Abstract
Clostridium perfringens is a Gram-positive anaerobic spore-forming bacterium that causes life-threatening gas gangrene and mild enterotoxaemia in humans, although it colonizes as normal intestinal flora of humans and animals. The organism is known to produce a variety of toxins and enzymes that are responsible for the severe myonecrotic lesions. Here we report the complete 3,031,430-bp sequence of C. perfringens strain 13 that comprises 2,660 protein coding regions and 10 rRNA genes, showing pronounced low overall G + C content (28.6%). The genome contains typical anaerobic fermentation enzymes leading to gas production but no enzymes for the tricarboxylic acid cycle or respiratory chain. Various saccharolytic enzymes were found, but many enzymes for amino acid biosynthesis were lacking in the genome. Twenty genes were newly identified as putative virulence factors of C. perfringens, and we found a total of five hyaluronidase genes that will also contribute to virulence. The genome analysis also proved an efficient method for finding four members of the two-component VirR/VirS regulon that coordinately regulates the pathogenicity of C. perfringens. Clearly, C. perfringens obtains various essential materials from the host by producing several degradative enzymes and toxins, resulting in massive destruction of the host tissues.
Clostridium perfringens is a Gram-positive anaerobic spore-forming bacterium known to be the most widely distributed pathogen in nature (1). It is commonly found in the environment (in soil and sewage) and in the intestines of animals and humans as a member of the normal flora (1, 2). C. perfringens strains are classified into five groups (types A–E) on the basis of their production of four major toxins (known as the alpha-, beta-, epsilon-, and iota-toxins) (1, 3). Type A strains cause gas gangrene (clostridial myonecrosis) as well as necrotic enteritis and mild diarrhea in humans (2).
During World War I, it was estimated that hundreds of thousands of soldiers died of gas gangrene as a result of battlefield injuries, and C. perfringens was widely recognized as being the most important causal organism of the disease. After the entry of vegetative cells or spores into the body through an injury, the organisms grow rapidly in the host tissue, producing various toxins and enzymes that cause massive destruction of the host tissues (4). In the worst case, the infection may lead to systemic toxemia, shock, and death unless prompt antibiotic and surgical treatment is given (4).
Among the toxigenic clostridial species, including Clostridium tetani, Clostridium botulinum, and Clostridium difficile, C. perfringens is the paradigm species for genetic studies, because of its oxygen tolerance, fast growth rate (8- to 10-min generation time in optimal conditions), and ability to be genetically manipulated (4). Many of the toxins and enzymes it produces have been studied, and most of their structural genes have been cloned and sequenced (4, 5). Other approaches, such as physical and genetic mapping, have also been used to elucidate the genomic structures of various C. perfringens strains (6, 7). Recently, several studies on the genetic regulation of the toxin genes have been reported (8) using the transformable C. perfringens strain 13 (9). However, understanding of the pathogenicity and physiology of C. perfringens is still poor compared with other well studied pathogenic bacteria.
Here we report the determination and analysis of the C. perfringens genomic sequence, the first Gram-positive anaerobic pathogen to be completely sequenced, to our knowledge. The whole genomic analysis and the comparative study with the genome of nonpathogenic Clostridium acetobutylicum (10) present interesting features of an anaerobic pathogen that will contribute to the understanding of the biology of pathogenic clostridia.
Methods
Genome Sequencing.
The whole genomic sequence was obtained from a combination of 52,198 end sequences (giving 9.1 × coverage) from a pUC18 genomic shotgun library, using an M13 universal primer and dye terminator chemistry on Megabase 1000 sequencing machines. Sequence assembly was accomplished by using phrap software (P. Green, unpublished data). Gap closure was performed by PCR direct sequencing, using the primers that were constructed to anneal to each end of the neighboring contigs. The neighboring contigs were estimated from the genetic map of the constructed C. perfringens strain 13 (11). Ten regions that contained genes for ribosomal RNAs were independently cloned from plasmid or phage genomic libraries (12), and the neighboring sequences were assembled with the contig sequences.
Informatics.
The DNA was compared with sequences in the GenBank database by using blastn and blastx (13). Transfer RNAs were predicted by tRNAscan-SE (14). Potential coding sequences (CDSs) were predicted by using the genome gambler program with start to stop prediction (15) and genehacker plus with Hidden Markov models (16), and the results were combined. The predicted protein sequences were searched against a nonredundant protein database by using blastp and FASTA3 (17). The protein sorting signal of each ORF was analyzed by a psort program (18). The results of all these analyses were assembled by using the genome gambler program and then used to inform a manual annotation of the sequence and predicted proteins. Annotation was based, wherever possible, on characterized proteins or genes. The complete genome sequence and annotation are available (http://w3.grt.kyushu-u.ac.jp/CPE/).
Comparative Genomics.
The genomic sequences of bacteria were collected through the web site of the National Center of Biological Information (NCBI, http://www.ncbi.nlm.nih.gov). The leading and lagging strands of the genomic sequences were identified by GC skew analysis (19). The percentage of ORFs on the leading strand of each bacterium was calculated from the protein table deposited in the NCBI web site.
Results
Sequencing and Genome Features.
C. perfringens strain 13 is a natural isolate from the soil. It is classified as a type A strain that commonly causes gas gangrene in humans (9). Some pathological studies have shown that this strain can establish experimental gas gangrene (myonecrosis) in a murine model (4). Nucleotide sequencing of the genome of strain 13 was performed by using a whole-genome shotgun strategy. A total of 52,198 PCR-amplified templates was finally sequenced and assembled into 194 contigs. Eighty-two gaps were subsequently filled by PCR direct sequencing, finally yielding one complete circular chromosome. During the sequencing process, one contig (contig 180) was derived from a 50-kb plasmid, as predicted in the mapping study (11).
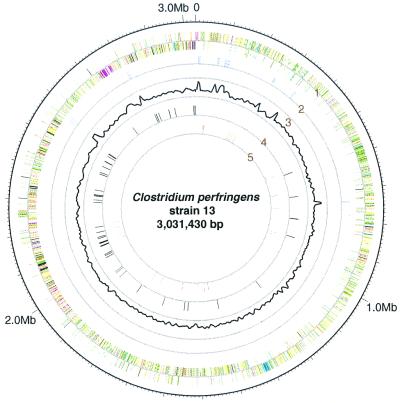
The chromosome of C. perfringens contained a 3,031,430-bp sequence that had a pronouncedly low G + C content (28.6%). We could not find any regions showing significantly higher G + C content, except portions encoding rRNA genes (Fig. 1). The chromosome has 10 rRNA genes (ref. 12; Fig. 1) and 96 species of tRNA genes. The replication origin of the C. perfringens chromosome was designated upstream of dnaA (CPE0001 in our nomenclature) on the basis of the gene distribution of the two strands (Fig. 1) and on the transition point in GC skew analysis (19). The plasmid (named pCP13) has a 54,310-bp nucleotide sequence, and the overall G + C content of the sequence of pCP13 is 25.5%, which is slightly lower than that of the chromosome.
Figure 1.
Circular map of the C. perfringens chromosome. The outer circle shows the scale. Ring 1 shows the coding sequence by strand (clockwise and counterclockwise); ring 2 shows the positions of rRNA genes; ring 3 shows G + C content (outer and inner rings correspond to 45% and 20%, respectively); ring 4 shows the positions of sporulation/germination genes; and ring 5 shows the positions of virulence-related genes. We used the following functional categories in ring 1: intermediary metabolism and fermentation (yellow), information pathways (pink), regulatory proteins (sky blue), conserved hypothetical proteins (orange), proteins of unknown function (light blue), insertion sequences and phage-related functions (blue), stable RNAs (purple), cell wall and cell processes (dark green), virulence (red), detoxification and adaptation (white), and sporulation and germination (black).
Annotation of C. perfringens Genes.
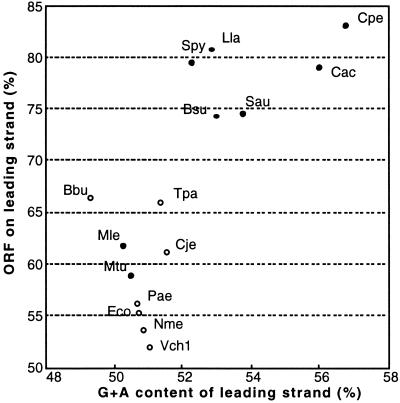
The chromosome contains 2,660 predicted ORFs that cover 83.1% of the whole chromosomal sequence with an average size of 946 bp, and the plasmid pCP13 contains 63 putative ORFs. We noticed that C. perfringens has a much stronger tendency to arrange genes such that their transcriptional orientation is the same as their replication direction than those observed in Mycoplasma species and Bacillus subtilis (ref. 20; Fig. 1). We compared the tendency among other bacteria whose leading and lagging strands are obviously identified by GC skew analysis on their sequenced genomes. Among many base composition skews, the content of G + A in the leading strands seems to have a correlation with the percentage of ORFs on the leading strands (Fig. 2). Furthermore, the plots could be divided into two major groups that were well related to Gram-negative and Gram-positive bacteria except for Mycobacterium leprae and Mycobacterium tuberculosis (Fig. 2). The majority of Gram-positive bacteria have strong tendencies to arrange their genes on the leading strands and to have high G + A content on the leading strands. Among the bacteria tested, C. perfringens is an extreme case together with C. acetobutylicum, in fine contrast to the Gram-negative bacteria Borrelia burgdorferi, whose G + C content (28.6%) is very similar to that of C. perfringens (Fig. 2).
Figure 2.
Relationship between the content of G + A on the leading strand and the percentage of ORFs on the leading strand in various bacteria. The data were calculated from the genomic sequences from B. burgdorferi (Bbu) (38), B. subtilis (Bsu) (26), C. acetobutylicum (Cac) (10),Campylobacter jejuni (Cje) (39), C. perfringens (Cpe), E. coli (Eco) (40), Lactococcus lactis (Lla) (41), M. leprae (Mle) (42), M. tuberculosis (Mtu) (43), Neisseria meningitidis (Nme) (44), Pseudomonas aeruginosa (Pae) (45), Staphylococcus aureus (Sau) (46), Streptococcus pyogenes (Spy) (47), Treponema pallidum (Tpa) (48), and Vibrio cholerae (Vch1) (49). Open and filled circles represent Gram-negative and Gram-positive bacteria, respectively.
In the C. perfringens chromosome, we assigned biological roles to 56.1% (1,492) of the ORFs; 18.9% (502) of the ORFs are similar to hypothetical genes of unknown function and 25.2% (666) are unique hypothetical genes with no significant similarities to putative or demonstrated genes in other organisms. All of the function-assigned ORFs were classified into the categories described by Riley and Labedan (21). There were few mobile genetic elements in the C. perfringens genome: there was 1 27-kb phage remnant that consisted of 37 ORFs (CPE1095 to 1131), and there were 11 transposase genes, 7 of which seem to be intact. Considering that two bacteriophages and three transposase genes exist in the C. acetobutylicum genome (10), it may be a common feature that the clostridial species have few mobile genetic elements in the chromosome.
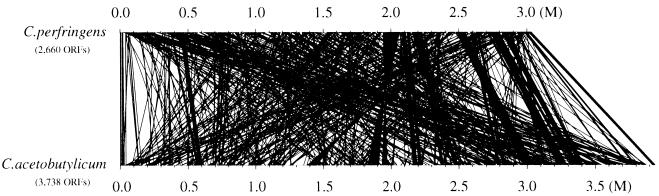
We checked the location of the homologous genes in the chromosomes of C. perfringens and C. acetobutylicum (10) to compare the genomic structure of the two species. The C. perfringens genes that showed high homologies to those of C. acetobutylicum were picked up and aligned in the chromosomes of both bacteria (Fig. 3). The majority of the homologous genes in C. perfringens appear to be located inversely in C. acetobutylicum genome (Fig. 3), whereas some of the C. perfringens gene clusters exist as the similar gene clusters in C. acetobutylicum (Fig. 3). It is speculated that the genome backbones of the two species are related to some extent but are still distinct. The repertoires of function-assigned genes in C. perfringens and C. acetobutylicum are very similar, and 732 genes of C. perfringens showed high homology scores (expect = 0 with the FASTA3 program) with those in the C. acetobutylicum genome. Briefly, the genes unique to C. perfringens are those encoding myoinositol catabolism proteins, α- and β-galactosidases, three extracytoplasmic function (ECF)-type sigma factors, α-mannosidase, components for V-type sodium ATP synthase, selenocysteine synthase, and various virulence-associated proteins. C. acetobutylicum has genes encoding chemotaxis/flagella-related proteins, nitrogen fixation proteins, β-mannanase, cellulosome-related proteins, and many amino acid biosynthesis proteins (hisABDFGHI, leuABCD, thrC, argCJ, glnA, trpABCDF, and ilvDN), which are not present in the C. perfringens genome.
Figure 3.
Genomic location of the homologous genes in C. perfringens and C. acetobutylicum.
The majority of the ORFs on pCP13 seem to encode proteins of unknown function; 16 ORFs showed similarities to hypothetical proteins from other organisms, and putative function could be assigned to 17 ORFs that had similarity to proteins of other organisms. Among the function-assigned genes in pCP13, we found the genes encoding Soj and ParB proteins involved in the partitioning of the plasmid, the genes for resolvases and type I topoisomerase, a total of five genes for truncated transposases, and many genes for proteins similar to plasmid- or phage-related proteins of other organisms.
Energy Metabolism.
As a strict anaerobe, C. perfringens has several features in its energy metabolism pathway as predicted from the genome sequence. C. perfringens has a complete set of enzymes for glycolysis and glycogen metabolism. We could not find any genes coding for tricarboxylic acid (TCA) cycle- or respiratory-chain-related proteins, in contrast to C. acetobutylicum, which has incomplete TCA cycle enzymes (10). Similar to C. acetobutylicum, we could construct a pathway map for anaerobic fermentation resulting in the production of lactate, alcohol, acetate, and butyrate, all of which have been commonly detected in C. perfringens cultures (22). In the fermentation pathway, pyruvate is converted into acetyl-CoA by pyruvate-ferredoxin oxidoreductase (CPE2061), producing CO2 gas and reduced ferredoxin. Electrons from the reduced ferredoxin are transferred to protons by hydrogenase (CPE2346), resulting in the formation of hydrogen molecules (H2) that are released from the cell together with CO2. This typical production of gases may contribute to the advantages in growth and survival of C. perfringens in host tissues, making a preferred anaerobic environment. Pyruvate is also converted to lactate by lactate dehydrogenase, whereas acetyl-CoA is converted into ethanol, acetate, and butyrate through various enzymatic reactions. This anaerobic glycolysis could be the main energy source for C. perfringens.
C. perfringens utilizes a variety of sugars such as fructose, galactose, glycogen, lactose, maltose, mannose, raffinose, starch, and sucrose (22), and various genes for glycolytic enzymes were found in the genome, such as α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, β-glucuronidase, β-fructofranosidase (sucrase), α-mannosidase, pullulanase, α-amylase, and endo-1,4-beta-xylanase. These enzymes will facilitate the utilization of a variety of sugar compounds by degrading the compounds into simpler sugars that can enter the main glycolytic pathway. We also found fermentation pathways that use serine and threonine. These amino acids are converted to propionate through propionyl-CoA, which results in energy production.
Amino Acid Biosynthesis.
In contrast to the variety of sugar-utilization enzymes, many genes of the orthologous enzymes required for amino acid biosynthesis are lacking in C. perfringens. Only 45 genes for enzymes in amino acid biosynthesis were identified, in fine contrast to C. acetobutylicum, which has a complete set of genes for amino acid biosynthesis (10). Many of the genes required for the biosynthesis of arginine, aromatic amino acids, branched-chain amino acids, glutamate, histidine, lysine, methionine, serine, and threonine are absent. These findings imply that C. perfringens cannot grow in the environment where an amino acid supply is limited.
Transporters.
C. perfringens has 211 transport-related genes for amino acids, cations/anions, carbohydrates, and nucleosides/nucleotides. Of the transporters, 52 ATP-binding cassette transporter systems (ABC transporters) could be constructed based on the similarities with those from E. coli (23), B. subtilis (24), and M. tuberculosis (25). The transporters could be divided into 26 importers and 26 exporters. Among the ABC importers, we identified three putative importers for amino acids, one oligopeptide importer, and five sugar importers. Other classes of amino acid transporters for branched amino acids, arginine, lysine, glutamate, aspartate, alanine, and spermidine/putrescine were also identified. As for sugar transporters, sugar-specific enzyme IIs of the phosphotransferase system (PTS) for mannose, glucose, trehalose, fructose, sucrose, and cellobiose were found in the genome along with PTS enzyme I (CPE2357) and the Hpr protein (CPE1669).
Sporulation and Germination.
We found 61 genes related to sporulation and germination, which were deduced from similarities with genes from the spore-forming bacteria B. subtilis (26), in the C. perfringens chromosome (Fig. 1). However, over 80 sporulation- and germination-related genes found in B. subtilis were absent in C. perfringens. The majority of the key genes encoding sporulation-related sigma factors and other stage-specific sporulation proteins are present, and we could confirm that sporulation can proceed to stage IV if the sporulation is initiated. However, many genes for spore coat proteins and some germination-related proteins are missing in the genome. In addition, the genes for the major phosphorelay system of the Spo0A protein required in the initiation process of sporulation, encoding such proteins as Spo0F, Spo0B, KinA to KinE, and Rap family phosphatases, are also missing in the C. perfringens genome. This feature seems to be common to the clostridial species because C. acetobutylicum also lacks similar orthologs in the genome (10). These facts suggest that the clostridial species have a unique sporulation process different from Bacillus species.
Extracellular Toxins and Enzymes.
When compared with the C. acetobutylicum genome (10), the most distinct feature of the C. perfringens genome is the existence of virulence-related genes. In addition to the known virulence genes, we found many virulence-associated genes in the C. perfringens chromosome. Five genes that encode putative hemolysins, other than alpha- and theta-toxins, were found, based on their similarity to hemolysins from other bacterial species (Table 1). We also identified four genes whose products are similar to enterotoxin from the Gram-positive pathogen Bacillus cereus (GenBank accession no. AF192766) (Table 1). Two types of putative fibronectin-binding protein genes with similarities to those from Listeria monocytogenes (27) and B. subtilis (26) were found (Table 1), implying their contribution to the pathogenicity of C. perfringens as colonization factors. Among many genes encoding proteinases, the product of CPE0846 had significant homology with that of the Clostridium histolyticum α-clostripain gene (28), a cysteine proteinase whose existence has not been identified in C. perfringens.
Table 1.
Virulence-related genes in C. perfringens
| Gene name | Product | Length, amino acids | Source of similar sequence |
|---|---|---|---|
| CPE0030 hlyA | Hemolysin-related protein | 421 | Thermotoga maritima |
| CPE0036 plc | Phospholipase C (alpha-toxin) | 398 | C. perfringens |
| CPE0163 pfoA | Perfringolysin O (theta-toxin) | 500 | C. perfringens |
| CPE0173 colA | Collagenase (kappa-toxin) | 1,104 | C. perfringens |
| CPE0191 nagH | Hyaluronidase (mu-toxin) | 1,628 | C. perfringens |
| CPE0378 | Myosin-crossreactive antigen | 597 | S. pyogenes |
| CPE0437 hlyB | Hemolysin | 445 | Streptococcus mutans |
| CPE0452 entC | Enterotoxin | 625 | B. cereus |
| CPE0553 nanJ | Exo-alpha-sialidase | 1,173 | Clostridium septicum |
| CPE0606 entD | Enterotoxin | 635 | B. cereus |
| CPE0725 nanH | Exo-alpha-sialidase | 694 | C. perfringens |
| CPE0737 | Fibronectin-binding protein | 220 | L. monocytogenes |
| CPE0846 | α-clostripain | 524 | Clostridium histolyticum |
| CPE0881 nagI | Hyaluronidase (mu-toxin) | 1,297 | C. perfringens |
| CPE1231 | Surface protein | 1,129 | Staphylococcus saprophytics |
| CPE1234 nagJ | Hyaluronidase (mu-toxin) | 1,001 | C. perfringens |
| CPE1258 entA | Enterotoxin | 955 | B. cereus |
| CPE1279 nagK | Hyaluronidase (mu-toxin) | 1,163 | C. perfringens |
| CPE1354 entB | Enterotoxin | 549 | B. cereus |
| CPE1474 hlyC | Hemolysin | 215 | Deinococcus radiodurans |
| CPE1523 nagL | Hyaluronidase (mu-toxin) | 1,127 | C. perfringens |
| CPE1818 hlyD | Hemolysin | 271 | Bacillus halodurans |
| CPE1847 | Fibronectin-binding protein | 575 | B. subtilis |
| CPE1915 hlyE | Hemolysin III | 213 | B. cereus |
| PCP17 cpb2 | beta 2 toxin | 265 | C. perfringens |
| PCP57 cna | Collagen adhesin | 1,368 | S. aureus |
Unexpectedly, the genome sequence revealed that five hyaluronidase genes are present in C. perfringens (Table 1). The products of these genes (designated nagH, nagI, nagJ, nagK, and nagL) differ in length (1,001–1,628 amino acids) (Table 1), but their N-terminal amino acid sequences are similar. psort analysis (18) revealed that all of the nag genes have putative N-terminal signal sequences, suggesting that they encode secreted enzymes. In addition to the chromosome-encoded virulence factors, we found the cpb2 gene encoding beta2 toxin (PCP17) in plasmid pCP13 (Table 1), which was identified as a novel virulence factor associated with digestive tract diseases of piglets and horses (29). We found another virulence-associated gene (PCP57) in pCP13 whose putative product showed similarity to a collagen adhesin (cna) from Staphylococcus aureus (Table 1). The cna gene might be involved in the attachment of C. perfringens cells to collagens in the host tissue and function as a colonization factor.
Interestingly, these virulence-associated genes found in C. perfringens do not form pathogenicity islands at all (Fig. 1). We searched for mobile genetic elements in the flanking regions of these genes but could not find any insertion sequence-, transposon-, or phage-related sequences. The G + C content of the virulence-related genes was not significantly different from those of other genes. In addition, the GeneHucker scores (16) calculated from the codon usage of the virulence genes were higher than the average score of all other genes in C. perfringens. These data imply that the horizontal transfer of the various virulence genes from other bacteria into the C. perfringens genome is not likely, or was not a recent event.
VirR/VirS Regulatory System.
C. perfringens has 48 genes that are classified into the bacterial two-component signal transduction system (30), including 28 sensor histidine kinases and 20 response regulators. Although little is known about the two-component systems in C. perfringens, the VirR/VirS regulatory system has been characterized only as regulating the expression of certain toxins (alpha-, kappa-, and theta-toxins) and proteins at the level of transcription (31–34). Because the response regulator protein VirR has been reported to bind to a repeated DNA sequence located upstream of the promoter of the theta-toxin gene (35), we tried to search for the VirR-binding sequence in the genome and found four previously unidentified putative VirR-binding sites (Fig. 4). One of these is located in the 5′ region of the hyp7 gene (CPE0957), which has been proven to be a member of the VirR/VirS regulon and to be a positive regulator for alpha- and kappa-toxin genes (32). Other VirR-binding motifs were found in regions adjacent to the genes for two hypothetical proteins (CPE0845 and CPE0920) and the newly found virulence factor, α-clostripain (Fig. 4). A comparison of the VirR-binding motifs revealed a consensus sequence, CCAnTT(n = 15)CCAGTT(n = 3)Cac, for a possible VirR-binding site (Fig. 4).
Figure 4.
VirR-binding motifs in the C. perfringens genome. The promoter region of the theta-toxin gene is on the top with consensus promoter sequences (−35 and −10) and the mRNA start point (31, 35). The putative VirR-binding sequences (shown in red) are aligned below, and deduced promoter sequences are underlined. The possible consensus VirR-binding motif is shown on the bottom.
Discussion
The complete genomic sequence of C. perfringens and the comparison with C. acetobutylicum genome reveal interesting features of this organism. C. perfringens has the highest tendency to arrange the genes on the leading strands (Fig. 1), which seems to be highly related to having the highest G + A content of the leading strands. This relationship seems to be common to other Gram-positive bacteria (Fig. 2). It is obvious that an excess of purines on the coding strands is a universal feature of genes in all bacteria, as proposed (20). However, the extent of the purine excess and gene arrangement on the leading strands seems to be different among Gram-positive and Gram-negative bacteria (Fig. 2), and Gram-positive bacteria show higher tendencies. However, M. leprae and M. tuberculosis, which are classified as Gram-positive bacteria, showed similar tendencies with Gram-negative bacteria (Fig. 2). Although the reason for this finding is unclear, their pronounced high G + C contents compared with those of other Gram-positive bacteria may result in a gene arrangement close to those of Gram-negative bacteria. The difference in strand compositional asymmetry may affect the overall gene expression of bacteria, because the orientation of highly expressed genes is arranged to be the same as their replication direction (20). Further analysis, including more extensive comparative studies of other not yet sequenced bacteria, will be required to elucidate the difference in their genetic backgrounds and the significance of the strand compositional asymmetry.
The genome information of C. perfringens and the comparison with C. acetobutylicum genome present us with a clear picture about its way of life as a pathogen. In an environment such as in soil, C. perfringens may not grow and survive in spore form. When the organism enters into human tissues through an injury and the preferred anaerobic environment is once established, it grows rapidly, using various host materials. C. perfringens can degrade large sugar compounds into simpler sugars by using various saccharolytic enzymes, and the resulting sugar is efficiently imported into the cells, where it is finally fermented by the anaerobic glycolysis pathway to generate energy and abundant gas. The anaerobic atmosphere will be enhanced by the production of gas, making the environment more suitable for C. perfringens. Five species of secreted hyaluronidases will facilitate the degradation of polysaccharides such as hyaluronic acid and chondroitin sulfate and help the organism to spread into deeper tissues. Hyaluronidase has been referred to as a spreading factor that decreases viscosity of ground substance in the connective tissue (36), and the variation in the C-terminal sequences of the hyaluronidases may represent their substrate specificities against various host polysaccharides.
Sialidases will also contribute to the degrading of sialic acid from host tissues and cells. The cytolytic toxins, including alpha-toxin, theta-toxin, and putative enterotoxins, may destroy the host cells to release various materials. The released proteins and host structural proteins are degraded by proteinases such as α-clostripain and collagenase. The resulting amino acids and/or peptides are efficiently imported into C. perfringens cells via various transporters, which is essential for the organism to synthesize proteins because of the lack of many enzymes for amino acid biosynthesis. Thus, C. perfringens actively degrades and imports various materials from the host tissues to grow and survive in the host, which in turn causes massive destruction of the host tissues and severe myonecrotic lesion. C. perfringens produces various toxins/enzymes during its early growth stage (early- to mid-exponential phase) (3), in contrast to many toxigenic pathogens that commonly produce toxins during the stationary phase (37). It is reasonable to assume that C. perfringens needs to quickly synthesize the degradative toxins/enzymes to gain nutritional sources efficiently through the degradation of the host material. Hence, the pathogenicity and nutritional acquisition must be highly coupled in C. perfringens infection, and this unique nutritional feature would be a possible target for the inhibition of growth and prevention of C. perfringens infection.
To date, the VirR/VirS system is the only two-component system known to regulate the transcription of many toxin genes and to contribute to the pathogenicity of C. perfringens (4, 8). Several genes are regulated by the VirR/VirS system, including genes for alpha-toxin (plc), theta-toxin (pfoA), kappa-toxin (colA), 2′,3′-cyclic nucleotide phosphodiesterase (cpd), protein tyrosine phosphatase (ptp), hypothetical 7-kDa protein (hyp7), cysteine synthase (cysK), and cystathionine gamma-synthase (metB) (31, 32). The search for the VirR-binding motif on the whole genomic sequence revealed that five genes have the consensus sequences to which the VirR protein may directly bind. This implies that the five genes including pfoA and hyp7 are directly regulated but that other genes are indirectly regulated by the VirR/VirS system. Because the hyp7 gene is thought to be a secondary transcriptional regulator mediating the signal from VirR/VirS to plc and colA (32), similar regulatory cascades might exist in C. perfringens to regulate other VirR/VirS-regulated genes. Further analysis of the genes found to have VirR-binding sites is required to elucidate the regulatory network of the VirR/VirS regulon, which develops effective strategies to overcome the life-threatening infection of C. perfringens.
There remain more questions than answers regarding the genetics and molecular biology of C. perfringens. Knowing and understanding all of the genomic information about C. perfringens will, with no doubt, open the door widely to future detailed studies on the pathogenicity and biology of this anaerobic pathogen and other pathogenic clostridia whose genomes are currently sequenced.
Acknowledgments
We thank S. T. Cole, Y. Fujinaga, T. Hayashi, T. Nakazawa, K. Oguma, M. Ohnishi, T. Shimizu, and H. Yoshikawa for their advice, support, and encouragement. We thank K. Furuya, K. Honjo, A. Kushibiki, S. Watanabe-Mekari, S. Ohshima, M. Saitoh, T. Sasaki, K. Shima, T. Ueda, and C. Yoshino for their technical assistance. This work was supported by Research for the Future Program from the Japan Society for the Promotion of Science (JSPS-RFTF00L01411) and by Grant-in-Aid 12206015 for Scientific Research on Priority Areas (C) “Genome Biology” from the Ministry of Education, Science, Sports and Culture of Japan. K.O. is a Research Fellow and H.H. is a Post Doctoral Fellow of the Japan Society for the Promotion of Science.
Footnotes
References
- 1.McDonel J L. Pharmacol Ther. 1980;10:617–635. doi: 10.1016/0163-7258(80)90031-5. [DOI] [PubMed] [Google Scholar]
- 2.Hatheway C L. Clin Microbiol Rev. 1990;3:66–98. doi: 10.1128/cmr.3.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petit L, Gibert M, Popoff M R. Trends Microbiol. 1999;7:104–110. doi: 10.1016/s0966-842x(98)01430-9. [DOI] [PubMed] [Google Scholar]
- 4.Rood J I. Annu Rev Microbiol. 1998;52:333–360. doi: 10.1146/annurev.micro.52.1.333. [DOI] [PubMed] [Google Scholar]
- 5.Rood J I, Cole S T. Microbiol Rev. 1991;55:621–648. doi: 10.1128/mr.55.4.621-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canard B, Cole S T. Proc Natl Acad Sci USA. 1989;86:6676–6680. doi: 10.1073/pnas.86.17.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katayama S-I, Dupuy B, Garnier T, Cole S T. J Bacteriol. 1995;177:5680–5685. doi: 10.1128/jb.177.19.5680-5685.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimizu T, Okabe A, Rood J I. In: The Clostridia: Molecular Biology and Pathogenesis. Rood J I, Songer G, McClane B A, Titball R W, editors. London: Academic; 1997. pp. 451–470. [Google Scholar]
- 9.Mahony D E, Moore T J. Can J Microbiol. 1976;22:953–959. doi: 10.1139/m76-138. [DOI] [PubMed] [Google Scholar]
- 10.Nolling J, Breton G, Omelchenko M V, Makarova K S, Zeng Q, Gibson R, Lee H M, Dubois J, Qiu D, Hitti J, et al. J Bacteriol. 2001;183:4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimizu T, Ohshima S, Ohtani K, Shimizu T, Hayashi H. Microbiol Immunol. 2001;45:179–189. doi: 10.1111/j.1348-0421.2001.tb01278.x. [DOI] [PubMed] [Google Scholar]
- 12.Shimizu T, Ohshima S, Ohtani K, Hoshino K, Honjo K, Hayashi H, Shimizu T. Syst Appl Microbiol. 2001;24:149–156. doi: 10.1078/0723-2020-00024. [DOI] [PubMed] [Google Scholar]
- 13.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe T M, Eddy S R. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakiyama T, Takami H, Ogasawara N, Kuhara S, Kozuki T, Doga K, Ohyama A, Horikoshi K. Biosci Biotechnol Biochem. 2000;64:670–673. doi: 10.1271/bbb.64.670. [DOI] [PubMed] [Google Scholar]
- 16.Yada T, Nakao M, Totoki Y, Nakai K. Bioinformatics. 1999;15:987–993. doi: 10.1093/bioinformatics/15.12.987. [DOI] [PubMed] [Google Scholar]
- 17.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakai K, Horton P. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 19.Lobry J R. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 20.McLean M J, Wolfe K H, Devine K M. J Mol Evol. 1998;47:691–696. doi: 10.1007/pl00006428. [DOI] [PubMed] [Google Scholar]
- 21.Riley M, Labedan B. In: Escherichia coli and Salmonella. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol; 1996. pp. 2118–2202. [Google Scholar]
- 22.Sneath P H A. In: Bergey's Manual of Systemic Bacteriology. Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Vol. 2. Baltimore: Williams & Wilkins; 1986. pp. 1104–1207. [Google Scholar]
- 23.Linton K J, Higgins C F. Mol Microbiol. 1998;28:5–13. doi: 10.1046/j.1365-2958.1998.00764.x. [DOI] [PubMed] [Google Scholar]
- 24.Quentin Y, Fichant G, Denizot F. J Mol Biol. 1999;287:467–484. doi: 10.1006/jmbi.1999.2624. [DOI] [PubMed] [Google Scholar]
- 25.Braibant M, Gilot P, Content J. FEMS Microbiol Rev. 2000;24:449–467. doi: 10.1111/j.1574-6976.2000.tb00550.x. [DOI] [PubMed] [Google Scholar]
- 26.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, et al. Nature (London) 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 27.Gilot P, Jossin Y, Content J. J Med Microbiol. 2000;49:887–896. doi: 10.1099/0022-1317-49-10-887. [DOI] [PubMed] [Google Scholar]
- 28.Dargatz H, Diefenthal T, Witte V, Reipen G, von Wettstein D. Mol Gen Genet. 1993;240:140–145. doi: 10.1007/BF00276893. [DOI] [PubMed] [Google Scholar]
- 29.Gibert M, Jolivet-Reynaud C, Popoff M R, Jolivet-Renaud C. Gene. 1997;203:65–73. doi: 10.1016/s0378-1119(97)00493-9. [DOI] [PubMed] [Google Scholar]
- 30.Hoch J A. Curr Opin Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 31.Ba-Thein W, Lyristis M, Ohtani K, Nisbet I T, Hayashi H, Rood J I, Shimizu T. J Bacteriol. 1996;178:2514–2520. doi: 10.1128/jb.178.9.2514-2520.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banu S, Ohtani K, Yaguchi H, Swe T, Cole S T, Hayashi H, Shimizu T. Mol Microbiol. 2000;35:854–864. doi: 10.1046/j.1365-2958.2000.01760.x. [DOI] [PubMed] [Google Scholar]
- 33.Lyristis M, Bryant A E, Sloan J, Awad M M, Nisbet I T, Stevens D L, Rood J I. Mol Microbiol. 1994;12:761–777. doi: 10.1111/j.1365-2958.1994.tb01063.x. [DOI] [PubMed] [Google Scholar]
- 34.Shimizu T, Ba-Thein W, Tamaki M, Hayashi H. J Bacteriol. 1994;176:1616–1623. doi: 10.1128/jb.176.6.1616-1623.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung J K, Rood J I. J Bacteriol. 2000;182:57–66. doi: 10.1128/jb.182.1.57-66.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hynes W L, Walton S L. FEMS Microbiol Lett. 2000;183:201–207. doi: 10.1111/j.1574-6968.2000.tb08958.x. [DOI] [PubMed] [Google Scholar]
- 37.Bassler B L. Curr Opin Microbiol. 1999;2:582–587. doi: 10.1016/s1369-5274(99)00025-9. [DOI] [PubMed] [Google Scholar]
- 38.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, et al. Nature (London) 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 39.Parkhill J, Wren B W, Mungall K, Ketley J M, Churcher C, Basham D, Chillingworth T, Davies R M, Feltwell T, Holroyd S, et al. Nature (London) 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 40.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 41.Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich S D, Sorokin A. Genome Res. 2001;11:731–753. doi: 10.1101/gr.169701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cole S T, Eiglmeier K, Parkhill J, James K D, Thomson N R, Wheeler P R, Honore N, Garnier T, Churcher C, Harris D, et al. Nature (London) 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 43.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, 3rd, et al. Nature (London) 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 44.Parkhill J, Achtman M, James K D, Bentley S D, Churcher C, Klee S R, Morelli G, Basham D, Brown D, Chillingworth T, et al. Nature (London) 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 45.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S, Hufnagle W O, Kowalik D J, Lagrou M, et al. Nature (London) 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 46.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, et al. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 47.Ferretti J J, McShan W M, Ajdic D, Savic D J, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov A N, Kenton S, et al. Proc Natl Acad Sci USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, et al. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 49.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, et al. Nature (London) 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]