Abstract
We have generated eight mAbs (MW1–8) that bind the epitopes polyglutamine (polyQ), polyproline (polyP), or the C terminus of exon 1 in huntingtin (htt) protein. In the brains of Huntington's disease (HD) mouse models, the anti-polyQ mAbs bind to various cytoplasmic compartments, whereas the anti-polyP and anti-C terminus mAbs bind nuclear inclusions containing htt. To use these mAbs as intracellular perturbation agents, we have cloned and expressed the antigen-binding domains of three of the mAbs as single-chain variable region fragment Abs (scFvs). In 293 cells cotransfected with htt exon 1 containing an expanded polyQ domain, MW1, MW2, and MW7 scFvs colocalize with htt exon 1. Moreover, these scFvs coimmunoprecipitate with htt exon 1 in cell extracts. In perturbation experiments, MW7 scFv, recognizing the polyP domains of htt, significantly inhibits aggregation as well as the cell death induced by mutant htt protein. In contrast, MW1 and MW2 scFvs, recognizing the polyQ stretch, stimulate htt aggregation and apoptosis. Therefore, these anti-htt scFvs can be used to investigate the role of the polyP and polyQ domains in HD pathogenesis, and antibody binding to the polyP domain has potential therapeutic value in HD.
Huntington's disease (HD), a fatal neurodegenerative disorder, is caused by abnormal expansion of CAG repeats that translate into an extended polyglutamine (polyQ) stretch in exon 1 of the protein huntingtin (htt) (1). Mutant htt with >40 CAG repeats gains a toxic function and induces death in subpopulations of neurons in the striatum and cortex (2–4). A hallmark of HD and other polyQ diseases is the formation of insoluble protein aggregates in affected neurons (5, 6). A major component of the aggregates in HD is the N terminus exon 1 of mutant htt (2, 5–8). Abnormal behavior and aggregate formation are also seen in transgenic mice expressing htt exon 1 with an expanded polyQ stretch (9–11).
Neuronal death in HD has been variously attributed to polyQ toxicity, activation of caspases, interference with transcriptional machinery, and sequestration/inactivation of wild-type htt and other important cellular factors (12–17). Several proteins that interact with exon 1 of htt have been identified (14, 18–23), and although the function of these proteins in the etiology of HD is unclear, the transcriptional coactivator CREB-binding protein (CBP) as well as proteins with WW domains have been implicated in the HD pathology (18–21). Binding of htt to CBP has been shown to repress CBP-mediated gene expression (18, 19). Moreover, ectopic expression of CBP appears to block htt-mediated toxicity, indicating that transcriptional dysregulation may contribute to HD pathogenesis (19, 24). Several different WW-containing proteins have been shown to interact with proline-rich domains in the C terminus of htt exon 1 (20, 21). These interactors include spliceosomes (HYPA and HYPC) and transcription factors (HYPB), which appear to have a higher affinity for expanded polyQ htt (20). By using specific antibody reagents, these WW domain proteins have been detected in postmortem brain sections associated with toxic htt N-terminal fragments (21). Such aberrant interactions may play a role in the pathology of HD.
Molecules that block the toxic effects of htt itself or the lethal consequences of its binding to other proteins may provide clues about HD pathogenesis and may also have potential as therapeutics. Intracellular expression of recombinant Abs is an approach to block the toxic effects of mutated proteins or other pathogenic agents with high selectivity (25). In fact, intracellular expression of an Ab against the N terminus of htt was shown to inhibit aggregate formation induced in cultured cells by mutant htt, although quantitative results on inhibiting htt toxicity were not reported (26). We have generated eight mAbs (MW1–8) that recognize polyQ, polyproline (polyP), or a unique epitope near the C terminus of htt exon 1 (27). Here we report that intracellular expression of some of these mAbs as recombinant, single-chain variable region fragments (scFvs) targeted to different regions of htt exon 1 can either block or enhance aggregation as well as the cell death induced by a mutant htt.
Materials and Methods
Molecular Cloning of Antigen-Binding Domains of MW1–8.
Total RNA was extracted from hybridoma cell lines secreting each of the anti-htt MW mAbs, and mRNA was purified by using oligo-dT columns (Qiagen, Valencia, CA). Complementary cDNA was produced for each mRNA pool by using random hexanucleotide primers. The cDNAs served as sources of DNA to amplify both variable region heavy (VH) and variable region light (VL) chains for each mAb by using primers complementary to the consensus sequences flanking each domain (Amersham Pharmacia) and PCR technology. To generate recombinant single-chain fragment Abs, the amplified VH and VL of each mAb were linked by a 45-mer nucleotide encoding Gly-Ser. These scFv genes were cloned into the M13 phagemid, pCANTBE5 (Amersham Pharmacia), and used to transform Escherichia coli, strain TG15, which supports production of recombinant phage. The amplified recombinant phage population was selected for binding on immunoblots to htt exon-1-glutathione S-transferase (GST)-containing a 67-polyQ repeat. Phage that specifically bound htt were eluted and used to reinfect TG15 E. coli. Individual clones were tested again for htt binding, and the nucleotide sequence of positive clones was determined by the dideoxynucleotide chain-termination method.
Cultured Cells.
293 cells were grown in DMEM supplemented with 10% heat inactivated bovine serum, 2 mM glutamine, 1 mM streptomycin and 100 international units of penicillin. Cells were grown in 6-well plates to ≈70% confluence and transfected with a total of 2 μg of DNA by using lipofectamine, following the manufacturer's recommendations (Invitrogen).
Expression and Evaluation of scFvs.
The reading frame for each scFv was subcloned into the mammalian plasmid pcDNA3.1 in frame with the Flag epitope for ease of detection (28). Selected clones were amplified and used to transfect 293 cells, as described above. Expression of scFvs was examined by Western blot analysis of the transfected cell extracts, by using an anti-Flag Ab (Sigma). For intracellular staining, transfected cells were fixed in 4% paraformaldehyde for 30 min at 4°C, permeabilized in 70% methanol at −20°C for 1 h, and incubated with anti-Flag Ab (1:1000) for 2 h. Cells expressing scFvs were detected by a goat anti-mouse Ab conjugated to Alexa 594 (Molecular Probes), and examined with a confocal microscope. To determine the effects of scFvs on cell viability, 293 cells were cotransfected with each scFv and a plasmid encoding enhanced green fluorescent protein (EGFP; CLONTECH) to readily detect which cells were transfected. Viable cells that expressed GFP were counted 4 days after transfection by using a fluorescence microscope.
For colocalization experiments, scFvs and htt exon 1, containing a 103-Q domain fused to EGFP, obtained from the Cure Huntington Disease Initiative Resource Bank (Univ. of California, Los Angeles; ref. 18), were cotransfected in 293 cells grown on coverslips. At 24 h after transfection, cells were fixed, stained, and examined as described. Depending on the experiment, 50–70% of the cells expressed EGFP. Coimmunoprecipitation experiments were performed with 293 cell lysates cotransfected as above. Briefly, cells were harvested 24 h after transfection and lysed by sonication in buffer A (25 mM Hepes, pH 7.4/2.5 mM MgCl2/50 mM NaCl/1 mM EDTA/1% Triton X-100). After clearing the lysates by centrifugation at 14,300 rpm (Eppendorf microcentrifuge) for 10 min at 4°C, 200 μg of each lysate was incubated at 4°C with rocking for 2 h with a 40-μl slurry of anti-Flag Ab coupled to protein A beads. The beads were then washed five times in buffer A by using centrifugation at 5,000 rpm, and the complexes were resolved on SDS/PAGE. For Western blotting, rabbit anti-HD1-17 (ref. 8; Vivian Hook) and anti-Flag (1:1000; Sigma) were used as the primary Abs. Secondary antibodies conjugated to horseradish peroxidase (HRP) were used to detect the reactive protein bands by enhanced chemiluminescence (Santa Cruz Biotechnology).
For aggregation studies, 293 cells cotransfected with mutant htt exon 1 and an scFv were harvested 48 h after transfection. Cells were lysed by sonication in buffer A. Lysates were centrifuged at 150,000 × g in an SW55 rotor (Beckman Instruments, Fullerton, CA) for 30 min (19). Pellets were dissolved in sample buffer containing 2% SDS, boiled and analyzed by SDS/PAGE, and transferred to nitrocellulose membranes for Western blotting. Aggregates were detected with anti-HD1–17 polyclonal antibody as described.
Terminal Deoxynucleotidyltransferase-Mediated dUTP Nick End Labeling (TUNEL) Assay.
Two days after double transfection as above, 293 cells were fixed as described and washed three times in PBS. The TUNEL reaction consisted of 25 units of terminal deoxynucleotidyltransferase and 1 mM dUTP conjugated to tetramethylrhodamine (Roche Molecular Biochemicals) in 1× buffer/2.5 mM CoCl2 in a final volume of 50 μl (according to manufacturer's instructions). Coverslips with fixed cells were laid over the reaction mixture and incubated at 37°C in a humidified incubator. Samples were washed four times with PBS, mounted on microscope slides, and examined by confocal microscopy. TUNEL-positive cells that were expressing mutant htt exon 1 were counted from at least 16 independent microscope fields with a ×20 objective lens in four separate experiments. The data were analyzed by using excel software to determine the standard deviation and the P value (t test).
Results
Cloning and Expression of Anti-htt scFvs.
We previously generated eight mAbs that bind the htt epitopes polyQ, polyP, or the C terminus of exon 1 (27). Although MW1 stains htt in the cytoplasm, MW2 displays vesicular staining of htt. Interestingly, MW7 stains htt in the perinuclear region as well in the nuclear inclusions formed by mutant htt (27). To examine the effects of these mAbs on the biological activities of htt exon 1, we generated scFvs for MW1 and MW2, which recognize the polyQ domain, and MW7, which recognizes the polyP domains of htt exon 1. The scFvs expressed in E. coli were tested for binding to htt on immunoblots (data not shown), and positive clones were selected for further characterization in mammalian cells.
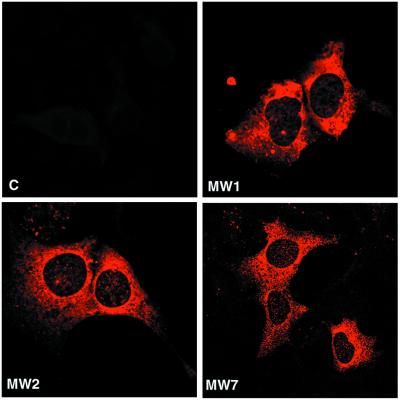
To test for scFv expression, 293 cells were transfected with the Flag-tagged scFvs and cell lysates were analyzed by Western blotting. Full-length proteins for MW1, -2, and -7 scFvs were detected by using an anti-Flag Ab (data not shown; see Fig. 2A). Histological examination of the transfected cells reveals that the scFvs have a predominantly cytoplasmic distribution (Fig. 1).
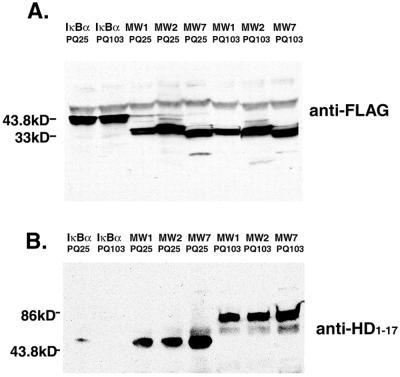
Figure 2.
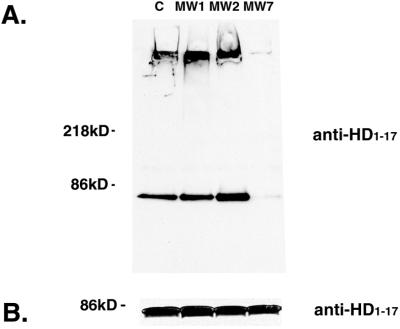
The scFvs immunoprecipitate htt exon 1. The 293 cells were cotransfected with htt exon 1-EGFP, either 25-residue polyQ (PQ25) or 103-residue polyQ (PQ103), and a Flag-scFv or Flag-IκBα. (A) An anti-Flag antibody was used to precipitate the expressed scFv proteins from lysates and for visualizing them on Western blots. The MW scFv proteins migrate with an apparent molecular mass of ≈35 kDa, whereas the control IκBα migrates as ≈43 kDa. (B) The same membranes were then stripped and reprobed with an anti-htt antibody to detect the presence of htt of ≈80 kDa (PQ103) or 50 kDa (PQ25). Precipitation of the MW scFvs results in coprecipitation of either normal or expanded-repeat htt, whereas precipitation of IκBα yields very little htt coprecipitation.
Figure 1.
Anti-htt scFvs are localized in the cytoplasm and have no obvious cytotoxic effects. The 293 cells were transfected with MW1, MW2, MW7 scFvs or an empty scFv vector (c). Two days after transfection, cells were fixed and stained with an anti-Flag antibody. The disposition of the scFvs and the appearance of the cells is very similar in each condition.
Because other proteins besides htt contain polyQ and polyP domains, it was of interest to test whether expression of the scFvs has an effect on cell viability. The 293 cells were transfected with scFvs along with a plasmid encoding EGFP as a transfection marker for evaluating cell viability (19). After 4 days of growth, the mean cell counts from at least 30 microscope fields in six wells each revealed no significant differences between the control (112 ± 6), and the scFvs for MW1 (97 ± 3), MW2 (117 ± 4), and MW7 (113 ± 5). Thus, by this measure, scFv expression does not affect cell growth or viability.
SCFVs Bind htt in Living Cells.
We used biochemical and histological methods to determine whether the scFvs interact with htt in living cells. Flag-tagged scFvs were coexpressed in 293 cells with htt exon 1 containing a polyQ domain of 103 repeats, fused to EGFP. The scFvs in Triton X-100 cell extracts were precipitated with anti-Flag Ab and analyzed for the presence of htt exon 1 by Western blotting. Similar amounts of each scFv were precipitated with the anti-Flag Ab (Fig. 2A; 33-kDa bands). Staining of the same membrane with an Ab to the N-terminal 17 aa of htt demonstrates that htt exon 1 coimmunoprecipitates with each scFv (Fig. 2B; 86-kDa bands). Similar results were obtained when htt exon 1 with a 25-Q stretch was used for transfection (data not shown). Expression of Flag-tagged IκBα was used as a negative control. Although IκBα was precipitated from the extract by the anti-Flag Ab (Fig. 2A; 44-kDa band), htt is not coprecipitated (Fig. 2B). The bands below 30 kDa in Fig. 2B likely represent nonspecific staining of the precipitating Ab.
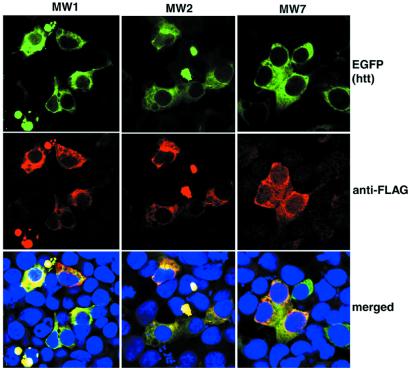
To confirm binding of the scFvs to htt and to localize the sites of interaction within cells, we used confocal microscopy to examine 293 cells cotransfected with 103 polyQ htt and each scFv. Although the scFvs are distributed throughout the cytoplasm in the absence of htt (Fig. 1), they are heavily concentrated in the perinuclear region in the presence of htt exon 1, and considerable colocalization is apparent (Fig. 3). These data indicate that anti-htt scFvs bind mutant htt in living cells.
Figure 3.
Anti-htt scFvs colocalize with htt. The 293 cells were cotransfected with mutant htt and an scFv. Twenty-four hours later, cells were fixed and stained with anti-Flag antibody (red), and htt was visualized by the fluorescence associated with its EGFP tag (green). Nuclei were visualized by a DNA stain (blue). In each case, scFvs colocalize with htt.
SCFVs Perturb htt-Induced Cell Death.
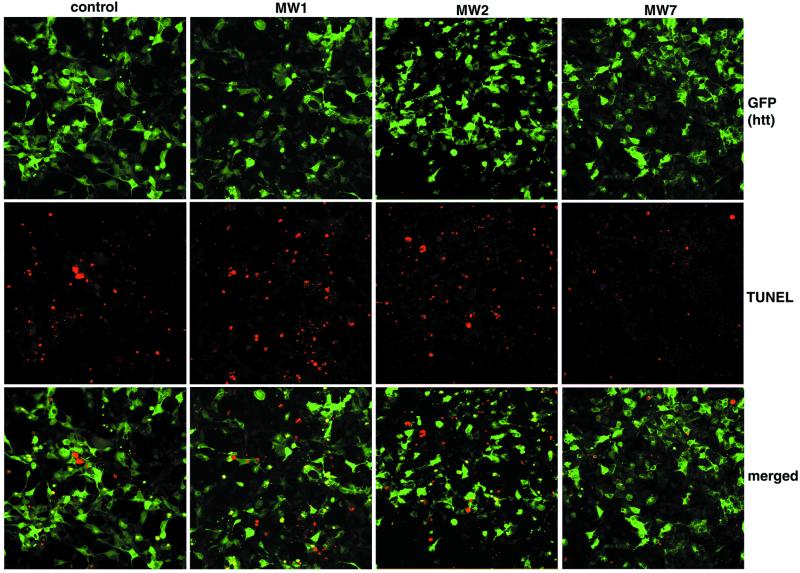
To evaluate the effect of scFvs on the toxic effects of mutant htt, we examined TUNEL staining of 293 cells cotransfected with 103-Q htt-EGFP and each scFv. Cells expressing mutant htt EGFP along with an empty scFv vector display significant TUNEL staining, and apoptotic bodies are observed starting about 12 h after transfection (Fig. 4, control column). Surprisingly, TUNEL staining is even more dramatic in the presence of MW1 or MW2 scFv and mutant htt (Fig. 4). Thus, scFv binding to the polyQ domain appears to accentuate the toxicity of mutant htt. In striking contrast, expression of MW7 scFv inhibits the toxicity of mutant htt (Fig. 4). These experiments were done under the same conditions as in Fig. 2A, which demonstrated equivalent expression of MW1, -2, and -7 scFvs in the cells.
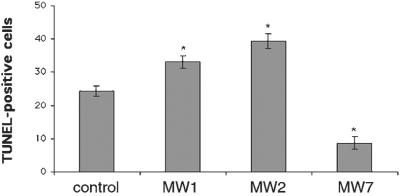
Figure 4.
MW7 inhibits htt-induced cell death. The 293 cells were transfected with htt exon 1-EGFP plus empty vector or one of the anti-htt scFvs. The transfected cells are visualized by GFP fluorescence (green), and dying cells by TUNEL staining (red). The presence of MW7 scFv decreases the number of TUNEL+ cells.
To quantify the effects of scFv expression on mutant htt toxicity, we counted TUNEL+ cells. The increase in mutant htt-induced TUNEL staining in the presence of MW1 and -2 scFvs is 38% and 67%, respectively (P < 0.05) (Fig. 5). In contrast, the number of TUNEL+ cells in the presence of MW7 scFv is reduced to 33% of the control (P < 0.05). Thus, although the anti-polyQ mAbs MW1 and -2 accentuate the toxicity of mutant htt, expression of the anti-polyP mAb MW7 inhibits the toxicity of mutant htt.
Figure 5.
MW7 inhibits htt-induced cell death whereas MW1 and -2 exacerbate it. Cultures such as those shown in Fig. 4 were used to count TUNEL+ cells. All three of the scFvs perturb the effect of htt toxicity, but only MW7 inhibits it.
MW7 scFv Reduces Aggregation of Mutant htt.
Given that the role of htt aggregation in HD is controversial, it was of interest to evaluate the effects of scFv expression on mutant htt aggregation in 293 cells. Aggregation was evaluated biochemically by examining the amount of htt that is precipitated from cell lysates by centrifugation at 150,000 × g for 30 min. The pellets contain aggregated htt (or htt that is bound to large structures) that can be solubilized by SDS treatment (Fig. 6A, 80-kDa bands), as well as htt that remains insoluble after boiling in SDS and cannot enter the gel (Fig. 6A, top of gel). Both such species of pelleted htt are detected in extracts of cells transfected with mutant htt exon 1 alone (Fig. 6A). Aggregation appears to be somewhat stronger when mutant htt exon 1 is coexpressed with MW1 or MW2 scFv. On the other hand, very little aggregated htt is found in the presence of MW7 scFv. Scanning the bands at the top of the gel yields values in arbitrary units of 68.8 for MW1, 54.3 for MW2, 0.2 for MW7, and 48.8 for no scFv. Thus, coexpression of MW7 scFv interferes with aggregation of mutant htt exon 1, and there is a qualitative correlation between the effects of the scFvs on htt aggregation and its toxicity. The expression of MW7 does not appear to cause a depletion in the level of soluble htt, however, as can be seen in Fig. 6B.
Figure 6.
MW7 scFv inhibits aggregation of mutant htt exon 1. (A) Lysates of 293 cells transfected with mutant htt and an scFv were subjected to high-speed centrifugation and were analyzed by Western blotting. The htt in the pellet that can be solubilized by SDS treatment is ≈80 kDa, whereas the htt that cannot be solubilized does not enter the gel and is visualized as a band at the top of the gel. Very little of either form of htt is found in lysates from cells transfected with MW7 scFv. In contrast, these forms of htt appear to be greater in lysates from cells transfected with MW1 or -2 scFvs than in lysates from control cells. (B) The level of soluble htt in the cleared lysates does not appear to be affected by scFv expression.
Discussion
We have cloned and expressed three recombinant antibodies targeted to either the polyQ or polyP motifs of htt. As determined by coimmunoprecipitation and histological colocalization, all three of scFvs bind htt in cells. In a functional perturbation study, MW7, recognizing the polyP domains of htt, inhibits cell death induced by mutant htt. Biochemical assays show that WM7 coexpression also strongly inhibits htt aggregation. A recent study of this type showed that intracellular expression of a mAb against the N terminus of htt also inhibits aggregate formation induced in cultured cells by mutant htt exon 1 (26). The latter paper did not, however, report the effects of this scFv on cell survival, and the connection between aggregates and toxicity remains controversial. It is therefore important, if surprising, that both MW1 and MW2, specific for the polyQ domain, accelerate the cell death as well as the aggregate formation induced in cultured cells by mutant htt. Because overexpression of a polyQ stretch itself is toxic (29), it might have been expected that blocking this domain in htt would lessen its toxicity. Moreover, it has been hypothesized that aggregation of htt involves stacking of β-sheets of polyQ, forming polar zippers (30). Therefore, molecules that bind to the polyQ domain should block the stacking of htt and therefore inhibit aggregate formation.
An alternative view is that the polyQ domain in htt is not directly toxic; rather, the extended polyQ stretch alters the binding of htt to other cell proteins, either positively or negatively, and this altered interaction(s) is toxic. It is also possible that the extended polyQ stretch results in the inhibition of htt degradation (31), which could cause toxicity. This scenario has not been observed in cultured lymphoblasts from HD patients, however (32). In these latter two scenarios, intracellular Abs that bind the extended polyQ domain could stabilize the mutant htt conformation, thereby increasing its toxicity (and its aggregation). In this regard, it will be of interest to determine the effect of MW1 and -2 scFv expression on the binding of htt to its known partners, such as CBP, which is a complex that enhances cell death (18, 19).
The inhibition of htt-induced cell death by MW7 targeted to polyP domains raises a number of questions. Such domains are binding sites for proteins with WW motifs (33). A potential mechanism contributing to HD pathogenesis is the recruitment of WW-rich proteins. In fact, several WW-containing proteins, including CA150, spliceosomes, and transcription factors, can interact with htt exon 1 (20–22). Moreover, interaction of some of the WW proteins with htt exon 1 is enhanced in htt with an expanded polyQ stretch (20, 21). Interestingly, the MW7 mAb selectively stains intranuclear aggregates formed in the transgenic mouse model (27). This finding implies that expanded polyQ makes the proline-rich motifs more available to binding to WW-containing proteins or to MW7. Intracellular expression of MW7 may therefore compete with the pernicious recruitment of other proteins that leads to cell death. It is also possible that binding of MW7 scFv to mutant htt may impose a conformation that enhances proteosome-mediated degradation.
We quantified htt aggregation by using a biochemical rather than a morphological method. The latter proved somewhat unreliable, as large aggregates containing htt were sometimes lost from the dishes during histological processing. It is notable that true nuclear aggregates of htt are not usually seen in this cell system (≈1% of the cells have clearly visible EGFP-labeled aggregates in the nucleus). A similar observation has been made for other tagged htt constructs expressed in 293 cells (18, 26, 34, 35) and in PC12 cells (E. Schweitzer, personal communication). One explanation for this observation is that the rapid cell death caused by the expanded repeat construct does not allow time for aggregates to form in the nucleus, yielding instead the large aggregates seen in Fig. 2. It is also possible that fusion of htt with GFP interferes with its nuclear aggregation. Biochemical analysis clearly shows, however, that mutant htt does aggregate in these cells and that expression of MW7 scFv strikingly reduces this aggregation. This result does not appear to be caused by lowering the concentration of soluble mutant htt in the cell, as this is not appreciably affected by MW7 expression. Alternatively, MW7 may stabilize mutant htt in a conformation that is less amenable to stacking by means of the polyQ stretch. It is striking that in transgenic mouse brain, the MW7 mAb strongly stains htt aggregates formed by mutant protein in the nucleus, as well as the perinuclear region, whereas it stains very little of normal htt and mutant htt found in various cytoplasmic locations (27). If such histological data apply to events in the living cell, it would argue that the MW7 scFv does not bind to mutant htt until it is in aggregates in the perinuclear region. This implies that MW7 scFv may act, not by inhibiting the formation of aggregates, but rather by enhancing their turnover.
In considering MW7 scFv as a potential therapeutic agent, a key question is whether its expression would disrupt the function of other polyP-containing proteins in the cell. We observed no deleterious effects on normal 293 cells 4 days after transfection with MW7 scFv. A more detailed analysis of cell physiology would be required for firm conclusions, however. The most important experiments regarding side effects of this potential therapeutic agent will need to be done in animal models.
Acknowledgments
We thank Vivian Hook, George Jackson, and George Lawless for generously providing antibodies and DNA constructs, and Doreen McDowell for administrative assistance. David Anderson provided camera and microscope access. Ethan Signer was an important source of information and facilitated obtaining reagents. The technical assistance of Sarah Teegarden and Melinda Owens is greatly appreciated. This work is supported by the Cure Huntington's Disease Initiative of the Hereditary Disease Foundation.
Abbreviations
- HD
Huntington's disease
- htt
huntingtin
- polyP
polyproline
- polyQ
polyglutamine
- scFv
single-chain variable region fragment
- EGFP
enhanced green fluorescent protein
- CBP
CREB-binding protein
- TUNEL
terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
References
- 1.Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Reddy P H, Williams M, Tangle D A. Trends Neurosci. 1999;22:248–255. doi: 10.1016/s0166-2236(99)01415-0. [DOI] [PubMed] [Google Scholar]
- 3.Zoghbi H Y, Orr H T. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 4.Tobin A J, Signer E R. Trends Cell Biol. 2000;10:531–536. doi: 10.1016/s0962-8924(00)01853-5. [DOI] [PubMed] [Google Scholar]
- 5.Ross C A. Neuron. 1997;19:1147–1150. doi: 10.1016/s0896-6273(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 6.Wanker E E. Biol Chem. 2000;81:937–942. doi: 10.1515/BC.2000.114. [DOI] [PubMed] [Google Scholar]
- 7.Cooper J K, Schilling G, Peters M F, Herring W J, Sharp A H, Kaminsky Z, Masone J, Khan F A, Delanoy M, Borchelt D R, et al. Hum Mol Genet. 1998;7:783–790. doi: 10.1093/hmg/7.5.783. [DOI] [PubMed] [Google Scholar]
- 8.Mende-Mueller L M, Toneff T, Hwang S R, Chesselet M F, Hook V Y. J Neurosci. 2001;21:1830–1837. doi: 10.1523/JNEUROSCI.21-06-01830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies S W, Turmaine M, Cozens B A, DiFiglia M, Sharp A H, Ross C A, Scherzinger E, Wanker E E, Mangiarini L, Bates G P. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 10.Sathasivam K, Hobbs C, Mangiarini L, Mahal A, Turmaine M, Doherty P, Davies S W, Bates G P. Philos Trans R Soc London B. 1999;354:963–969. doi: 10.1098/rstb.1999.0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brouillet E, Conde F, Beal M F, Hantraye P. Prog Neurobiol. 1999;59:427–468. doi: 10.1016/s0301-0082(99)00005-2. [DOI] [PubMed] [Google Scholar]
- 12.Wellington C L, Leavitt B R, Hayden M R. J Neural Transm Suppl. 2000;58:1–17. doi: 10.1007/978-3-7091-6284-2_1. [DOI] [PubMed] [Google Scholar]
- 13.Li X J. Mol Neurobiol. 1999;20:111–124. doi: 10.1007/BF02742437. [DOI] [PubMed] [Google Scholar]
- 14.Cattaneo E, Rigamonti D, Goffredo D, Zuccato C, Squitieri F, Sipione S. Trends Neurosci. 2001;24:182–188. doi: 10.1016/s0166-2236(00)01721-5. [DOI] [PubMed] [Google Scholar]
- 15.Chen M, Ona V O, Li M, Ferrante R J, Fink K B, Zhu S, Bian J, Guo L, Farrell L A, Hersch S M, et al. Nat Med. 2000;6:797–801. doi: 10.1038/77528. [DOI] [PubMed] [Google Scholar]
- 16.Leavitt B R, Guttman J A, Hodgson J G, Kimel G H, Singaraja R, Vogl A W, Hayden M R. Am J Hum Genet. 2001;68:313–324. doi: 10.1086/318207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez I, Xu C J, Juo P, Kakizaka A, Blenis J, Yuan J. Neuron. 1999;22:623–633. doi: 10.1016/s0896-6273(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 18.Steffan J S, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu Y Z, Gohler H, Wanker E E, Bates G P, Housman D E, Thompson L M. Proc Natl Acad Sci USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nucifora F C, Jr, Sasaki M, Peters M F, Huang H, Cooper J K, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson V L, et al. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 20.Faber P W, Barnes G T, Srinidhi J, Chen J, Gusella J F, MacDonald M E. Hum Mol Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 21.Passani L A, Bedford M T, Faber P W, McGinnis K M, Sharp A H, Gusella J F, Vonsattel J P, MacDonald M E. Hum Mol Genet. 2000;9:2175–2182. doi: 10.1093/hmg/9.14.2175. [DOI] [PubMed] [Google Scholar]
- 22.Holbert S, Denghien I, Kiechle T, Rosenblatt A, Wellington C, Hayden M R, Margolis R L, Ross C A, Dausset J, Ferrante R J, Neri C. Proc Natl Acad Sci USA. 2001;98:1811–1816. doi: 10.1073/pnas.041566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boutell J M, Thomas P, Neal J W, Weston V J, Duce J, Harper P S, Jones A L. Hum Mol Gen. 1999;8:1647–1655. doi: 10.1093/hmg/8.9.1647. [DOI] [PubMed] [Google Scholar]
- 24.Cha J H. Trends Neurosci. 2000;23:387–392. doi: 10.1016/s0166-2236(00)01609-x. [DOI] [PubMed] [Google Scholar]
- 25.Worn A, Pluckthun A. J Mol Biol. 2001;305:989–1010. doi: 10.1006/jmbi.2000.4265. [DOI] [PubMed] [Google Scholar]
- 26.Lecerf J M, Shirley T L, Zhu Q, Kazantsev A, Amersdorfer P, Housman D E, Messer A, Huston J S. Proc Natl Acad Sci USA. 2001;98:4764–4769. doi: 10.1073/pnas.071058398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ko J, Ou S, Patterson P H. Brain Res Bull. 2001;56:319–329. doi: 10.1016/s0361-9230(01)00599-8. [DOI] [PubMed] [Google Scholar]
- 28.Chiang C M, Roeder R G. Pept Res. 1993;6:62–64. [PubMed] [Google Scholar]
- 29.Marsh J L, Walker H, Theisen H, Zhu Y Z, Fielder T, Purcell J, Thompson L M. Hum Mol Genet. 2000;9:13–25. doi: 10.1093/hmg/9.1.13. [DOI] [PubMed] [Google Scholar]
- 30.Perutz M F. Mol Med. 1995;1:718–721. [PMC free article] [PubMed] [Google Scholar]
- 31.Sherman M Y, Goldberg A L. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 32.Persichetti F, Carlee L, Faber P W, McNeil S M, Ambrose C M, Srinidhi J, Anderson M, Barnes G T, Gusella J F, MacDonald M E. Neurobiol Dis. 1996;3:183–190. doi: 10.1006/nbdi.1996.0018. [DOI] [PubMed] [Google Scholar]
- 33.Staub O, Rotin D. Structure (London) 1996;15:495–499. doi: 10.1016/s0969-2126(96)00054-8. [DOI] [PubMed] [Google Scholar]
- 34.Li L, Li X-J. Hum Mol Genet. 1998;7:777–782. doi: 10.1093/hmg/7.5.777. [DOI] [PubMed] [Google Scholar]
- 35.Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker E E. Mol Biol Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]