Abstract
A high-density transposon mutagenesis strategy was applied to the Haemophilus influenzae genome to identify genes required for growth or viability. This analysis detected putative essential roles for the products of 259 ORFs of unknown function. Comparisons between complete genomes defined a subset of these proteins in H. influenzae having homologs in Mycobacterium tuberculosis that are absent in Saccharomyces cerevisiae, a distribution pattern that favors their use in development of antimicrobial therapeutics. Three genes within this set are essential for viability in other bacteria. Interfacing the set of essential gene products in H. influenzae with the distribution of homologs in other microorganisms can detect components of unrecognized cellular pathways essential in diverse bacteria. This genome-scale phenotypic analysis identifies potential roles for a large set of genes of unknown function.
The availability of complete microbial genome sequences allows large-scale functional analysis of genes known only by their DNA sequences. A growing number of high-throughput approaches have been developed to use this information. Expression profiling by DNA microarrays (1) measures gene expression in a highly parallel fashion and can identify possible functional relationships between genes by evaluation of coexpression (2). Two-hybrid protein interaction technologies determine which proteins can physically interact providing a possible means of linking genes to known phenotypes. Construction of specific gene-knockouts on a genomic scale was initiated in Escherichia coli (3) and Bacillus subtilis (4). This strategy provides a direct means to assess which genes are required for a given phenotype yet it is laborious and generally impractical for individual laboratories. Several alternative approaches have been recently used. In Saccharomyces cerevisiae, genetic footprinting applied to chromosome V detected ≈53 essential genes (5). Sequencing of randomly generated mutants in Mycoplasma genitalium and Mycoplasma pneumoniae has been shown to allow rapid identification of putatively nonessential genes (6), a technique that is feasible for laboratories with access to high-throughput DNA sequencing capabilities. This approach can potentially identify essential genes with a confidence level that depends on characterizing enough transposon junctions to cover the complete genome at a high level of redundancy (6). Several random genetic screens are applicable to essential gene identification. A transposon-based conditional expression system was used to identify 16 essential genes in Vibrio cholerae (7). A screen based on inhibition of gene function by antisense RNA applied to Staphylococcus aureus ascribed roles in viability to ≈150 genes (8). These approaches offer the ability to easily identify the subset of essential genes amenable to conditional expression or inhibition. However, these methods are stochastic and, therefore, provide information only for those genes encountered during the screen.
We have described an approach (9) for detecting essential genes that applies high-density in vitro transposon mutagenesis and genetic footprinting to discrete chromosomal segments of genomes of naturally transformable organisms. This approach rapidly detected known and previously unknown genes essential for growth and viability in vitro. We now report application of this method to the complete genome of H. influenzae, a common human pathogen causing a range of diseases including otitis media and pneumonia (10, 11). The data obtained from this study indicate that ≈54% of essential genes lack defined functional roles. Sequence comparison detects potential orthologs of a large number of these putatively essential genes in other bacterial species. This information is relevant to defining both the common essential pathways of life and potential targets for development of antimicrobial therapeutics. This genome-scale database provides the first experimental evidence of putative biological roles for a large group of genes of unknown function in H. influenzae.
Methods
Plasmid Construction.
To further increase the efficiency of the in vitro transposon mutagenesis procedure we constructed a mariner-based minitransposon containing an H. influenzae DNA uptake consensus sequence (US) that increases transformation frequency (12). A KpnI site in pENT3 (9) was removed by digestion with SpeI and SapI followed by blunt-end creation with T4 polymerase, religation, and cloning of a derivative containing a unique KpnI site within the transposon, pENT3kpnI. Primers (US1-BA, 5′-GGTACCAAAACCAGTTTCAGGTCAGCAAG-3′; and US2-BA, 5′-GGTACCTCGCCTTTGTTCCTTGTGTGCC-3′) were used to PCR-amplify a 177-bp segment of the H. influenzae chromosome containing a strong match to the US consensus that is distributed throughout the genome (13). This US segment was cloned into the KpnI site of pENTkpnI to create pENTUS carrying the Tnmagellan4 minitransposon.
Mutagenesis of H. influenzae Genomic Segments and Genetic Footprinting.
H. influenzae Rd genome (ATCC #9008) was mutagenized by in vitro transposition, using a series of overlapping ≈10-kb PCR products as targets (see Fig. 1 for overview). Primers were designed to create overlapping ≈10-kb PCR products representing the complete H. influenzae genome sequence (14). Automated primer design was conducted with the macvector program with the desired melting point parameter set at a minimum of 62°C. The resulting primer set was filtered by using Microsoft excel to select primers that produced ≈10-kb products with ≈5-kb of overlap. This procedure produced a set of primers to generate targets for in vitro transposition and for querying each 2.5-kb segment of the chromosome by genetic footprinting. In vitro transposition reactions were conducted essentially as described (9) and contained 500 ng of each 10-kb PCR product, 100 ng of pENTUS, and purified hyperactive C9 mutant (15) Himar1 transposase, a gift of D. Lampe (Duquesne Univ., Pittsburgh). The C9 mutant transposase increases the efficiency of this mutagenesis procedure by 10–50-fold (15). Each reaction was transformed into competent H. influenzae (16) outgrown for 1 h in brain heart infusion (BHI) supplemented with hemin (10 μg/ml) and nicotinamide adenine dinucleotide [10 μg/ml (BHIs)], and selected for colony formation after 36 h of growth at 37°C on BHIs agar containing 20 μg/ml of kanamycin sulfate. Transformants were pooled in BHIs with 15% glycerol and stored as an ordered set at −80°C. Dilutions of each frozen stock were plated to obtain at least 2,000 colonies. Colonies were pooled and adjusted to an approximate OD600 of 0.1, and 1 μl from each was added to PCR reactions containing a chromosomal primer corresponding to the mutagenized region and a primer specific to the Himar1 inverted repeat at both ends of Tnmagellan4, Marout (5′-CCGGGGACTTATCAGCCAACC-3′). Genetic footprinting was conducted essentially as described (9) except that the following reaction cycle was used: 94°C for 30 s followed by 68°C for 5 min + 15 s per cycle for 30 cycles. A GIBCO/BRL 1-kb DNA ladder was used as a size marker for gel electrophoresis.
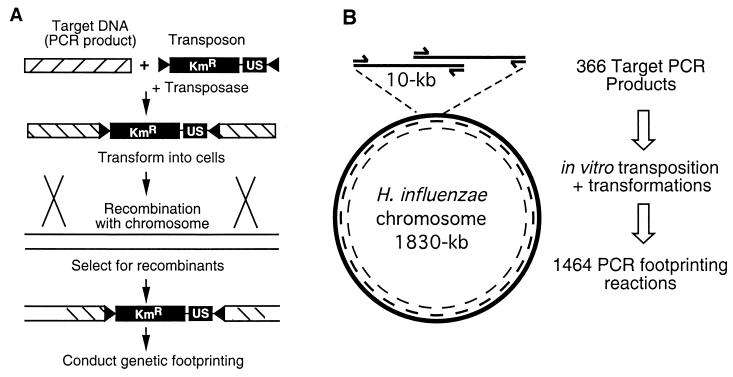
Figure 1.
Strategy for whole-genome mutagenesis by in vitro mariner mutagenesis and functional analysis by genetic footprinting. (A) In vitro transposon mutagenesis with Tnmagellan4 conducted on PCR products corresponding to regions of the H. influenzae chromosome. Each mutagenized DNA segment was introduced separately into H. influenzae by natural transformation and homologous recombination. The Tnmagellan4 element encodes kanamycin resistance (KmR) and contains a DNA US, both of which are flanked by transposon-inverted repeats (black triangles). (B) PCR products (10 kb) were generated as targets for mutagenesis with ≈5 kb of overlap between adjacent segments. These products were mutagenized (as in A) to generate 366 individual pools of transposon mutants. The same primers were used to obtain genetic footprinting data for every ≈2.5 kb of the genome, and each primer was used on two independent pools of mutants.
Results
Generation of an Ordered Collection of Transposon Insertion Mutants Encompassing the H. influenzae Genome.
Our previous analysis of H. influenzae by the genome analysis and mapping by in vitro transposition (GAMBIT) procedure indicated that this approach could be applied to the complete genome to define functional roles of genes based on DNA sequence information as the sole input data (9). Genes essential for viability under a defined growth condition (in this case, the ability to form colonies on BHIs agar medium) sustain a markedly reduced frequency of transposon insertions as detected by genetic footprinting. Results of this approach correlate well with independent methods for assigning essentiality (9, 17) and, therefore, genes that cannot sustain transposon insertions are designated as putatively essential. Adjacent nonessential genes or intergenic regions in each DNA segment serve as internal controls that can sustain insertions. Extended-length PCR reactions were used to amplify 366 ≈10-kb DNA segments representing the H. influenzae genome. The segments were designed to overlap by ≈5-kb (Fig. 1). This design achieves two objectives. (i) Mutations within most segments of the genome are represented in at least two independent pools of transposon mutants. (ii) Primers within the set are complementary to sites on the H. influenzae chromosome spaced, on average, every 2.5 kb. Analysis of each gene in two independent pools provides verification. The distance of 2.5 kb between primer binding sites allows us to obtain overlapping data from some primers because the average resolution limit of our electrophoresis gels is ≈4 kb.
Each PCR product was mutagenized by in vitro transposition in reactions containing the hyperactive C9 mutant of the Himar1 transposase and a mariner-based minitransposon (Tnmagellan4) carrying an H. influenzae DNA uptake signal and a gene encoding kanamycin resistance. Mutagenized DNA was transformed into H. influenzae and selected for colony formation on BHIs agar containing kanamycin (Methods). The majority (351/366) of reactions each yielded between 80 to 5,000 colonies with an average of 564. Six reactions yielded fewer than 50 colonies and 9 target PCR products that differed in size from the predicted segments based on the genome sequence were omitted. Variation in numbers of colonies obtained, in part, reflects that many 10-kb stretches of DNA contain different proportions of essential and nonessential genes. For example, when 4 of the 6 regions that yielded fewer than 50 colonies were analyzed by means of adjacent overlapping segments, they were found to contain a relatively high proportion of putatively essential genes (22 of the 27 genes examined). Therefore, a relative scarcity of nonessential genes within these regions could account for recovery of fewer colonies. The other two regions that yielded few colonies were adjacent segments corresponding to putative phage gene products homologous to those of phage Mu (14). The significance of reduced insertional mutagenesis frequency in this Mu-like locus is not clear.
Analysis of Transposon Insertion Mutants in Each Subgenomic Chromosomal Region.
Mutants representing each chromosomal region were pooled and analyzed by genetic footprinting as described (9). Distances from each chromosome-specific primer were calculated, and the map positions of each ORF with respect to the primer were directly superimposed onto the gel images as shown in Fig. 2. Each region was scored and color-coded (Fig. 2) according to whether insertions could be detected. The intensity of the bands on the footprinting gels, although not precisely quantitative, provides an indication of the relative number of mutants containing insertion mutations at each chromosomal position. As previous observations (5, 9) indicated that essential genes are able to sustain insertion mutations in the 3′ end, we exclusively scored the 5′ 75% of the length of each ORF by these criteria. Genes sustaining no insertions in this region were designated as putatively essential for growth or viability. Those capable of sustaining insertions in a single apparent position are also likely to be essential because some essential genes encode nonessential domains or can be divided into complementing ORFs (although some bands might represent insertions in multiple adjacent sites depending on the resolution of the gel system). We scored genes as containing single permissive insertion sites only when we could resolve bands to within 10% of the length of the gene. Sixty-five ORFs contained insertions apparently limited to two locations within the coding region. This group includes both genes known to be essential (e.g., secA; ref. 9) and those demonstrated to be nonessential (such as the hktE gene encoding catalase; ref. 18) and, therefore, such information was considered equivocal. Genes encoding untranslated RNA were not scored. Small genes (≈300 bp or less) were not scored unless they were located sufficiently close to analysis primers to allow accurate resolution of insertion site positions.
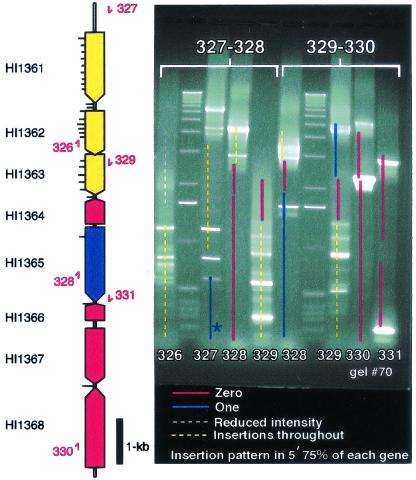
Figure 2.
Scoring system for genetic footprinting of the H. influenzae genome. Maps of ORF locations with respect to binding sites of each primer were superimposed on gel images. Dashed (putative nonessential regions) or solid (putative essential regions) lines on the gel image represent predicted segments of H. influenzae coding sequences. Primer reference numbers (red) are listed near primer positions on the gene map (Left) and on the gel image at the bottom of each lane. Tick marks on the gene map represent approximate positions where insertions were detected. Pools of mutants derived from independent transposition reactions are named by the primer pairs used to generate targets for in vitro transposon mutagenesis (top, brackets). The gel image shows analysis of two overlapping ≈10-kb regions corresponding to mutant pools 327–328 and 329–330. Colors denote the summarized results. Red , no insertions; blue, one insertion site; white, insertion mutations partially reduce growth; and yellow, evenly distributed insertions/putative nonessential gene. Absence of insertions within 300 bp of the end of a target PCR product was not considered significant because of the minimum distance required for homologous recombination (e.g., blue line marked with asterisk in lane 327).
The overall numbers of results and distribution in the known vs. unknown functional categories are listed in Table 1. Zero or one insertion was detected in 38% and 15%, respectively, of the total 1,272 ORFs that were scored. Thus, we identified 670 putative essential genes of which 478 genes contained no insertions detected by this method. The complete list of putative essential genes is available in Table 3, which is published as supporting information on the PNAS web site, www.pnas.org.
Table 1.
Summary of putative essential and nonessential genes
| Functional designation | No. |
|---|---|
| Essential gene candidates (no insertions) | 478 |
| Hypothetical genes (unknown functions) | 259 |
| Genes with known functions | 219 |
| Nonessential genes (insertions distributed along ORF) | 538 |
| One insertion detected | 191 |
| Two insertions detected | 65 |
| Not scored (too far from primers) | 453 |
Genome Analysis and Mapping by in Vitro Transposition (GAMBIT) Analysis of Gene Products Homologous to Genes with Assigned Functions in Other Organisms.
Many of the putative essential genes we detected in H. influenzae have essential homologs in other organisms. These include cell division proteins, murein synthesis enzymes, enzymes of one-carbon metabolism, and many others (19–21) (Table 3). Essentiality of several gene products was initially unexpected based on the functional annotation provided with the genome sequence. HI0119 was detected as an essential gene in our screen, yet was annotated as fimA, encoding a major subunit of the fimbrial surface structure that is nonessential for growth in many organisms. However, HI0119 was shown by Lu et al. (22) to encode a protein critical for zinc transport. Deletion of this ORF in H. influenzae confers a severe growth defect under aerobic conditions in the absence of supplementation with a high concentration of zinc (22). Large-scale functional screens such as GAMBIT can aid in reconciling genome annotation with subsequent experimental data in the literature by detecting such apparent inconsistencies.
We also detected putatively essential genes in H. influenzae that are nonessential in other bacteria. The smpB gene of H. influenzae was identified as essential in our screen yet is nonessential in E. coli and B. subtilis (23, 24). Transposon insertions in the smpB of M. pneumoniae have also been detected (6). SmpB is essential for activity of the tmRNA (23, 25). The tmRNA functions as an mRNA and as a tRNA that adds a short peptide tag to prematurely terminated translation products, targeting them for degradation. Although tmRNA is nonessential in E. coli it is essential for viability in Neisseria gonorrhoeae (26). H. influenzae and N. gonorrhoeae are both obligate human parasites with relatively small genomes (≈1.8 and ≈2.2-Mbp, respectively) and could potentially lack factors present in E. coli that suppress the growth defect of an smpB mutant.
Identification of Putative Essential Genes of Unknown Functions.
Comparison of the list of essential genes (Table 3) with current genome annotation revealed 259 putatively essential genes lacking defined functional roles. At least 159 of these were initially found to be conserved in other organisms (14), a number that increases as additional genomes are sequenced. Genes that are conserved in evolutionarily divergent genera potentially encode components of basic cellular processes. Essential genes of otherwise unknown function within this category will be important to characterize to better understand microbial cell biology and also for defining optimal bacterial targets for the development of antimicrobial therapeutic agents. We determined which putatively essential gene products with otherwise unknown functions were conserved in a phylogenetically distant pathogen, Mycobacterium tuberculosis. To exclude proteins that could potentially be present in humans, those with homologs in S. cerevisiae were removed. Absence of these proteins in humans was evaluated by tblastn comparisons of the list (Table 2) to the human genome sequence database (September 2001). Table 2 notes roles in viability for several conserved genes (homologs of HI0004, HI0767, HI1282, HI1608, and HI1654) that have been examined in B. subtilis (ref. 4; http://locus.jouy.inra.fr/cgi-bin/genmic/madbase_home.pl), E. coli (27), M. genitalium (6), or Pseudomonas aeruginosa (17). Both of the putatively essential H. influenzae genes in the list that are conserved in M. genitalium are potentially essential in M. genitalium. Homologs of the HI1608 gene product were essential in two other organisms in which they were examined, B. subtilis and E. coli. This gene was characterized independently by several other groups during the course of our study and shown to encode a component of an essential isoprenoid biosynthesis pathway in E. coli (27–29). It is possible that other genes identified by our analysis encode previously unrecognized components of similarly well characterized cellular pathways. Conversely, highly conserved products of another gene HI1654 were putatively essential in Mycoplasma spp. but not in B. subtilis, E. coli, or P. aeruginosa. Products of HI1654, therefore, could participate in a potential essential pathway for which some bacteria produce functionally redundant components. Although comparisons of this kind are subject to the specific experimental conditions and methods used, the results indicate that the requirement for a given gene for viability varies between organisms.
Table 2.
H. influenzae putative essential genes conserved in M. tuberculosis and absent in S. cerevisae
| Hin | Mtu | Bsu | Mge | Cpn | Ctr | Nme | Cje | Hpy | Eco | Pmu | Pae | Vch | Bbu | Tpa | PROSITE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HI0004 | + | + | +E | + | + | + | + | + | +N | + | + | + | + | + | UPF0054 |
| HI0175 | + | +N | + | + | + | + | + | + | + | + | + | ||||
| HI0303 | + | + | + | + | + | + | + | + | + | + | + | ||||
| HI0338 | + | + | + | + | + | + | + | + | Predicted transmembrane | ||||||
| HI0466 | + | + | + | + | + | + | |||||||||
| HI0570 | + | + | + | + | + | ||||||||||
| HI0767 | + | +N | + | + | + | + | + | +N | + | + | + | + | + | N6_MTASE | |
| HI1086 | + | + | + | + | + | + | PROKAR_LIPOPROTEIN | ||||||||
| HI1282 | + | +N | + | + | + | +N | + | + | + | + | + | ||||
| HI1608 | + | +E | + | + | + | + | +E | + | + | + | + | ||||
| HI1654 | + | +N | +E | + | + | + | +N | + | +N | + | + | + | UPF0011 | ||
| HI1731 | + | + | + | + | + | + |
Presence (+) or absence (blank) of homologs to putatively essential H. influenzae proteins in a range of other bacteria. Homologs of H. influenzae proteins included or excluded from the table yielded PAM250 DP values > 80 and P values <10−3 based on precomputed blastp comparisons of 37 microbial genomes (http://mbgd.genome.ad.jp/). Growth phenotypes previously determined for mutants deficient in homologs in other species (see text) are summarized as essential (E) or nonessential (N) when data are available. H. influenzae (hin), M. tuberculosis (mtu), B. subtilis (bsu), Chlamydia pneumoniae (cpn), Chlamydia trachomatis (ctr), N. meningitidis (nme), Campylobacter jejuni (cje), H. pylori (hpy), E. coli (eco), Pasteurella multocida (pmu), P. aeruginosa (pae), V. cholerae (vch), Borrelia burgdorferi (bbu), and Treponema pallidum (tpa).
Discussion
Based on our results, we estimate that 38% of H. influenzae genes are critical for growth or viability of colonies on rich medium. Putatively essential genes were no more likely to have been previously characterized than genes with nonessential functions. In several cases, putatively essential genes that we have identified are nonessential in other organisms. Studies in H. influenzae can potentially detect essential cellular pathways that have several alternate mechanisms in organisms with larger genomes. Alternatively, H. influenzae may contain additional factors that become lethal when the essential gene is depleted. Moreover, organisms with larger genome sizes compared with those with smaller coding capabilities may have an increase in gene functions that seem redundant under a given growth condition.
An estimate of essential gene content of 55–73% was generated for Mycoplasma spp. based on a large-scale sequencing project that identified genes containing transposon insertions (6). Two small-scale surveys examining a total of 53 selected genes in E. coli indicated that much lower proportions (≈22%) of the genes examined were essential (6/27 or 6/26, respectively) (27, 30). In fact, the true proportion in E. coli could potentially be even lower as these authors chose targets conserved in divergent bacterial species in an attempt to favor detection of essential genes. Analysis of chromosome V of S. cerevisiae indicated that ≈20% of the genes examined was essential (5). The causes for the presence of a greater proportion of essential genes in H. influenzae and Mycoplasma spp. are likely to be complex. It is likely that because these obligate parasites are more reliant on nutrients from the host, they have fewer biosynthetic capabilities with overlapping functions compared with bacteria that have environmental niches in water or soil. It will also be possible to compare our H. influenzae data with the list of ≈150 critical genes detected in the Gram-positive pathogen, S. aureus, when the exact gene identifiers for genes mediating antisense RNA-inhibited phenotypes in this bacterium become available (8). However, it is important to note that these experiments with diverse bacteria were conducted with different culture conditions and growth media, factors predicted to have a major impact on requirements for viability.
A number of “postgenomic” approaches have been advanced to refine the list of potential targets for antimicrobial drug development to small numbers of genes that can be examined by directed methods. A common approach focuses on proteins that are conserved in many organisms and are therefore more likely to mediate basic cellular functions such that compounds acting on them could give rise to broad spectrum therapeutic agents (27, 30). Others have analyzed genes unique to an organism, often in an effort to identify targets for narrow-spectrum antibiotics (31). However, as more genomes are sequenced, it is likely that fewer genes will be considered organism-specific. Although directed approaches examining few or single genes can produce a high confidence level that a given gene is needed for growth, the low output of essential gene candidates makes finding ideal candidates less likely. To obtain the most reliable assessment of the critical biological pathways in an organism requires studies in a natural environment such as during growth of a pathogen within an infected host. Our studies have identified a comprehensive list of factors that are essential in vitro under a standard growth condition. These results provide information that can be used to identify genes that are also essential during infection. We have completed a functional genomic analysis of H. influenzae and we provide a publicly available database of putatively essential and nonessential genes in this organism (see Table 3). Detailed analysis of each gene is required to confirm their specific functions, yet the list provides a starting point for targeted analysis of subsets of genes interesting to investigators examining critical biological processes. For example, a kinetic approach to monitoring loss of mutants from a pool directly after in vitro transposon mutagenesis can provide a rapid means of initial verification of essentiality (5, 32). In addition, insertions in specific DNA regions can be more precisely mapped by PAGE to examine genes that were not evaluated (17, 33). This study integrates genomic sequence data with genome-scale mutagenesis. Other global analyses such as genomic expression profiling can be combined with these results to further define genetic functions. Genome analysis and mapping by in vitro transposition (GAMBIT) and genetic footprinting results from this study show that essentiality of bacterial genes in cultures grown on complex medium does not follow simple patterns. Essentiality is not predicted exclusively by conservation across species, and genes needed in one organism are not necessarily needed in another. However, comparing essential gene databases in divergent organisms can be used to prioritize genetic and biochemical characterization of genes of unknown function.
Supplementary Material
Acknowledgments
This work was supported by a University of Michigan Rackham Faculty Research grant (to B.J.A.), and by National Institutes of Health Grants AI-02137 and AI-48704 (to E.J.R.) and AI-26289 (to J.J.M.).
Abbreviations
- US
uptake consensus sequence
- BHI
brain heart infusion
References
- 1.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mori H, Isono K, Horiuchi T, Miki T. Res Microbiol. 2000;151:121–128. doi: 10.1016/s0923-2508(00)00119-4. [DOI] [PubMed] [Google Scholar]
- 4.Ogasawara N. Res Microbiol. 2000;151:129–134. doi: 10.1016/s0923-2508(00)00118-2. [DOI] [PubMed] [Google Scholar]
- 5.Smith V, Chou K N, Lashkari D, Botstein D, Brown P O. Science. 1996;274:2069–2074. doi: 10.1126/science.274.5295.2069. [DOI] [PubMed] [Google Scholar]
- 6.Hutchison C A, Peterson S N, Gill S R, Cline R T, White O, Fraser C M, Smith H O, Venter J C. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 7.Judson N, Mekalanos J J. Nat Biotechnol. 2000;18:740–745. doi: 10.1038/77305. [DOI] [PubMed] [Google Scholar]
- 8.Ji Y, Zhang B, Van S F, Horn, Warren P, Woodnutt G, Burnham M K, Rosenberg M. Science. 2001;293:2266–2269. doi: 10.1126/science.1063566. [DOI] [PubMed] [Google Scholar]
- 9.Akerley B J, Rubin E J, Camilli A, Lampe D J, Robertson H M, Mekalanos J J. Proc Natl Acad Sci USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein J O. Pediatr Infect Dis J. 1997;16:S5–S8. doi: 10.1097/00006454-199702001-00002. [DOI] [PubMed] [Google Scholar]
- 11.Bartlett J G, Breiman R F, Mandell L A, File T M., Jr Clin Infect Dis. 1998;26:811–838. doi: 10.1086/513953. [DOI] [PubMed] [Google Scholar]
- 12.Danner D B, Deich R A, Sisco K L, Smith H O. Gene. 1980;11:311–318. doi: 10.1016/0378-1119(80)90071-2. [DOI] [PubMed] [Google Scholar]
- 13.Smith H O, Tomb J F, Dougherty B A, Fleischmann R D, Venter J C. Science. 1995;269:538–540. doi: 10.1126/science.7542802. [DOI] [PubMed] [Google Scholar]
- 14.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 15.Lampe D J, Akerley B J, Rubin E J, Mekalanos J J, Robertson H M. Proc Natl Acad Sci USA. 1999;96:11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herriott R M, Meyer E M, Vogt M. J Bacteriol. 1970;101:517–524. doi: 10.1128/jb.101.2.517-524.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong S M, Mekalanos J J. Proc Natl Acad Sci USA. 2000;97:10191–10196. doi: 10.1073/pnas.97.18.10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bishai W R, Howard N S, Winkelstein J A, Smith H O. Infect Immun. 1994;62:4855–4860. doi: 10.1128/iai.62.11.4855-4860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutkenhaus J, Mukhergee A. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik W S, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Vol. 2. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1615–1626. [Google Scholar]
- 20.Heijenoort J V. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik W S, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1025–1034. [Google Scholar]
- 21.Matthews R G. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik W S, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 600–611. [Google Scholar]
- 22.Lu D, Boyd B, Lingwood C A. J Biol Chem. 1997;272:29033–29038. doi: 10.1074/jbc.272.46.29033. [DOI] [PubMed] [Google Scholar]
- 23.Karzai A W, Susskind M M, Sauer R T. EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiegert T, Schumann W. J Bacteriol. 2001;183:3885–3889. doi: 10.1128/JB.183.13.3885-3889.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wower I K, Zwieb C W, Guven S A, Wower J. EMBO J. 2000;19:6612–6621. doi: 10.1093/emboj/19.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang C, Wolfgang M C, Withey J, Koomey M, Friedman D I. EMBO J. 2000;19:1098–1107. doi: 10.1093/emboj/19.5.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freiberg C, Wieland B, Spaltmann F, Ehlert K, Brotz H, Labischinski H. J Mol Microbiol Biotechnol. 2001;3:483–489. [PubMed] [Google Scholar]
- 28.Luttgen H, Rohdich F, Herz S, Wungsintaweekul J, Hecht S, Schuhr C A, Fellermeier M, Sagner S, Zenk M H, Bacher A, Eisenreich W. Proc Natl Acad Sci USA. 2000;97:1062–1067. doi: 10.1073/pnas.97.3.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lange B M, Croteau R. Proc Natl Acad Sci USA. 1999;96:13714–13719. doi: 10.1073/pnas.96.24.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arigoni F, Talabot F, Peitsch M, Edgerton M D, Meldrum E, Allet E, Fish R, Jamotte T, Curchod M L, Loferer H. Nat Biotechnol. 1998;16:851–856. doi: 10.1038/nbt0998-851. [DOI] [PubMed] [Google Scholar]
- 31.Chalker A F, Minehart H W, Hughes N J, Koretke K K, Lonetto M A, Brinkman K K, Warren P V, Lupas A, Stanhope M J, Brown J R, Hoffman P S. J Bacteriol. 2001;183:1259–1268. doi: 10.1128/JB.183.4.1259-1268.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reich K A, Chovan L, Hessler P. J Bacteriol. 1999;181:4961–4968. doi: 10.1128/jb.181.16.4961-4968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubin E J, Akerley B J, Novik V N, Lampe D J, Husson R N, Mekalanos J J. Proc Natl Acad Sci USA. 1999;96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.