Abstract
Hemoglobin degradation is a metabolic process that is central to the growth and maturation of the malaria parasite Plasmodium falciparum. Two aspartic proteases that initiate degradation, plasmepsins (PMs) I and II, have been identified and extensively characterized. Eight additional PM genes are present in the P. falciparum genome. To better understand the enzymology of hemoglobin degradation, it is necessary to determine which of these genes are expressed when hemoglobin degradation is occurring, which encode active enzymes, and which gene products are found in the food vacuole where catabolism takes place. Our genome-wide analysis reveals that PM I, II, and IV and histo-aspartic protease encode hemoglobin-degrading food vacuole proteases. Despite having a histidine in place of one of the catalytic aspartic acids conserved in other aspartic proteases, histo-aspartic protease is an active hydrolase.
Each year malaria afflicts an estimated 500 million people and kills nearly 2 million, mostly children (1). Plasmodium falciparum, the parasite that causes the deadliest form of malaria, is becoming increasingly resistant to drugs, making it essential to identify new antimalarial agents. Proteases are attractive candidates for drug development because they play vital roles in parasite metabolism (2).
Intraerythrocytic stages of Plasmodium rely on red blood cell hemoglobin as a nutrient source, consuming vast amounts of it within a short time. In P. falciparum, several proteases have been implicated in the process of hemoglobin degradation. Hemoglobin is endocytosed from the erythrocyte cytosol and trafficked to an acidic food vacuole (FV), where it is degraded by four parasite proteases proposed to act in a semiordered fashion (3, 4). Two homologous aspartic proteases, called plasmepsins I and II (PM I and II) initiate the degradative process by cleaving the native hemoglobin molecule in a highly conserved hinge region (5, 6). A cysteine protease, falcipain-2, and a metalloprotease, falcilysin, act further downstream in the pathway to degrade hemoglobin to small peptides (7–10). Inhibitors of these proteases kill parasites in culture and animal models, suggesting that hemoglobin-degrading proteases are valid targets for chemotherapy (11–17).
Two proteins, known as histo-aspartic protease (HAP) and PM IV, are homologous to the previously characterized PM I and II (18). HAP is ≈60% identical to PM I and II, but has several substitutions, including replacement of a catalytic aspartate with a histidine and changes in a conserved flap region that lies over the binding cleft (19). It has been unclear if HAP is an inactive homolog of the hemoglobin-degrading PMs, or if it is an active protease with a novel catalytic mechanism.
As the P. falciparum genome sequencing effort nears completion, it has become apparent that the parasite contains at least 10 aspartic protease genes, PM I, II, and IV–X and HAP (20). To determine which of these genes are important for hemoglobin degradation, we have investigated PM expression, localization in parasites, and enzymatic activities. Our data indicate that HAP and PM IV are additional proteases involved in the metabolism of hemoglobin. Furthermore, HAP displays a unique active-site and inhibitor sensitivity.
Materials and Methods
Reagents.
All reagents were obtained from Sigma, unless otherwise specified.
Culture of Parasites.
P. falciparum clone HB3 (a gift of W. Trager, Rockefeller Univ., New York) was grown as described by Trager and Jensen, with Albumax instead of serum (21).
Antibody Production and Specificity.
Monoclonal antibodies to PM I (1C6–24) and HAP (3E10–19, 3G2–5, 3F10–18) were generated as described (22). Recombinant PM I and HAP were used as immunogens. Both were prepared by using the protocol for PM I (23). Large amounts of antibody were purified from mouse ascites by using Protein G Sepharose (Amersham Pharmacia). Monoclonal antibodies to PM IV (13.9.2) and PM V (23.1.2) were generated by the St. Louis University Hybridoma Development Service. Again, recombinant proteins were used as immunogens. Polyclonal antibodies to PM IV (1323), IX (1455), and X (1463) were made by Cocalico Biologicals (Reamstown, PA), with use of MAP peptides SENDSIELDDVANLMFYGEGQI (amino acids 1–22 of mature PM IV), KREKASDNKS (amino acids 109–118 of putative mature PM IX), and RRSFIEKNLH (amino acids 67–76 of putative mature PM X). Polyclonal antibody to PM II (737) was generated as described (24).
PM I, II, and IV and HAP antibodies were analyzed for their ability to recognize recombinant proteins on immunoblots or in vitro translated proteins in immunoprecipitation assays. This study uses only those antibodies determined to be specific for the protein they were raised against (data not shown).
Immunofluorescence Microscopy.
Infected RBCs were smeared onto coverslips and fixed in acetone for 10 min at room temperature or in 1% formaldehyde/PBS for 30 min at room temperature. Samples were immunolabeled with mouse anti-PM I (1C6–24, 1:20), rabbit anti-PM II (737, 1:1,000), mouse anti-HAP (3G2–5, 1:20), mouse anti-PM IV (13.9, undiluted hybridoma supernatant), mouse anti-PM V (23.1.2, undiluted supernatant), affinity-purified rabbit anti-PM IX (1455, 1:10), and affinity-purified rabbit anti-PM X (1463, 1:50) followed by Texas red- or Alexa-conjugated goat anti-rabbit or goat anti-mouse IgG. Coverslips were mounted in Vectashield (Vector Laboratories) and viewed by using a Zeiss axioskope microscope. Localization of PM I, II, and IV and HAP was optimal after acetone fixation, whereas localization of PM V, IX, and X was unsuccessful in acetone-treated samples and was therefore performed by using formaldehyde fixation.
Immunoelectron Microscopy.
Infected RBCs were fixed in 4% paraformaldehyde/0.5% glutaraldehyde in 100 mM Pipes/0.5 mM MgCl2, pH 7 for 1 h at 4°C. Samples were then embedded in 10% gelatin and infiltrated with 2.3 M sucrose/20% polyvinyl pyrrolidone in Pipes/MgCl2 at 4°C. Samples were trimmed, frozen in liquid nitrogen, and sectioned with a RMC MT7/CR21 cryo-ultramicrotome. Sections (70–90 nm) were immunolabeled with affinity-purified rabbit anti-PM I (574, 1:100), affinity-purified rabbit anti-PM II (737, 1:20), mouse anti-HAP (3E10–19, 1:10), or mouse anti-PM IV (13.9.2, 1:2) followed by 18-nm colloidal gold-conjugated goat anti-mouse or goat anti-rabbit IgG. Sections were stained with 0.3% uranyl acetate/2% polyvinyl alcohol and allowed to air dry. Controls omitting the primary antibody were consistently negative at the concentration of gold-conjugated secondary antibodies used in these studies.
Immunoblotting.
Parasite lysates were fractionated by SDS/PAGE (25), and transferred to Protran nitrocellulose (Schleicher and Schuell). Blots were incubated with primary antibody followed by horseradish peroxidase-conjugated secondary antibody (Amersham Pharmacia) and developed by using enhanced chemiluminescence Western blotting detection reagents (Amersham Pharmacia). Primary antibodies used were: mouse anti-PM I (1C6–24, 1:10,000), rabbit anti-PM II (737, 1:5,000), mouse anti-HAP (3F10–18, 1:5,000), and rabbit anti-PM IV (1323, 1:1,000).
For stage-specific expression experiments, a synchronized culture of early-ring-stage parasites was divided equally into five plates. One plate was harvested at each of the following stages: rings, young trophozoites, middle-stage trophozoites, mature trophozoites, and schizonts. Cultures were more than 95% homogeneous.
Expression, Refolding, and Purification of Recombinant PM IV.
The full-length gene for PM IV was amplified from P. falciparum HB3 genomic DNA by PCR. Primers used were PM4-NcoI (5′-GATCCCATGGCTCTTACCGTTAAAGAAGAAGAA-3′) and PM4CtermStop (5′-CCGCTCGAGTTATAAATTTTTAGCTACTGCAAAACCAACACT-3′). The product was cloned into pET15b (Novagen), sequenced, and transformed into Escherichia coli BL21 (DE3) Codon Plus cells (Stratagene). Expression clones were grown at 37°C in LB broth containing 0.1 mg/ml ampicillin and 34 μg/ml chloramphenicol. At OD600 of 0.6, isopropyl β-D-thiogalactoside was added to a final concentration of 0.4 mM, and cultures were grown for 3 h at 37°C. PM IV was solubilized from inclusion bodies, refolded, and purified as described for PM I and II (23).
Activity Assays.
Globin and hemoglobin degradation.
Three micrograms of human globin or hemoglobin (Sigma) was incubated with enzyme in 100 mM citrate-phosphate buffer, pH 5.0 (PM II), pH 5.4 (PM IV), or pH 5.7 (HAP) (40 μl total volume). Reactions were stopped by mixing with electrophoresis sample buffer and boiling for 5 min. Products were separated by SDS/PAGE and detected by silver staining (26).
[14C]globin assay.
Human globin was labeled by reductive methylation with [14C]formaldehyde (27). Labeled globin (0.5 μl; ≈17,000 cpm) was mixed with enzyme and 100 mM citrate-phosphate buffer, pH 5.7 (HAP) or 5.4 (PM IV), in a total volume of 40 μl and incubated at 37°C. To stop the reaction, 100 μl of 2 mg/ml unlabeled BSA and then 150 μl 8% trichloroacetic acid were added. Samples were mixed, incubated on ice for 30 min, and centrifuged at 13,000 × g for 30 min. The supernatant was assayed for radioactivity in a scintillation counter.
Fluorogenic substrate.
Samples were incubated with 2.5 μM quenched fluorescence substrate α33–34 corresponding to residues 30–37 of the α chain of human hemoglobin in 100 mM citrate-phosphate buffer, pH 5.4 (PM IV) or pH 5.7 (HAP), at 37°C. This assay has been described (23). The concentrations of active HAP and PM IV were determined by titration with the inhibitor pepstatin A. Km and kcat were calculated by using sigmaplot. Ki was determined by fitting data to the Morrison equation (28). Ki values for recombinant PM I and II were measured for comparison. Recombinant proteins were expressed and purified as described (23, 29). To determine the pH sensitivities of the enzymes, assays were performed in 100 mM citrate-phosphate buffer from pH 3 to 6.6 or in 100 mM [bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane buffer at pH 7.0.
Purification of Native HAP.
Immunoaffinity purification.
Two different immunoaffinity columns were used. One milligram of anti-HAP mAb (3E10–19 or 3G2–5) was coupled to 500 μl of Protein A Sepharose (Amersham Pharmacia) as described (30). Food vacuoles were isolated as described (5) except that the low-speed pellet after sorbitol lysis was harvested twice with 1.5 mM MgCl2/PBS, and the final Percoll gradient centrifugation step was omitted. Two milliliters of packed food vacuoles was resuspended in 6 ml of lysis buffer (PBS/1% Triton X-100/10 μM E64). The cells were lysed by means of 10 strokes in a Dounce homogenizer, incubated on ice for 1 h, and then centrifuged at 25,000 × g for 1 h to pellet insoluble material. The supernatant was passed three times over the antibody column. The column was washed with 20 column volumes of 1 M NaCl/PBS and two column volumes of preelution buffer (10 mM phosphate, pH 8.0) and eluted with six column volumes of elution buffer (100 mM triethylamine, pH 11.5). Eluant fractions were neutralized with 1/10 volume of 1 M sodium phosphate, pH 5.5. All chromatography was performed at 4°C.
HPLC chromatography.
Lysates from crude FVs were prepared as above and fractionated on a 4-ml DEAE-Sepharose (Fast Flow) column (Amersham Pharmacia) equilibrated in 20 mM [bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane, pH 7, and eluted with a stepwise gradient of 0–1 M NaCl. Active fractions were pooled, diluted with 50 mM Mes (pH 6.7), and loaded onto a Mono S PC 1.6/5 HPLC column (Amersham Pharmacia). Bound proteins were eluted with a linear gradient of 0–2 M NaCl in 50 mM Mes (pH 6.7) at 0.1 ml/min for 50 min.
Samples containing HAP were identified by Western blotting and injected onto a Superdex-200 HPLC column (Amersham Pharmacia) run isocratically at 0.5 ml/min in PBS. Fractions of 250 μl were collected and assayed for activity.
Trichloroacetic acid precipitation of elution fractions.
Fractions of 400 μl containing activity were combined with 200 μl of protease-free BSA solution (final concentration, 0.5 mg/ml) and 400 μl of 8% trichloroacetic acid (final concentration, 3.2%). Samples were mixed, incubated on ice for 30 min, and centrifuged at 13,000 × g for 30 min. The pellets were washed once in acetone, resuspended in nonreducing sample buffer, and boiled before loading onto 10% gels. Gels were stained with silver (26) revealing a single band migrating at 37 kDa, or the proteins were transferred to nitrocellulose for immunodetection.
Results
Expression Analysis of 10 PMs.
To determine which PMs are expressed in asexual stage parasites, reverse transcription–PCR with gene-specific primers was performed on total RNA from asynchronous intraerythrocytic parasites. PCR products of the expected size could be detected for seven of the 10 PM genes (data not shown). PM VI, VII, and VIII could not be detected with two different primer pairs for each gene, with random or poly(T)-primed cDNA. All PCR primer pairs were used successfully to amplify appropriate sequences from genomic DNA (data not shown).
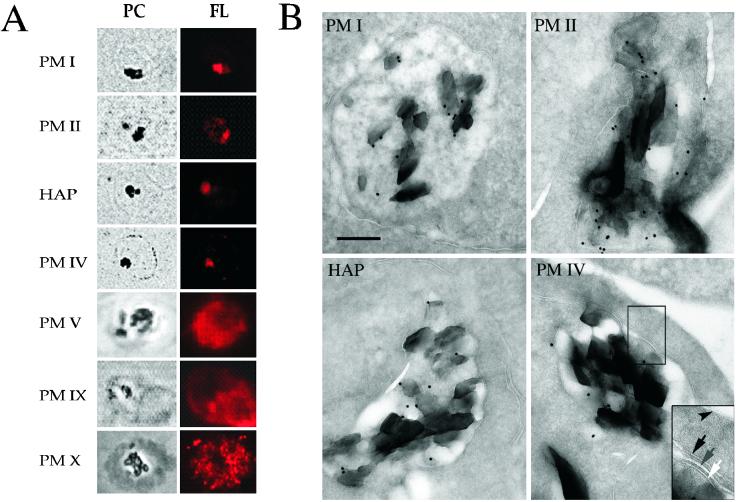
Specific antibodies recognizing HAP and PM I, II, IV, V, IX, and X were used to localize the proteins in trophozoite stage parasites by immunofluorescence microscopy (Fig. 1A). PM II, PM IV, and HAP colocalized with hemozoin in the FV, similar to results reported for PM I (11). In contrast, PM V, IX, and X had more diffuse patterns and were excluded from the FV. Immunoelectron microscopy confirmed that PM I, II, and IV and HAP are in the FV (Fig. 1B). Gold labeling was found exclusively within the FV.
Figure 1.
PM localization in intraerythrocytic stages. (A) Indirect immunofluorescence microscopy analysis of PMs. Phase/contrast (PC) and fluorescence (FL) images of trophozoites stained with antibodies to PM I, II, IV, V, IX, and X and HAP are shown. The dark regions in the phase/contrast images are hemozoin crystals within the FV. (B) Immunoelectron microscopy of trophozoites using anti-PM I, II, and IV and HAP antibodies. All four panels are shown at the same magnification. (Scale bar = 0.25 μm.) (Inset) Close-up of membranes surrounding the FV; arrowhead, RBC membrane; black arrow, parasitophorous vacuolar membrane; gray arrow, parasite plasma membrane; white arrow, FV membrane. The dark crystals within the FV are hemozoin.
PMs I, II, and IV and HAP Are FV Proteases with High Sequence Similarity.
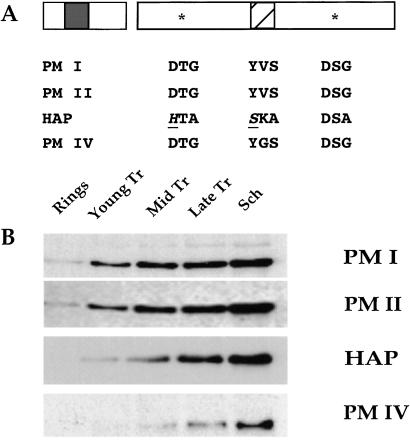
We concentrated our further analysis on the PMs found in the FV. Salient features are conserved in PM I, II, and IV and HAP (Fig. 2A). Each is predicted to be a 51-kDa proenzyme with a hydrophobic transmembrane domain within its prosegment. All four proteins apparently are processed at a conserved site, yielding 37-kDa mature forms (unpublished work). HAP, as reported, contains substitutions of key residues in the active site and in the flap that lies over the substrate-binding cleft (19). PM I, II, and IV and HAP lie in a cluster on chromosome 14. The genes span a region of 20 kb, with ≈4 kb separating one ORF from the next. The predicted coding sequences are 50–70% identical with each other at the amino acid level (19). High levels of sequence identity in both pro and mature regions suggest that PM I, II, and IV and HAP have arisen through relatively recent gene duplication events. PM V–X, in contrast, are not clustered and share much lower sequence similarity with PM I, II, and IV and HAP (20).
Figure 2.
PM I, II, and IV and HAP are similar in sequence and expression pattern. (A) Schematic diagram of PM I, II, and IV and HAP features. Each protein contains pro and mature regions. A transmembrane domain (shaded box) is present in the prosegment. Two active-site motifs (*) and the loop, known as the flap (hatched box), are shown in the mature proteins. Active-site amino acid substitutions in HAP are evident, most notably D32H and Y75S (underlined and italics). (B) Stage-specific expression of PMs. Blots containing equal numbers of ring, young, mid or late trophozoite (Tr), and schizont (Sch) stage parasites were probed with antibodies to PM I, II, and IV and HAP.
The stage-specific expression of PM I, II, and IV and HAP was determined by immunoblotting (Fig. 2B). When equal numbers of ring, trophozoite, and schizont stage parasites were analyzed, all four proteins were detectable in trophozoites and persisted to schizogony. In ring-stage parasites, PM I and II were detectable, but HAP and PM IV were not.
Native HAP Isolation and Characterization.
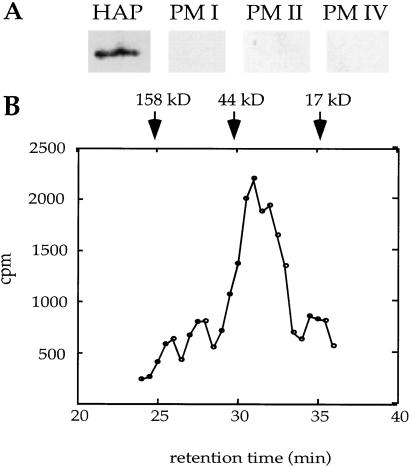
Our lab and others (19) have found that recombinant HAP expressed in E. coli is not active, possibly because of improper refolding from inclusion bodies and/or inability to autoactivate. To determine whether HAP is an active protease, we purified the native enzyme from parasite food vacuoles by using three different methods: HPLC chromatography and immunoaffinity chromatography with two different specific anti-HAP mAbs. All three purification methods yielded identical activities that migrated as a single band of 37 kDa by SDS/PAGE (data not shown). By Western analysis, the purified protein was recognized only by anti-HAP antibodies and not by antibodies to other PMs (Fig. 3A). On gel filtration chromatography, HAP activity migrated at a molecular weight of about 35,000, suggesting that the enzyme is active as a monomer (Fig. 3B).
Figure 3.
Characterization of purified HAP. (A) Active fractions from a 3G2–5 HAP immunoaffinity column contain only HAP. Eluant was trichloroacetic acid-precipitated, resolved by SDS/PAGE, blotted, and probed separately with antibodies to HAP and PM I, II, and IV. (B) Purified HAP migrates as a monomer of 35 kDa on gel filtration chromatography. HAP purified by DEAE and Mono S chromatography was injected onto a Superdex-200 column. Fractions were assayed for ability to cleave [14C]globin. The migration of molecular weight standards are indicated.
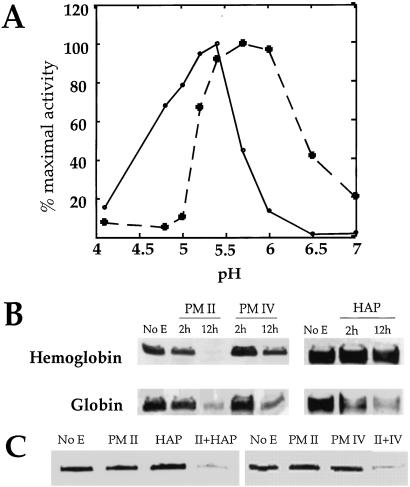
HAP activity was completely inhibited by the aspartic protease inhibitor pepstatin A (1 μM), and by a serine protease inhibitor, PMSF (1 mM). Other inhibitors of serine (3,4-dichloroisocoumarin, leupeptin, 7-amino-1-chloro-3-tosylamido-3-heptanone, L-1-tosylamido-2-phenylethyl chloromethyl ketone, 10–100 μM) cysteine (E64, leupeptin, 10 μM), or metalloproteases (EDTA, 5 mM) had no effect on HAP activity. The pH optimum of HAP was near 6, almost an entire pH unit higher than the optima of PM I and II. Activity decreased beyond pH 6 (Fig. 4A).
Figure 4.
Properties of HAP and PM IV. (A) pH sensitivity of HAP and PM IV activity. Cleavage of α33–34 by each enzyme was monitored in 100 mM citrate-phosphate buffers ranging from pH 3.5 to 6.6. At pH 7.0, 100 mM [bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane was used. A saturating substrate concentration (2.5 μM) was used. Dashed line, HAP; solid line, PM IV. (B) Hemoglobin and globin cleavage by HAP and PM IV. Human hemoglobin or acid-denatured globin were incubated without enzyme (No E) or with 50 nM recombinant PM II, recombinant PM IV, or native HAP for 2 or 12 h in 100 mM citrate-phosphate, pH 5.0 (PM II), pH 5.4 (PM IV), or pH 5.7 (HAP). The amount of substrate remaining was determined by silver staining. (C) A combination of enzymes degrades hemoglobin efficiently. Human hemoglobin was incubated at 37°C for 2 h without enzyme (No E) or with 50 nM recombinant PM II, recombinant PM IV, native HAP, or a combination of enzymes. Assays were performed in 100 mM citrate-phosphate, pH 5.7 (HAP and PM II) or pH 5.4 (PM IV and PM II). Gels were developed with silver staining.
HAP cleaved a fluorogenic peptide, α33–34 (Fig. 4A), 14C-labeled globin (Fig. 3B), and unlabeled acid-denatured globin (Fig. 4B). HAP did not efficiently cleave native human hemoglobin (Fig. 4B). However, a combination of HAP and PM II cleaved native hemoglobin faster than either enzyme alone (Fig. 4C).
The amount of active HAP was titrated with pepstatin A, and kinetic parameters for cleavage of α33–34 were determined (Table 1). HAP has a Km of 0.29 μM, revealing a slightly higher affinity for the peptide substrate than demonstrated by PM I and II (23). However, HAP has a slow kcat (0.05 s−1), resulting in a kcat/Km of 1.8 × 105 M−1⋅s−1 and suggesting that HAP is about 20-fold less efficient than PM I or II at cleaving this substrate (23). HAP binds pepstatin A with high affinity (Ki = 81 pM), like the other PMs.
Table 1.
Kinetic parameters for PM I, II, HAP, and PM IV cleavage of α33-34
| Km (μM) | kcat (s−1) | kcat/Km | Pepstatin Ki (pM) | |
|---|---|---|---|---|
| PM I | 0.49 ± 0.12 | 2.3 | 4.7 × 106 | 393 ± 0.25 |
| PM II | 2.6 ± 0.9 | 11 | 4.3 × 106 | 25 ± 0.09 |
| HAP | 0.29 ± 0.04 | 0.05 ± 0.002 | 1.8 × 105 | 81 ± 0.03 |
| PM IV | 0.33 ± 0.04 | 1.05 ± 0.03 | 3.2 × 106 | 313 ± 0.09 |
PM I and II Km and kcat values are from ref. 23. kcat/Km in M−1⋅s−1.
Recombinant PM IV Characterization.
Unlike recombinant HAP, recombinant PM IV was active. The recombinant protein was expressed as the proform and autoactivated at acidic pH. N-terminal sequence of the activated recombinant protein was determined to be FKSGYA, a site that is 12 aa upstream of the predicted mature N terminus in vivo. Recombinant PM II also autoprocesses itself 12 residues upstream of the known in vivo mature N terminus. Like a typical aspartic protease, PM IV was inhibited by pepstatin A (100% inhibition at 1 μM) but not by inhibitors of other enzyme classes, including 1 mM PMSF. Its pH optimum is 5.4 (Fig. 4A), slightly higher than the optima of PM I and II (29, 31) but lower than that of HAP.
Kinetic properties for PM IV cleavage of peptide substrate α33–34 were determined. PM IV showed relatively high affinity for this substrate (Km = 0.33 μM) and a catalytic efficiency greater than HAP, but similar to native PM I and II (Table 1). Equal amounts of recombinant PM II and IV were used to cleave acid-denatured globin and native hemoglobin (Fig. 4B). PM IV prefers globin over hemoglobin and is less active than PM II against these protein substrates. Overnight incubation was required for PM IV to cleave the same amount of hemoglobin cleaved by PM II in 2 h. A combination of PM IV and PM II cleaved hemoglobin faster than either enzyme alone (Fig. 4C).
Discussion
Of the 10 PMs in the P. falciparum genome, three (PM VI, VII, and VIII) are not expressed in intraerythrocytic parasites and may function in insect or exoerythrocytic stages. Three others (PM V, IX, and X) are expressed in intraerythrocytic stages but do not seem to function in the FV. Only PM I, II, and IV and HAP localize to the FV and participate in hemoglobin degradation. PM I and II have been characterized extensively (11, 23, 24, 31, 32). PM IV is inhibited by pepstatin A, like most aspartic proteases, and has a pH optimum and kinetic constants similar to those of PM I and II. In contrast, HAP has a unique inhibitor profile and lacks one of the catalytic aspartic acid residues; it may represent a novel class of protease.
We present several lines of evidence that our native HAP preparations are not contaminated by other PMs. First, HAP purified by using conventional HPLC chromatography or either of two monoclonal immunoaffinity protocols yielded enzyme with identical properties. The purified protein reacted only with antibodies to HAP and not with antibodies recognizing the closely related PM I, II, or IV. HAP is less efficient than PM I or II at cleaving the peptide substrate α33–34 as well as native hemoglobin. HAP also has a pH optimum near 6 and is inhibited by PMSF. In contrast, recombinant PM I, II, and IV have more acidic pH optima and are completely insensitive to PMSF (data not shown). Finally, HAP activity migrated as a monomer in gel filtration chromatography, ruling out the possibility that HAP is forming heterodimers with other PMs under these conditions.
The catalytic mechanism of HAP remains unclear. Pepstatin A binds competitively in the active site of aspartic proteases and makes hydrogen bonds with the two catalytic aspartates. HAP apparently has high affinity for pepstatin A, with a Ki similar to those of other PMs. We therefore propose that HAP may work through an aspartic protease-like mechanism, with a putative catalytic dyad comprising histidine and aspartic acid. In classical aspartic proteases, one aspartate residue acts as a general acid, whereas the other is in the ionized form and acts as a general base to extract a proton from a water molecule (33). This activated water molecule is the nucleophile that attacks the scissile bond. In HAP, at its pH optimum near 6, the active-site histidine may function in the protonated state and act as a general acid, if its pKa remains near 6–7, the pKa range of free histidine. Alternatively, the pKa of the histidine may be lower in the microenvironment of the active site, causing the histidine to be unprotonated and a general base near pH 6. A precedent exists for a single aspartic acid being sufficient for catalysis. The nodavirus coat protein is an aspartic protease that may have an aspartic acid and an asparagine in its catalytic dyad (34). With a single active-site aspartic acid, dimerization is a possible mode of bringing two aspartic acids together, as in retroviral proteases. Dimerization is unlikely in HAP, because the purified active protein migrated as a monomer in gel filtration chromatography.
It is also unclear why HAP is sensitive to PMSF, unlike other aspartic proteases. HAP is different from the PMs and most other aspartic proteases in that it has a serine (S75) in place of a conserved tyrosine protruding into the active site. PMSF, a compound that usually binds to the active-site serine of serine proteases, might react with S75 or bind noncovalently in the catalytic pocket. Other serine protease inhibitors (DCI, leupeptin, 7-amino-1-chloro-3-tosylamido-3-heptanone, L-1-tosylamido-2-phenylethylchloromethyl ketone) did not inhibit HAP.
Similar active-site changes have been observed in some pregnancy-associated glycoproteins of ungulate mammals. Pregnancy-associated glycoproteins are aspartic protease homologs; those with active-site substitutions are presumed to be inactive but are still capable of binding peptides and the inhibitor pepstatin A (35–37). The function of pregnancy-associated glycoproteins remains unknown. They are abundantly secreted by the placenta, may account for the principal transcripts in mature placenta, and may number more than 100 genes in cattle and sheep (38).
Despite its unusual active-site amino acids, HAP seems to be a reasonably active enzyme. It is ≈20-fold less efficient than PM I, II, and IV at cleaving α33–34 (23), not an optimized substrate for HAP, because the peptide is based on the initial PM I cleavage site in native hemoglobin, whereas HAP prefers to cleave denatured globin at unknown sites. Indeed, HAP is an order of magnitude faster at cleaving globin than the other PMs. Comparable amounts of globin were cleaved by 2 nM HAP and 50 nM PM II or IV (data not shown). Also, 50 nM HAP cleaved as much globin in 2 h as did 50 nM PM II or IV in 12 h (Fig. 4B). The fact that both HAP and PM IV prefer to cleave globin over native hemoglobin, and are first expressed slightly later in parasite maturation than PM I and II, suggests that HAP and PM IV act later in the hemoglobin degradation pathway, further indicating the ordered nature of hemoglobin degradation within the FV. A combination of HAP and PM II, or PM IV and PM II cleaved hemoglobin much faster than each enzyme alone. It is likely that PM I, PM II, HAP, and PM IV have different substrate specificities and act upon diverse sites in globin; this synergism may facilitate efficient degradation of the molecule.
The identification of two additional proteases that participate in hemoglobin degradation within the FV highlights the complexity of this vital biochemical pathway. It is of great therapeutic interest to understand these proteases on a molecular level. The unique properties of HAP make it an especially attractive protein to target for antimalarial drug development.
Acknowledgments
We thank A. Lingnau for extensive help with mAb production, A. Oksman for assistance with parasite culturing and FV harvesting, C. Murata for generosity with her FV preparations, P. Siripurkpong for assistance with recombinant PM production, D. McCourt for N-terminal sequencing, J. Dant for assistance with immunoelectron microscopy, and E. Istvan for critical review of this manuscript. This work was supported by National Institutes of Health Grant AI-47798. D.E.G. is a recipient of the Burroughs Wellcome Fund Scholar Award in Molecular Parasitology. PM sequence information was obtained from The Institute for Genomic Research and Sanger Center databases of the Malaria Genome Project Consortium. Funding for the consortium is provided by the Wellcome Trust, Burroughs Wellcome Fund, National Institute of Allergy and Infectious Diseases, and the Department of Defense.
Abbreviations
- PM
plasmepsin
- FV
food vacuole
- HAP
histo-aspartic protease
References
- 1.Trigg P I, Kondrachine A V. Malaria: Parasite Biology, Pathogenesis and Protection. Washington, DC: Am. Soc. Microbiol.; 1998. [Google Scholar]
- 2.Rosenthal P J. Adv Parasitol. 1999;43:105–159. doi: 10.1016/s0065-308x(08)60242-0. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee R, Goldberg D E. In: Antimalarial Chemotherapy: Mechanisms of Action, Resistance, and New Directions in Drug Discovery. Rosenthal P J, editor. Totowa, NJ: Humana; 2000. pp. 43–63. [Google Scholar]
- 4.Goldberg D E. Semin Cell Biol. 1993;4:355–361. doi: 10.1006/scel.1993.1042. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg D E, Slater A F G, Beavis R, Chait B, Cerami A, Henderson G B. J Exp Med. 1991;173:961–969. doi: 10.1084/jem.173.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gluzman I Y, Francis S E, Oksman A, Smith C E, Duffin K L, Goldberg D E. J Clin Invest. 1994;93:1602–1608. doi: 10.1172/JCI117140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenthal P J, McKerrow J H, Aikawa M, Nagasawa H, Leech J H. J Clin Invest. 1988;82:1560–1566. doi: 10.1172/JCI113766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenthal P J, Nelson R G. Mol Biochem Parasitol. 1992;51:143–152. doi: 10.1016/0166-6851(92)90209-3. [DOI] [PubMed] [Google Scholar]
- 9.Francis S E, Gluzman I Y, Oksman A, Banerjee D, Goldberg D E. Mol Biochem Parasitol. 1996;83:189–200. doi: 10.1016/s0166-6851(96)02772-7. [DOI] [PubMed] [Google Scholar]
- 10.Eggleson K K, Duffin K L, Goldberg D E. J Biol Chem. 1999;274:32411–32417. doi: 10.1074/jbc.274.45.32411. [DOI] [PubMed] [Google Scholar]
- 11.Francis S E, Gluzman I Y, Oksman A, Knockerbocker A, Mueller R, Bryant M L, Sherman D R, Russell D G, Goldberg D E. EMBO J. 1994;13:306–317. doi: 10.1002/j.1460-2075.1994.tb06263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dianni Carroll C, Patel H, Johnson T O, Guo T, Orlowski M, He Z, Cavallaro C L, Guo J, Oksman A, Gluzman I Y, et al. Bioorg Med Chem Lett. 1998;8:2315–2320. doi: 10.1016/s0960-894x(98)00419-3. [DOI] [PubMed] [Google Scholar]
- 13.Jiang S, Prigge S T, Wei L, Gao Y, Hudson T H, Gerena L, Dame J B, Kyle D E. Antimicrob Agents Chemother. 2001;45:2577–2584. doi: 10.1128/AAC.45.9.2577-2584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenthal P J, Olson J E, Lee G K, Palmer J T, Klaus J L, Rasnick D. Antimicrob Agents Chemother. 1996;40:1600–1603. doi: 10.1128/aac.40.7.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olson J E, Lee G K, Semenov A, Rosenthal P J. Bioorg Med Chem. 1999;7:633–638. doi: 10.1016/s0968-0896(99)00004-8. [DOI] [PubMed] [Google Scholar]
- 16.Rosenthal P J, Lee G K, Smith R E. J Clin Invest. 1993;91:1052–1056. doi: 10.1172/JCI116262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holland K P, Elford H L, Bracchi V, Annis C G, Schuster S M, Chakrabarti D. Antimicrob Agents Chemother. 1998;42:2456–2458. doi: 10.1128/aac.42.9.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humphreys M J, Moon R P, Klinder A, Fowler S D, Rupp K, Bur D, Ridley R G, Berry C. FEBS Lett. 1999;463:43–48. doi: 10.1016/s0014-5793(99)01597-5. [DOI] [PubMed] [Google Scholar]
- 19.Berry C, Humphreys M J, Matharu P, Granger R, Horrocks P, Moon R P, Certa U, Ridley R G, Bur D, Kay J. FEBS Lett. 1999;447:149–154. doi: 10.1016/s0014-5793(99)00276-8. [DOI] [PubMed] [Google Scholar]
- 20.Coombs G H, Goldberg D E, Klemba M, Berry C, Kay J, Mottram J C. Trends Parasitol. 2001;17:532–537. doi: 10.1016/s1471-4922(01)02037-2. [DOI] [PubMed] [Google Scholar]
- 21.Trager W, Jensen J B. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 22.Niebuhr K, Lingnau A, Frank R, Wehland J. Cell Biology: A Laboratory Handbook. New York: Academic; 1998. pp. 398–403. [Google Scholar]
- 23.Luker K E, Francis S E, Gluzman I Y, Goldberg D E. Mol Biochem Parasitol. 1996;79:71–78. doi: 10.1016/0166-6851(96)02651-5. [DOI] [PubMed] [Google Scholar]
- 24.Francis S E, Banerjee R, Goldberg D E. J Biol Chem. 1997;272:14961–14968. doi: 10.1074/jbc.272.23.14961. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Wray W, Boulikas T, Wray V P, Hancock R. Anal Biochem. 1981;118:197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- 27.Dottavio-Martin D, Ravel J M. Anal Biochem. 1978;87:562–565. doi: 10.1016/0003-2697(78)90706-6. [DOI] [PubMed] [Google Scholar]
- 28.Copeland R A. Enzymes, A Practical Introduction to Structure, Mechanism, and Data Analysis. New York: Wiley; 2000. pp. 307–313. [Google Scholar]
- 29.Moon R P, Tyas L, Certa U, Rupp K, Bur D, Jacquet C, Matile H, Loetscher H, Grueninger-Leitch F, Kay J, et al. Eur J Biochem. 1997;244:552–560. doi: 10.1111/j.1432-1033.1997.00552.x. [DOI] [PubMed] [Google Scholar]
- 30.Harlow E, Lane D. Using Antibodies, A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 311–325. [Google Scholar]
- 31.Dame J B, Reddy G R, Yowell C A, Dunn B M, Kay J, Berry C. Mol Biochem Parasitol. 1994;64:177–190. doi: 10.1016/0166-6851(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 32.Moon R, P, Bur D, Loetscher H, D'Arcy A, Tyas L, Oefner C, Grueninger-Leitch F, Mona D, Rupp K, Dorn A, et al. In: Aspartic Proteinases. James M, editor. New York: Plenum; 1998. pp. 397–406. [DOI] [PubMed] [Google Scholar]
- 33.Fischer G. In: Enzyme Mechanisms. Page M I, Williams A, editors. Cambridge, U.K.: R. Soc. Chem.; 1987. , Chap. 13. [Google Scholar]
- 34.Johnson J E, Schneemann A. In: Handbook of Proteolytic Enzymes. Barrett A J, Rawlings N D, Woessner J F, editors. San Diego: Academic; 1998. pp. 963–967. [Google Scholar]
- 35.Xie S, Green J, Beckers J, Roberts R M. Gene. 1995;159:193–197. doi: 10.1016/0378-1119(94)00928-l. [DOI] [PubMed] [Google Scholar]
- 36.Xie S, Low B G, Nagel R J, Kramer K K, Anthony R V, Zoli A P, Beckers J, Roberts R M. Proc Natl Acad Sci USA. 1991;88:10247–10251. doi: 10.1073/pnas.88.22.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guruprasad K, Blundell T L, Xie S, Green J, Szafranska B, Nagel R J, McDowell K, Baker C B, Roberts R M. Protein Eng. 1996;9:849–856. doi: 10.1093/protein/9.10.849. [DOI] [PubMed] [Google Scholar]
- 38.Xie S, Green J, Bixby J B, Szafranska B, DeMartini J C, Hecht S, Roberts M. Proc Natl Acad Sci USA. 1997;94:12809–12816. doi: 10.1073/pnas.94.24.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]