defective molecular clones pNL4–3/PR− and pNL4–3/PR−/PTAP− (8). WT MLV was expressed by using pSVψ−MLV-env− (ref. 39; obtained through the National Institutes of Health AIDS Research and Reference Reagent Program from N. Landau). The M-PMV molecular clone pSARM4 was generously provided by E. Hunter (Univ. of Alabama, Birmingham, AL). The TSG-5′ expression vector (28) was obtained from Z. Sun (Stanford Univ., Stanford, CA). The 15A NC mutant (40) was a kind gift of J. Luban (Columbia Univ., New York).
Transfections, Metabolic Labeling, Radioimmunoprecipitation, Western Blotting, and Electron Microscopy.
Transfections and metabolic labeling were performed as described (41). In brief, HeLa cells were transfected in six-well dishes. In all transfections, DNA concentrations were held constant with vector plasmid. Two days after transfection, cells were metabolically labeled overnight in 500 μCi (1 Ci = 37 GBq) per well [35S]Met/Cys. Cell and virion lysates were prepared and immunoprecipitated as described (41). Anti-HIV-1, MLV, and M-PMV immunoprecipitations were performed with anti-HIV-1 Ig (obtained through the NIH AIDS Research and Reference Reagent Program); goat anti-MLV Gag p30 (ViroMed Biosafety Laboratories, Camden, NJ), and goat anti-M-PMV Gag p27 (ViroMed Biosafety Laboratories), respectively. Anti-HA antibodies were obtained from Zymed, CLONTECH, or Sigma. Western blotting was performed as detailed (42). Quantitative Western blotting was performed with a fluorchem analyzer (Alpha Innotech, San Leandro, CA) by using the manufacturer's specifications. Virus-expressing HeLa cells were prepared and examined by transmission electron microscopy (EM) as described (43).
Results
TSG-5′ Inhibits HIV-1 Particle Budding.
If TSG101 plays a physiologically important role in HIV-1 assembly and release, expression of the N-terminal region of TSG101, which contains the E2-like domain, might interfere with HIV-1 particle production in a transdominant negative manner. To test this possibility, we used a vector that expresses the E2-like domain of TSG101 and a portion of the downstream Pro-rich motif [TSG-5′ (28); Fig. 1A]. We cotransfected HeLa cells with pNL4–3 and TSG-5′ at varying input DNA ratios (pNL4–3:TSG-5′ ratios 1:1, 1:2, and 1:4). We then evaluated the effect on virus particle production by radioimmunoprecipitation analysis. The results indicated that TSG-5′ expression induced a marked and dose-responsive inhibition of virus particle production. At a 1:1 DNA ratio, virus production was reduced by 60%; at a 1:4 ratio a 5-fold inhibition of virus production was observed (Fig. 1 B and C). Similar data were obtained in transfected 293T cells (data not shown). A severe defect in the kinetics of virus release in the presence of TSG-5′ was also observed by pulse–chase analysis (data not shown).
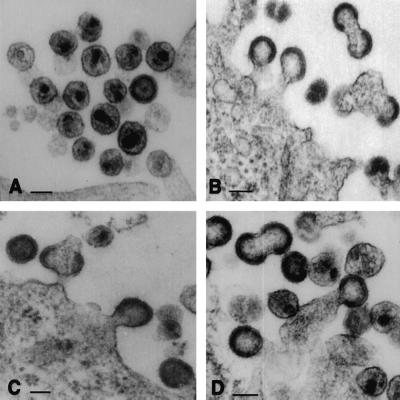
To identify the step in virus production that is inhibited by TSG-5′ expression, we performed EM on HeLa cells singly transfected with pNL4–3 or cotransfected with pNL4–3 + TSG-5′ (Fig. 2). In pNL4–3-transfected cells, numerous released mature virus particles were observed (Fig. 2A). In contrast, cells cotransfected with pNL4–3 and the TSG-5′ expression vector displayed an abundance of particles tethered to the plasma membrane (Fig. 2 B–D). Doublet particles were also frequently observed. This defect closely resembles that induced by mutations in the p6 L domain (7, 8, 38), strongly suggesting that TSG-5′ inhibits budding by blocking L domain function.
Figure 2.
TSG-5′ blocks HIV-1 budding. Cells transfected with pNL4–3 alone (A) or a 1:1 ratio of pNL4–3 + TSG-5′ (B–D) were analyzed by EM. Note in B–D the numerous tethered structures and doublet particles. (Bar = 100 nm.)
TSG-5′ Does Not Inhibit the Release of L-Domain-Defective p6 Mutants.
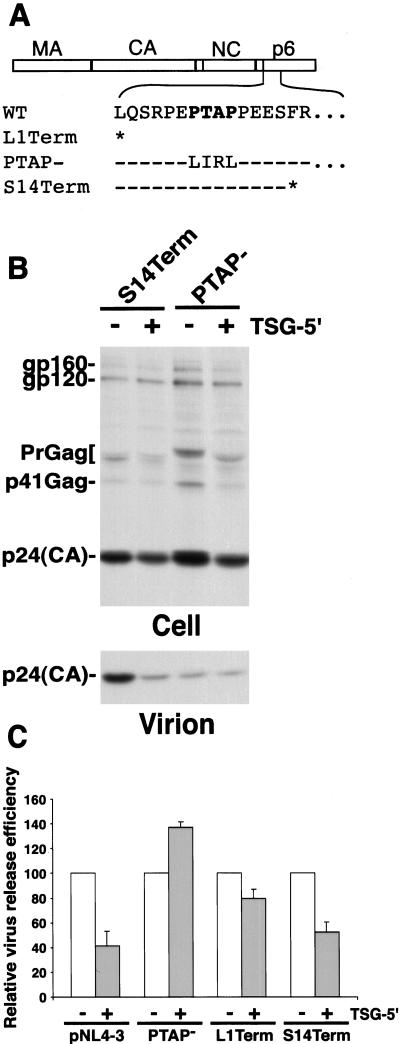
The EM data presented above suggest that TSG-5′ disrupts p6 L domain function. Thus, the release of p6 mutants lacking a functional L domain should not be diminished further by TSG-5′ expression. To test this prediction, we examined the impact of TSG-5′ on the release of several Gag mutants (Fig. 3A): p6/L1Term, which contains a stop codon at p6 position 1 and therefore lacks all of p6 (8); p6/PTAP−, which contains mutations in all four residues of the PTAP motif (8); and S14Term, which bears a stop codon at p6 position 14 but is release-competent (38). TSG-5′ expression reduced p6/L1Term particle release only slightly (20%) and caused a small but reproducible increase in the efficiency of p6/PTAP− release (Fig. 3 B and C). In contrast, release of the L-domain-competent p6 mutant S14Term was inhibited to an extent comparable with WT.
Figure 3.
The inhibition of virus budding by TSG-5′ depends on the presence of an intact p6 L domain. (A) p6 mutations used in the study. The major Gag domains are depicted at Top, under which is shown the amino acid sequence of the N terminus of WT HIV-1 p6. The p6 L domain motif (PTAP) is shown in bold. Sequence of the L1Term, PTAP− (8), and S14Term (38) mutants is provided. Asterisks denote stop codon; dashes denote sequence identity with WT. (B) Radioimmunoprecipitation analysis of cell and viral lysates obtained from HeLa cells transfected with the indicated p6 mutants alone or cotransfected with TSG-5′ at a 1:1 DNA ratio. Positions of proteins are indicated as in Fig. 1 B and C. Quantitative analysis of the effect of TSG-5′ on the production of virion-associated Gag, determined by PhosphorImager analysis as in Fig. 1C. The virus release efficiency in the absence of TSG-5′ is set at 100% for each clone. Data are averages from at least three independent experiments, ± SE. Note that the virus release efficiency of the PTAP− and L1Term molecular clones in the absence of TSG-5′ is markedly lower than that of WT (8), whereas S14Term virus release efficiency is similar to that of WT (38).
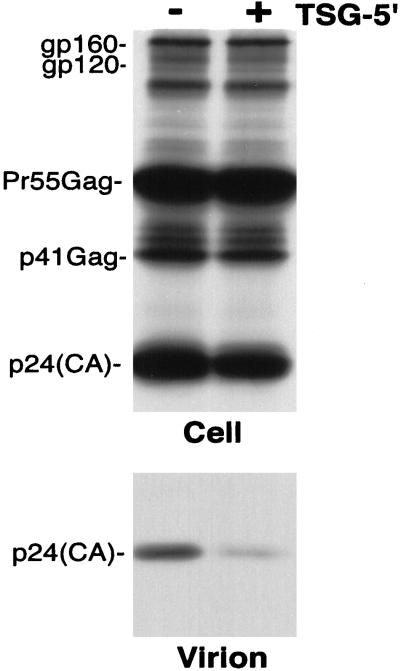
Because the L1Term and PTAP− p6 mutations severely impair virus release relative to WT, we wished to test another mutant that is similarly defective in virus production yet contains an intact L domain. Inhibition of the release of such a mutant by TSG-5′ would validate the conclusion that L domain mutations and TSG-5′ block the same step in the assembly/release pathway. For this purpose, we used the 15A NC mutant, in which all basic residues in NC were mutated to Ala (40). The 15A mutant displays a 10- to 20-fold defect in virus production relative to WT (40; data not shown), reportedly because of a defect in Gag assembly. In contrast to the insensitivity to TSG-5′ observed with the L-domain-defective mutants, the release of 15A was inhibited to the same extent as WT (Fig. 4). Together, these results strongly suggest that TSG-5′ disrupts virus release by specifically inhibiting p6 L domain function.
Figure 4.
TSG-5′ inhibits the release of the 15A NC mutant. Radioimmunoprecipitation analysis of cell and viral lysates obtained from HeLa cells transfected with the 15A molecular clone (40) or cotransfected with 15A and TSG-5′ at a 1:1 DNA ratio. Positions of proteins are indicated as in Fig. 1B. In this experiment, TSG-5′ expression caused a 60% reduction in virus release efficiency.
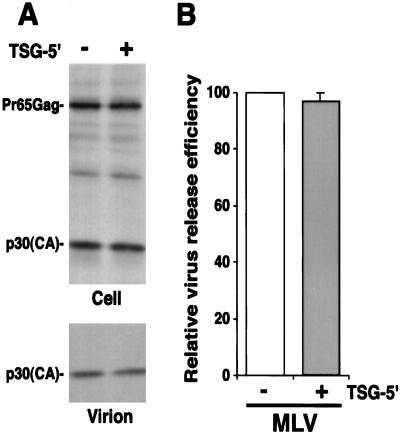
TSG-5′ Does Not Inhibit the Release of MLV or M-PMV Particles.
If the virus-budding defect imposed by TSG-5′ is specific for the HIV-1 p6 L domain, we would expect that the release of other retroviruses would be insensitive to TSG-5′ expression. To test this expectation, we examined the impact of TSG-5′ expression on MLV particle production. Unlike HIV-1, which contains a PTAP L domain, MLV contains an L domain (in p12-Gag) of the PPPY-type (44). HeLa cells were transfected with the MLV expression vector pSVψ−MLV-env− (39) or cotransfected with pSVψ−MLV-env− + TSG-5′. Cell- and virion-associated proteins were detected by radioimmunoprecipitation analysis (Fig. 5). In contrast to results obtained with WT HIV-1, MLV particle production was not inhibited by TSG-5′. We also tested the effect of TSG-5′ expression on M-PMV particle release. Like MLV, M-PMV contains a PPPY-type L domain (12). We observed that M-PMV particle budding was unaffected by TSG-5′ (data not shown). These results further support the conclusion that TSG-5′ specifically inhibits HIV-1 p6 L domain function.
Figure 5.
TSG-5′ does not inhibit the budding of MLV virions. HeLa cells were transfected with pSVψ−MLV-env− (39) alone or at a 1:1 DNA ratio with TSG-5′. (A) Radioimmunoprecipitation analysis of cell and viral lysates. The positions of the MLV Gag precursor Pr65Gag and the mature CA protein p30 are indicated at the left. (B) Quantitative analysis of the effect of TSG-5′ on the production of virion-associated Gag, determined by PhosphorImager analysis as in Fig. 1C. Data are averages from three independent experiments, ± SE.
TSG-5′ Is Incorporated into Virions in a Manner That Depends on an Intact p6 L Domain.
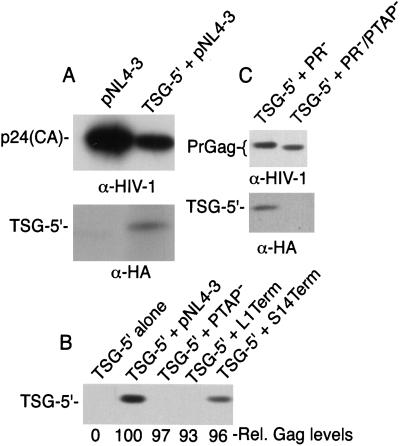
To probe the interaction between TSG-5′ and Gag, we sought to determine whether TSG-5′ is incorporated into HIV-1 virions. HeLa cells were transfected with pNL4–3 alone or pNL4–3 + TSG-5′. Transfected cells were metabolically labeled, and cell and virion lysates were immunoprecipitated with either anti-HIV-1 Ig or, because TSG-5′ contains an N-terminal HA epitope tag (28), with anti-HA antiserum. A band of the expected size (≈30 kDa) was clearly detected in virion lysates produced from cells cotransfected with pNL4–3 and TSG-5′ but not in lysates of virions produced from cells expressing pNL4–3 alone (Fig. 6A).
Figure 6.
TSG-5′ is incorporated into WT but not L-domain-deficient HIV-1 virions. (A) Radioimmunoprecipitation analysis of virions derived from cells transfected with pNL4–3 alone or pNL4–3 + TSG-5′ (1:1 DNA ratio). Immunoprecipitations were performed with either anti-HIV Ig or anti-HA (CLONTECH). Levels of cell-associated Gag expression were equivalent in each lysate (data not shown). (B) Western blot analysis of virions produced from HeLa cells transfected with TSG-5′ alone, or a 1:1 ratio of TSG-5′ + pNL4–3, pNL4–3/PTAP−, pNL4–3/L1Term, or pNL4–3/S14Term. Western blotting was performed with anti-HA antibody (Zymed) or anti-HIV Ig. The anti-HIV-1 Ig blots were subjected to quantitative fluorchem analysis (see Materials and Methods); the relative amounts of virion-associated Gag are indicated under each lane (Rel. Gag levels). Levels of cell-associated Gag and TSG-5′ expression were comparable in each lysate transfected with an HIV-1 molecular clone (data not shown). (C) Western blot analysis of virions produced from cells cotransfected with a 1:1 DNA ratio of TSG-5′ and either pNL4–3/PR− or pNL4–3/PR−/PTAP− (8). Western blotting was performed with anti-HIV-1 Ig or anti-HA antibody (Zymed). Levels of cell-associated Gag and TSG-5′ were comparable in each lysate (data not shown).
We demonstrated above (Fig. 3) that the release of p6 mutants lacking a functional L domain (e.g., L1Term and PTAP−) was not significantly inhibited by TSG-5′ expression, whereas release of the S14Term mutant was markedly impaired by TSG-5′. These results suggested that mutational inactivation of the p6 L domain might prevent the interaction of TSG-5′ with Gag and the subsequent incorporation of TSG-5′ into virions. To test this possibility, we evaluated the level of TSG-5′ in L1Term, PTAP−, and S14Term virions. To increase the sensitivity of the analysis, we used Western blotting to detect both Gag and TSG-5′. The results (Fig. 6B) indicated that L1Term and PTAP− mutant virions failed to incorporate detectable levels of TSG-5′. In contrast, TSG-5′ was readily detected in WT and S14Term virions. Cells transfected with TSG-5′ alone did not release pelletable TSG-5′ into the medium. The levels of virion-associated Gag were measured by quantitative Western blotting analysis (see Materials and Methods); the relative amounts of virion Gag in each sample are shown under each lane in Fig. 6B. We also examined TSG-5′ incorporation into WT and p6-mutant virions by using the PR− molecular clones pNL4–3/PR− and pNL4–3/PR−/PTAP−. The use of PR− clones simplifies the detection of Gag, because only the Gag precursor is produced. Consistent with the data presented in Fig. 6B, virions produced upon expression of WT Gag readily incorporated TSG-5′, whereas virions lacking the PTAP L domain motif failed to incorporate detectable levels of TSG-5′ (Fig. 6C).
Discussion
In this study, we demonstrate that overexpression of the N-terminal region of TSG101 (TSG-5′) markedly inhibits HIV-1 particle production by blocking efficient virus budding from the plasma membrane. The insensitivity of L-domain-defective mutants and heterologous retroviruses (MLV and M-PMV) to inhibition by TSG-5′ strongly suggests that TSG-5′ acts by blocking HIV-1 p6 L domain function. Virion incorporation data indicate that TSG-5′ interacts with WT HIV-1 Gag but not Gag mutants lacking the PTAP L domain motif.
The data presented here strongly support the hypothesis that TSG101 plays a physiologically relevant role in HIV-1 budding. Although precisely what role TSG101 plays in modulating HIV-1 release remains to be determined, consideration of the roles TSG101 and other E2-like proteins play in the cell (see the introduction) might offer some clues. Two general classes of models seem most likely: TSG101 could influence the ubiquitination of Gag itself or the ubiquitination of a host factor; or TSG101 could alter the localization or sorting of a ubiquitinated host protein. (i) The reported ability of TSG101 and other E2-like proteins to inhibit or modify the target specificity of ubiquitination (29, 45) raises the possibility that binding of TSG101 by p6 could alter the extent to which Gag becomes ubiquitinated. Whether ubiquitination of Gag itself plays a role in virus budding remains to be established. Direct fusion of Ub to Rous sarcoma virus Gag reverses the impact of proteasome inhibitors on particle release but does not eliminate the requirement for the L domain in budding (20). Removal of the sites of p6 ubiquitination (p6 residues Lys27 and Lys33) by amino acid substitution (46) or p6 truncation [e.g., S14Term (38)] has no effect on the efficiency of virus budding. Furthermore, S14Term remains sensitive to TSG-5′ inhibition, and incorporates TSG-5′ protein into virions, despite the fact that it lacks the sites of p6 ubiquitination. However, the latter observations do not exclude a possible role for Gag ubiquitination upstream of the p6 domain. Alternatively, ubiquitination of Gag itself may be an irrelevant consequence of the recruitment of ubiquitination machinery to the site of budding. Thus, the ubiquitination of unidentified host factor(s) could be the driving force in stimulating virus budding. Because ubiquitination plays a role both in endosomal sorting (33) and in the internalization of plasma membrane proteins (47), the modulation of host protein ubiquitination could alter the protein composition of the plasma membrane at the site of budding. (ii) Recent evidence (33) indicates that TSG101 plays a role in the sorting of ubiquitinated proteins into the endosomal pathway. Cellular machinery normally involved in budding reactions at intracellular sites (e.g., the multivesicular body) could be recruited to the site of budding at the plasma membrane through interactions with the L domain. The protein whose sorting is affected by TSG101 could be a host factor that directly or indirectly modulates budding. Our recent observation of a link between L domain function and rafts (48) suggests that the hypothetical host protein(s) whose expression or localization is affected by TSG101 may be raft localized.
If TSG101 plays a central role in HIV-1 budding, how might TSG-5′ inhibit particle release? TSG-5′ could bind p6 and prevent it from interacting with its endogenous ligand (perhaps full-length TSG101). Alternatively, TSG-5′ could interact with and disrupt cellular machinery, such as the high molecular weight ESCRT-1 complex in which TSG101/Vps23 is localized (ref. 33; see the introduction) thereby preventing it from functioning in HIV-1 particle release. Regardless of its mechanism of action, further investigations into the ability of TSG-5′ to disrupt particle budding will likely provide new insights into this crucial step in the HIV-1 life cycle. The high degree of specificity displayed by TSG-5′ in blocking HIV-1 budding raises the possibility that TSG-5′, or other TSG101 derivatives, may be useful in gene therapy strategies to inhibit HIV-1 replication in vivo. Alternatively, small molecule inhibitors that disrupt the p6/TSG101 interaction may display antiviral activity.
After submission of this manuscript, two articles were published that support the hypothesis that TSG101 plays a crucial role in HIV-1 budding. Garrus et al. (49) reported an interaction between HIV-1 p6 and TSG101; this interaction was blocked by mutations in the PTAP motif of p6. The functional relevance of the p6/TSG101 interaction was demonstrated by the finding that transient inhibition of TSG101 expression induced a budding defect similar to that observed with p6 L domain mutants. Martin-Serrano et al. (50) reported that both HIV-1 p6 and Ebola virus VP40 (which also contains a PTAP motif) interact with TSG101. Recruitment of TSG101 in trans to the site of HIV-1 budding could substitute for a functional L domain. Thus, with use of complementary approaches, these two studies, together with our report, highlight the importance of TSG101 in HIV-1 release.
Acknowledgments
We thank Z. Sun, J. Luban, and E. Hunter for plasmids. The following reagents were obtained through the National Institutes of Health AIDS Research and Reference Reagent Program: pSVψ−MLV-env− (from N. Landau) and anti HIV-Ig (from Nabi, Rockville, MD).
Abbreviations
- TSG101
tumor susceptibility gene 101
- MA
matrix
- CA
capsid
- NC
nucleocapsid
- MLV
murine leukemia virus
- M-PMV
Mason–Pfizer monkey virus
- L
late
- Ub
ubiquitin
- EM
electron microscopy
- PR
protease
- WT
wild type
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Swanstrom R, Wills J W. Synthesis, Assembly, and Processing of Viral Proteins. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [PubMed] [Google Scholar]
- 2.Freed E O. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 3.Harty R N, Brown M E, Wang G, Huibregtse J, Hayes F P. Proc Natl Acad Sci USA. 2000;97:13871–13876. doi: 10.1073/pnas.250277297. . (First Published November 28, 2000; 10.1073/pnas.250277297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craven R C, Harty R N, Paragas J, Palese P, Wills J W. J Virol. 1999;73:3359–3365. doi: 10.1128/jvi.73.4.3359-3365.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harty R N, Paragas J, Sudol M, Palese P. J Virol. 1999;73:2921–2929. doi: 10.1128/jvi.73.4.2921-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang M, Orenstein J M, Martin M A, Freed E O. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puffer B A, Parent L J, Wills J W, Montelaro R C. J Virol. 1997;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan B, Li X, Goff S P. EMBO J. 1999;18:4700–4710. doi: 10.1093/emboj/18.17.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang Y, Cameron C E, Wills J W, Leis J. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yasuda J, Hunter E. J Virol. 1998;72:4095–4103. doi: 10.1128/jvi.72.5.4095-4103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayer B J, Eck M J. Curr Biol. 1995;5:364–367. doi: 10.1016/s0960-9822(95)00073-x. [DOI] [PubMed] [Google Scholar]
- 14.Ohno H, Stewart J, Fournier M C, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino J S. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- 15.Sudol M. Prog Biophys Mol Biol. 1996;65:113–132. doi: 10.1016/s0079-6107(96)00008-9. [DOI] [PubMed] [Google Scholar]
- 16.Garnier L, Wills J W, Verderame M F, Sudol M. Nature (London) 1996;381:744–745. doi: 10.1038/381744a0. [DOI] [PubMed] [Google Scholar]
- 17.Puffer B A, Watkins S C, Montelaro R C. J Virol. 1998;72:10218–10221. doi: 10.1128/jvi.72.12.10218-10221.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogt V M. Proc Natl Acad Sci USA. 2000;97:12945–12947. doi: 10.1073/pnas.97.24.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ott D E, Coren L V, Copeland T D, Kane B P, Johnson D G, Sowder R C, 2nd, Yoshinaka Y, Oroszlan S, Arthur L O, Henderson L E. J Virol. 1998;72:2962–2968. doi: 10.1128/jvi.72.4.2962-2968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patnaik A, Chau V, Wills J W. Proc Natl Acad Sci USA. 2000;97:13069–13074. doi: 10.1073/pnas.97.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schubert U, Ott D E, Chertova E N, Welker R, Tessmer U, Princiotta M F, Bennink J R, Krausslich H G, Yewdell J W. Proc Natl Acad Sci USA. 2000;97:13057–13062. doi: 10.1073/pnas.97.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strack B, Calistri A, Accola M A, Palu G, Gottlinger H G. Proc Natl Acad Sci USA. 2000;97:13063–13068. doi: 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kikonyogo A, Bouamr F, Vana M L, Xiang Y, Aiyar A, Carter C, Leis J. Proc Natl Acad Sci USA. 2001;98:11199–11204. doi: 10.1073/pnas.201268998. . (First Published September 18, 2001; 10.1073/pnas.201268998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.VerPlank L, Bouamr F, LaGrassa T J, Agresta B, Kikonyogo A, Leis J, Carter C A. Proc Natl Acad Sci USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. . (First Published June 26, 2001;, 10.1073/pnas.131059198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Cohen S N. Cell. 1996;85:319–329. doi: 10.1016/s0092-8674(00)81111-3. [DOI] [PubMed] [Google Scholar]
- 26.Xie W, Li L, Cohen S N. Proc Natl Acad Sci USA. 1998;95:1595–1600. doi: 10.1073/pnas.95.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hittelman A B, Burakov D, Iniguez-Lluhi J A, Freedman L P, Garabedian M J. EMBO J. 1999;18:5380–5388. doi: 10.1093/emboj/18.19.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Z, Pan J, Hope W X, Cohen S N, Balk S P. Cancer. 1999;86:689–696. doi: 10.1002/(sici)1097-0142(19990815)86:4<689::aid-cncr19>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Liao J, Ruland J, Mak T W, Cohen S N. Proc Natl Acad Sci USA. 2001;98:1619–1624. doi: 10.1073/pnas.98.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Kane T, Tipper C, Spatrick P, Jenness D D. Mol Cell Biol. 1999;19:3588–3599. doi: 10.1128/mcb.19.5.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babst M, Odorizzi G, Estepa E J, Emr S D. Traffic. 2000;1:248–258. doi: 10.1034/j.1600-0854.2000.010307.x. [DOI] [PubMed] [Google Scholar]
- 32.Bishop N, Woodman P. J Biol Chem. 2001;276:11735–11742. doi: 10.1074/jbc.M009863200. [DOI] [PubMed] [Google Scholar]
- 33.Katzmann D J, Babst M, Emr S D. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 34.Koonin E V, Abagyan R A. Nat Genet. 1997;16:330–331. doi: 10.1038/ng0897-330. [DOI] [PubMed] [Google Scholar]
- 35.Varshavsky A. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 36.Murakami T, Freed E O. Proc Natl Acad Sci USA. 2000;97:343–348. doi: 10.1073/pnas.97.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demirov D G, Orenstein J M, Freed E O. J Virol. 2002;76:105–117. doi: 10.1128/JVI.76.1.105-117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landau N R, Littman D R. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cimarelli A, Luban J. J Virol. 2000;74:6734–6740. doi: 10.1128/jvi.74.15.6734-6740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freed E O, Martin M A. J Virol. 1994;68:2503–2512. doi: 10.1128/jvi.68.4.2503-2512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiernan R E, Ono A, Englund G, Freed E O. J Virol. 1998;72:4116–4126. doi: 10.1128/jvi.72.5.4116-4126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freed E O, Orenstein J M, Buckler-White A J, Martin M A. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan B, Campbell S, Bacharach E, Rein A, Goff S P. J Virol. 2000;74:7250–7260. doi: 10.1128/jvi.74.16.7250-7260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hofmann R M, Pickart C M. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 46.Ott D E, Coren L V, Chertova E N, Gagliardi T D, Schubert U. Virology. 2000;278:111–121. doi: 10.1006/viro.2000.0648. [DOI] [PubMed] [Google Scholar]
- 47.Hicke L. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 48.Ono A, Freed E O. Proc Natl Acad Sci USA. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garrus J E, von Schwedler U K, Pornillos O W, Morham S G, Zavitz K H, Wang H E, Wettstein D A, Stray K M, Cote M, Rich R L, et al. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 50.Martin-Serrano J, Zang T, Bieniasz P D. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]