Abstract
Trichromacy in humans and other Old World primates evolved from a dichromatic color vision system ≈30–40 million years ago. One essential part of this evolution was the duplication and divergence of sequences on the X chromosome to create the present-day red and green cone pigment genes. Earlier work demonstrated that a locus control region (LCR) located upstream of these genes is essential for their expression. In the present work, we have generated a variety of modified human red and green pigment gene arrays that direct the expression of distinguishable histochemical reporters from each gene promoter. Transgenic mice carrying a single copy of each modified array were studied to define the role of three variables in producing mutually exclusive expression of red and green pigment transgenes: the distance between the promoter and the LCR, the identity of the visual pigment promoter, and LCR copy number. The results support a model in which the mutually exclusive expression of these genes in their respective cone types is controlled by competition between visual pigment promoters for pairing with the LCR, and they suggest a facile mechanism for the evolution of trichromacy after visual pigment gene duplication.
The evolution of a new dimension of chromatic discrimination requires (i) the production of a visual pigment with a novel absorbance spectrum, (ii) the expression of the new visual pigment in a distinct class of photoreceptors, and (iii) patterns of neural connectivity that can extract chromatic information by comparing the degree of excitation of the new and preexisting classes of photoreceptors. The present work addresses the second of these three requirements as it relates to the most recent step in the evolution of human color vision.
The human red and green pigment genes reside on the X chromosome in a head-to-tail tandem array in which a single red pigment gene lies 5′ to one or more nearly identical green pigment genes (1–3). A locus control region (LCR) resides between 3.1 and 3.7 kb 5′ of the start site of red pigment gene transcription (4, 5). The LCR is highly conserved among diverse mammals and, unlike the visual pigment gene transcription units, seems not to have been duplicated in the human genome. As a result of X-inactivation in females and X chromosome hemizygosity in males, the X chromosomal location of these genes implies that, for any given cone photoreceptor, the decision to express either a red or a green pigment gene needs to be made on only one allele.
Two contrasting models, which we shall refer to as “standard” and “stochastic”, might account for the mutually exclusive expression of the red and green pigment genes in their respective cone types. The standard model envisions red and/or green cone-specific transcriptional activators or repressors acting to coordinate cell type-specific gene expression. By contrast, the stochastic model posits that differential expression of the red and green pigment genes reflects a choice between two stable and mutually exclusive configurations of cis-acting DNA sequences and identical trans-acting factors. In its simplest form, the stochastic model assumes that red and green cones have identical sets of transcriptional regulatory proteins, and that they have no intrinsic molecular differences other than their visual pigments. One prediction of the stochastic model is that a mammal with a single X-linked pigment gene might possess the requisite transcriptional regulators to effect mutually exclusive expression of an introduced human red and green pigment gene array. Recent work has confirmed this prediction, although the efficiency of mutually exclusive transgene expression in those experiments was only ≈75%, and the mechanism by which it occurred was unexplored (6). Here, we present evidence that the LCR plays a critical role in this process.
Materials and Methods
Embryonic Stem (ES) Cells and Blastocyst Injection.
R1 cells (a gift of Janet Rossant, University of Toronto, and Se-jin Lee, Johns Hopkins University, Baltimore) were electroporated with the various constructs linearized at a unique NotI site at the 5′ edge of the array, and colonies were selected in G418. For each construct, ≈200 colonies were screened by Southern blot hybridization: EcoRI + HindIII digests were hybridized with a probe derived from the mouse protamine 1 gene (present at the 3′ end of each reporter cassette) to determine the copy number of the transgene array and then were hybridized with a probe derived from the LCR to confirm the integrity of the 5′ end of the array. Colonies that were judged to carry single, unrearranged transgenic arrays were injected into blastocysts to derive chimeric mice. Progeny showing germ-line transmission were crossed to C57/Bl6 and the loxP-flanked neo marker excised by further crossing to germ-line cre mice (7).
Histochemistry.
X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and X-Phos (5-bromo-4-chloro-3-indolyl-phosphate)/nitro-blue tetrazolium (NBT) histochemical reactions were performed essentially as described (6) after brief glutaraldehyde fixation of intact eyes or isolated retinas. X-Gal staining was performed for 1 day at 37°C, and then the tissue was fixed further, heated to 65°C to eliminate endogenous alkaline phosphatase activity, stained with X-Phos/NBT, cryoprotected in sucrose, frozen in OCT compound, and sectioned. In some cases, the tissue was sectioned before the X-Phos/NBT reaction.
Results
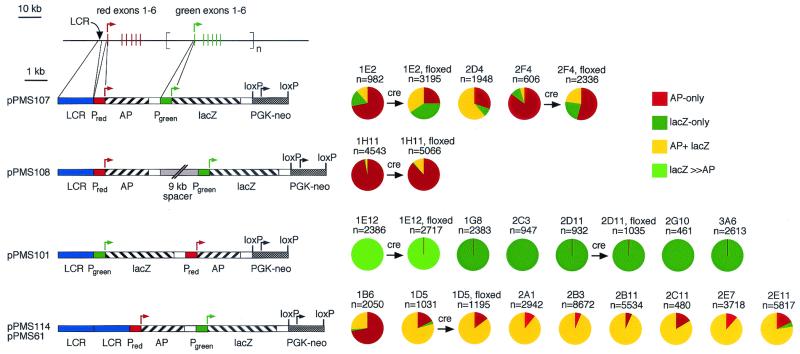
Four derivatives of the human red and green pigment gene array were tested in mice (Fig. 1). Each derivative consists of a red pigment gene-promoter driving an AP-reporter and a green pigment gene-promoter driving an E. coli β-galactosidase reporter (lacZ). In the wild-type arrangement (pPMS107), an LCR at the 5′ end is followed by the AP and lacZ transcription units. Two variations on this construct were designed to examine whether the absolute or relative distances between individual promoters and the LCR affects the number of cones expressing either reporter. In one variation, the AP and lacZ transcription units were switched (pPMS101); in the second, a 9-kb spacer was inserted between the AP and lacZ transcription units (pPMS108). A fourth pair of constructs tested the effect of duplicating the LCR (pPMS61 and pPMS114). If promoter choice involves the formation of a complex between one promoter and the LCR, then duplicating the LCR might permit simultaneous pairing with both promoters and, therefore, a higher frequency of coexpression of AP and lacZ reporters. Each construct includes at its 3′ end a PGK-neo gene in the same orientation as the two reporter genes.
Figure 1.
Human red and green pigment transgene arrays and patterns of reporter-gene expression. (Top Left) The normal human red and green pigment gene array showing the locations of the LCR, transcription units, and exons. (Lower Left) Modified visual pigment gene arrays shown at a 10-fold enlarged scale. The start sites and direction of transcription are shown by arrows. LCR, 1.6 kb from 3,009 to 4,564 bp 5′ of the red pigment gene-initiator codon; Pred, human red pigment gene promoter (496 bp immediately 5′ of the red pigment gene-start codon); Pgreen, human green pigment gene promoter (496 bp immediately 5′ of the green pigment gene-start codon); AP, human placental alkaline phosphatase; lacZ, E. coli β-galactosidase. Each reporter enzyme-coding region is followed by a 0.5 kb fragment containing an intron and poly(A) site from the mouse protamine-1 gene. The 9-kb “spacer” DNA is an EcoRI fragment encompassing the 3′ half of the human red pigment gene (1). pPMS61 is identical to pPMS114, except that the phosphoglycerate kinase (PGK)-neo marker is not flanked by loxP sites. (Right) Pie charts showing the fraction of transgene-expressing cells that express only AP (red), only lacZ (dark green), both AP and lacZ (yellow), or lacZ≫AP (light green) in chimeric or germ-line-transmitted mice for the constructs at Left. Cell counts from different mice derived from the same ES cell line were pooled to produce a single pie chart; variation in the ratios of expressing cell types was minimal among animals derived from the same ES cell line. Cell counts for those mouse lines from which the PGK-neo marker was excised by crossing to germ-line cre mice are shown immediately to the right of the pie chart for the corresponding parental line.
To generate mice carrying a single copy of each construct, individual DNAs were electroporated into ES cells, and G418-resistant colonies were screened by Southern blotting to identify those carrying a single intact copy of the transgene (6). Under the conditions used, 1–3% of ES colonies were found to be of this type. Twenty of these ES lines were used to create chimeric mice, of which six transmitted the transgene through the germ line. The expression of AP and lacZ reporters in the retina was studied histochemically by staining with X-Phos and X-Gal (6), which produce purple and blue deposits, respectively. For each ES line, retinas were analyzed either from multiple chimeric founders or from multiple progeny that had stably inherited the transgene. In the latter case, the analysis was performed both before and after crossing to germ-line cre mice (7) to remove the loxP-flanked PGK-neo cassette. The expression patterns were analyzed only in mice that were hemizygous for the transgene array, so that patterns of transgene expression reflect the activity of only one locus.
Mice derived from 18 of 20 ES lines were found to express the transgene array in the retina, and in every case expression was found exclusively in cone photoreceptors, as judged by cell location and morphology. This high frequency of expression in vivo may reflect a preselection in ES cells for integration sites that are permissive for PGK-neo expression. In general, only a subset of cones express each transgene, presumably because of position-effect variegation (which is commonly seen for transgenes expressed in photoreceptors; refs. 5, 8, and 9) and, in founder mice, tissue chimerism.
Transgene-expressing cells were scored as AP-only, lacZ-only, AP+lacZ, or lacZ≫AP (Table 1). The spatially distinct localization of β-galactosidase in the cytosol and placental alkaline phosphatase in the plasma membrane facilitates the visualization of both activities in the same cell. As a consequence, the AP+lacZ category includes cells with ratios of staining intensities that span a range of at least 10-fold.
Table 1.
Numbers of transgene-expressing cells (AP-only, lacZ-only, AP+lacZ)
| Construct | ES line | Chimera | Nonfloxed | Floxed |
|---|---|---|---|---|
| pPMS107 | 1E2 | 707, 161, 114 | 798, 1292, 1105 | |
| 2D4 | 590, 167, 1191 | |||
| 2F4 | 517, 62, 27 | 1263, 539, 534 | ||
| pPMS108 | 1H11 | 4402, 34, 107 | 4447, 68, 551 | |
| 2A12 | 0, 0, 0 | |||
| pPMS101 | 1E12 | 0, 2386, 0* | 2, 2715, 0* | |
| 1G8 | 1, 2382, 0 | |||
| 2C3 | 0, 947, 0 | |||
| 2D11 | 2, 930, 0 | 5, 1030, 0 | ||
| 2E10 | 0, 0, 0 | |||
| 2G10 | 0, 461, 0 | |||
| 3A6 | 8, 2605, 0 | |||
| pPMS114 | 1B6 | 1479, 46, 525 | ||
| 1D5 | 189, 30, 812 | 173, 4, 1018 | ||
| pPMS61 | 2A1 | 321, 1, 2620 | ||
| 2B3 | 534, 2, 8136 | |||
| 2B11 | 352, 22, 5160 | |||
| 2C11 | 77, 2, 401 | |||
| 2E7 | 427, 3, 3288 | |||
| 2E11 | 1041, 273, 4503 |
For line pPMS101 1E12, the central number indicates lacZ≫AP. In this line, >99% of expressing cones show intense lacZ activity and extremely low levels of AP activity. Faint AP staining is detectable only in the outer segments, a region of high-membrane content that is largely devoid of lacZ activity (Fig. 2H).
As seen in Fig. 1 and Fig. 2 A and C, the wild-type construct, pPMS107, produces mutually exclusive expression from red and green pigment promoters in 65–95% of expressing cones. Inserting a 9-kb spacer between the two transcription units (pPMS108) leads to a large shift from lacZ- to AP-expressing cells (≈1% lacZ-only; Fig. 1 and Fig. 2D) relative to the wild-type construct, suggesting the general idea that proximity to the LCR increases the efficiency of gene activation. In support of this idea, exchanging the locations of the two transcription units (pPMS101) leads to lacZ-only or lacZ≫AP in more than 99% of expressing cones (Fig. 1, Fig. 2 F and H). Importantly, a small number of AP-only cells were observed in pPMS101 lines 1E12 (2 of 2,715 in floxed mice; e.g., Fig. 2H), 3A6 (8 of 2,613), 2D11 (2 of 932 in nonfloxed mice, and 5 of 1,035 in floxed mice), and 1G8 (1 of 2,383), indicating that, in these mice, the AP-transcription unit is intact but largely silenced.
Figure 2.
Histochemical visualization of reporter-enzyme activity in transgenic retinas. Retinas were doubly stained with X-Gal and X-Phos (A–F, H) or with X-Gal and naphthol phosphate/fast red (G) to reveal lacZ-expressing cells (blue) and AP-expressing cells (purple/brown or red), respectively. Flat mounts: (A) pPMS107 1E2; (B) pPMS114 1D5. Vertical sections: (C) pPMS107 1E2; (D) pPMS108 1H11; (E) pPMS114 1D5; (F) pPMS101 1E12; (G) pPMS61 2B3; (H) pPMS101 1E12, showing a rare AP-only cone in a lacZ≫AP retina. All retinas except for that shown in G are from floxed mice.
Duplicating the LCR in pPMS61 and pPMS114 leads to a large increase in the relative frequency of AP+lacZ cones in 7 of 8 lines (Fig. 1, Fig. 2 B, E, and G). The only exception to this pattern, line pPMS114 1B6, shows a high percentage of AP-only cones, perhaps because of an effect of sequences flanking the integration site. Interestingly, the relative intensities of the AP and lacZ histochemical reaction products in AP+lacZ cones in the double LCR lines seem less variable than in the AP+lacZ cones observed in single-LCR lines. These data show that the presence of two LCRs favors the formation of a chromatin configuration in which both promoters are activated, whereas the presence of a single LCR favors the activation of only a single promoter. Cre-mediated excision of the 3′-flanking PGK-neo gene produced changes of up to several-fold in the frequency of reporter-gene expression, the largest effect being seen in line pPMS107 1E2, in which the frequency of AP-only cells declined 3-fold, and the frequencies of lacZ-only and AP+lacZ cells increased 2.5- and 3-fold, respectively. Overall, the effect of the PGK-neo selectable marker seems to be smaller in this setting than in some of the experimental arrangements reported by others (10–12).
Discussion
The observations reported here have a number of interesting implications for the physiology and evolution of trichromacy in higher primates, including humans. First, they demonstrate that the presence of tandem visual pigment genes and an adjacent LCR leads to a high frequency of mutually exclusive expression in a mammal that has never before confronted this genetic regulatory challenge. We note that this mechanism may be of general relevance to the evolution of divergent patterns of gene expression and function after tandem duplication. Presumably, the ancestral primate in which the X chromosome visual pigment gene duplication first occurred similarly experienced a segregation (or partial segregation) of the two pigments into separate cone populations. If this original gene duplication occurred by means of an unequal crossing-over between spectrally distinct alleles—a plausible scenario given the high frequency of within-species allelic variability of just this type in many present-day lower primates (13)—then this ancestral primate would have acquired, in a single genetic event, the essential photoreceptor components for trichromatic vision (Fig. 3A).
Figure 3.
Schematic model of the ancestral visual pigment gene duplication and mutually exclusive visual pigment gene expression in red and green cones. (A) Hypothesized ancestral duplication event in which nonhomologous recombination during a female meiosis occurred on opposite sides of two spectrally distinct alleles of the X-linked visual pigment gene. (B) Hypothesized mechanism of selective visual pigment gene transcription via LCR-promoter pairing in present-day Old World primates.
Second, these data suggest that the green pigment gene promoter is intrinsically more effective than the red pigment gene promoter in some property that offsets their different distances from the LCR (42 kb vs. 3 kb). This property can be understood most simply as an ability to compete for contact with the LCR (Fig. 3B). Analogous models have been proposed for promoter-LCR interactions in the β-globin system (reviewed in refs. 14 and 15). Precisely tuning the strengths of the red and green pigment gene promoters in this competition may account for both the ratio of red and green cones in the human retina, which ranges roughly from 1:1 to 4:1 (16–19), and for the absence of transcripts from the “extra” green pigment genes in more distal repeat units (20, 21).
Third, the high frequency of AP-only and lacZ-only cones in pPMS107 mice supports the plausibility of the stochastic model in determining the choice of red or green pigment gene expression and, hence, cone identity (6). Finally, these data reveal one of the special advantages of X-linkage as an adjunct to gene regulation. As noted above, X-inactivation in females and X chromosome hemizygosity in males implies that, within each cone, the choice of red or green pigment gene expression needs to be made on only one chromosome. By eliminating the need to coordinate the decision at a second allele, red vs. green cone identity can be determined by a single molecular event. A simple extension of this idea suggests that a single X-linked or imprinted LCR might serve an analogous role in other systems where expression of only one member of a gene family is required in each cell. Such a system is seen, for example, among mammalian olfactory receptors.
Acknowledgments
We thank Mitra Cowan, Chip Hawkins, and Diane Blesh for blastocyst injection, and Drs. Tudor Badea, Amir Rattner, and Randy Reed for helpful comments on the manuscript. This work was supported by the Howard Hughes Medical Institute.
Abbreviations
- LCR
locus control region
- ES cell
embryonic stem cell
- AP
human placental alkaline phosphatase
- PGK
phosphoglycerate kinase
References
- 1.Nathans J, Thomas D, Hogness D S. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 2.Vollrath D, Nathans J, Davis R W. Science. 1988;240:1669–1671. doi: 10.1126/science.2837827. [DOI] [PubMed] [Google Scholar]
- 3.Feil R, Aubourg P, Helig R, Mandel J L. Genomics. 1990;6:367–373. doi: 10.1016/0888-7543(90)90578-i. [DOI] [PubMed] [Google Scholar]
- 4.Nathans J, Davenport C M, Maumenee I H, Lewis R A, Hejtmancik J F, Litt M, Lovrien E, Weleber R, Bachynski B, Zwas F, et al. Science. 1989;245:831–838. doi: 10.1126/science.2788922. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Macke J P, Merbs S L, Klaunberg B, Bennett J, Zack D, Gearhart J, Nathans J. Neuron. 1992;9:429–440. doi: 10.1016/0896-6273(92)90181-c. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Smallwood P M, Cowan M, Blesh D, Lawler A, Nathans J. Proc Natl Acad Sci USA. 1999;96:5251–5656. doi: 10.1073/pnas.96.9.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Gorman S, Dagenais N A, Qian M, Marchuk Y. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zack D J, Bennett J, Wang Y, Davenport C, Klaunberg B, Gearhart J, Nathans J. Neuron. 1991;6:187–199. doi: 10.1016/0896-6273(91)90355-4. [DOI] [PubMed] [Google Scholar]
- 9.Lem J, Applebury M L, Falk J D, Flannery J G, Simon M I. Neuron. 1991;6:201–210. doi: 10.1016/0896-6273(91)90356-5. [DOI] [PubMed] [Google Scholar]
- 10.Olson E N, Arnold H-H, Rigby P W J, Wold B J. Cell. 1996;85:1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- 11.Ramirez-Solis R, Zheng H, Whiting J, Krumlauf R, Bradley A. Cell. 1993;73:279–294. doi: 10.1016/0092-8674(93)90229-j. [DOI] [PubMed] [Google Scholar]
- 12.Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin D I K, Enver T, Ley T J, Groudine M. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs G H. Invest Ophthalomol Visual Sci. 1998;39:2205–2216. [Google Scholar]
- 14.Grosveld F. Curr Opin Genet Dev. 1999;9:152–157. doi: 10.1016/S0959-437X(99)80023-9. [DOI] [PubMed] [Google Scholar]
- 15.Li Q, Harju S, Peterson K R. Trends Genet. 1999;15:403–408. doi: 10.1016/s0168-9525(99)01780-1. [DOI] [PubMed] [Google Scholar]
- 16.Rushton W A H, Baker H D. Vision Res. 1964;4:75–85. doi: 10.1016/0042-6989(64)90034-3. [DOI] [PubMed] [Google Scholar]
- 17.Dartnall H J A, Bowmaker J K, Mollon J D. Proc R Soc London Ser B. 1983;220:115–130. doi: 10.1098/rspb.1983.0091. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs G H, Neitz J. In: Color Vision Deficiencies XI. Drum B, editor. Dordrecht, The Netherlands: Kluwer; 1993. pp. 107–112. [Google Scholar]
- 19.Roorda A, Williams D R. Nature (London) 1999;397:520–522. doi: 10.1038/17383. [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi T, Motulsky A, Deeb S S. Hum Mol Genet. 1997;6:981–990. doi: 10.1093/hmg/6.7.981. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi T, Motulsky A G, Deeb S S. Nat Genet. 1999;22:90–93. doi: 10.1038/8798. [DOI] [PubMed] [Google Scholar]