Abstract
ADP-glucose pyrophosphorylase catalyzes a rate-limiting reaction in prokaryotic glycogen and plant starch biosynthesis. Despite sharing similar molecular size and catalytic and allosteric regulatory properties, the prokaryotic and higher plant enzymes differ in higher-order protein structure. The bacterial enzyme is encoded by a single gene whose product of ca. 50,000 Da assembles into a homotetrameric structure. Although the higher plant enzyme has a similar molecular size, it is made up of a pair of large subunits and a pair of small subunits, encoded by different genes. To identify the basis for the evolution of AGPase function and quaternary structure, a potato small subunit homotetrameric mutant, TG-15, was subjected to iterations of DNA shuffling and screened for enzyme variants with up-regulated catalytic and/or regulatory properties. A glycogen selection/screening regimen of buoyant density gradient centrifugation and iodine vapor colony staining on glucose-containing media was used to increase the stringency of selection. This approach led to the isolation of a population of AGPase small subunit homotetramer enzymes with enhanced affinity toward ATP and increased sensitivity to activator and/or greater resistance to inhibition than TG-15. Several enzymes displayed a shift in effector preference from 3-phosphoglycerate to fructose-6 phosphate or fructose-1,6-bis-phosphate, effectors used by specific bacterial AGPases. Our results suggest that evolution of AGPase, with regard to quaternary structure, allosteric effector selectivity, and effector sensitivity, can occur through the introduction of a few point mutations alone with low-level recombination hastening the process.
Keywords: starch‖metabolic engineering‖allosteric regulation‖DNA shuffling
The ADP-glucose pyrophosphorylase catalyzes a key regulatory reaction in glycogen biosynthesis in prokaryotic cells and starch biosynthesis in higher plants. The catalytic activity of the enzyme is modulated by small effector molecules whose nature reflects the major carbon assimilatory pathway of the organism. Many bacterial enzymes are activated by intermediates of glycolysis or the Entner Douderoff pathway [e.g., pyruvate, fructose-6-phosphate (Fru-6-P), fructose-1,6-bisphosphate (Fru-1,6-bisP2)] and inhibited by AMP (1). The enzymes from blue–green algae and higher plants are activated by 3-phosphoglyceric acid and inhibited by Pi, key intermediates in CO2 assimilation by the C3 pathway.
Despite carrying out the same reaction and being subjected to allosteric regulation, the prokaryotic and higher plant enzymes have distinct quaternary structures. Compared with the simple homotetrameric structure of the bacterial AGPase, the higher plant enzyme is much more complex having a heterotetrameric arrangement of related large and small subunits, each having different roles in enzyme function (2, 3). The small subunit (SS) is capable of forming a homotetrameric enzyme exhibiting normal catalytic properties but is defective in allosteric regulatory properties (4, 5). The SS enzyme requires more than 30-fold greater amounts of 3-phosphoglyceric acid (3-PGA) for activation and is more sensitive to Pi inhibition as compared with the heterotetrameric enzyme. The large subunit, which is incapable of forming an active enzyme, increases the allosteric regulatory response of the SS to effectors.
Directed evolution of protein function has been used to obtain enzymes with specific enhancement of properties (6–8). The utilization of these enabling technologies in structure/function studies of enzymes has been slow to be realized. DNA shuffling (9, 10), which combines low-level point mutation and in vitro recombination in the search of protein sequence space, was used to further assay the ability of SS AGPase to form a functional “prokaryotic-like” homotetrameric enzyme (11). By combining a buoyant density shift in glycogen-accumulating bacteria and iodine vapor-staining sensitivity on medium containing decreasing concentrations of glucose, both point mutants and combinatorial mutants that display altered allosteric sensitivity and selectivity for metabolic effectors were identified. These results not only identify regions of the primary sequence involved in allosteric effector recognition and binding, but they also demonstrate that the allosteric effector site exhibits significant plasticity to recognize different effectors when altered by a single or few point mutations. The ability of the SS to accept amino acid changes, mediated through point mutation and low-level recombination, indicates that the evolutionary pathway from prokaryotic homotetramer to heterotetrameric subunit is neither lengthy nor complex for the SS.
Experimental Protocol
DNA Shuffling.
The 1.4-kb SS homotetrameric mutant TG-15 gene, which contains the double mutations L48F and V59I (11), or Devo derivatives (shuffled mutants) were digested from pTG-15 with NcoI and SacI or amplified from pTG-15 with the primers Devo-amp S: 5′-TAAGAAGGAGATATATCCATGG-3′ complementary to the small subunit expression vector (pML-10) (4) sequence upstream of the NcoI site and Devo-amp AS: 5′-ATCTGAATTCGAGCTCATCAG-3′ complementary to the pML-10 sequence downstream of the SacI site. The PCR mixture A contained 0.2 mM of each dNTP/3 mM MgCl2/10 mM KCl/10 mM NH4(SO4)2/20 mM Tris⋅HCl, pH 8.8/0.1% Triton-X-100 with Taq DNA polymerase. A PCR program of 30 cycles of 96°C for 60 s, 55°C for 60 s, and 72°C for 90 s was used to amplify the TG-15 coding sequence with primers Devo-amp S and AS described above. The PCR product was purified with a QIAquick PCR purification kit (Qiagen, Chatsworth, CA) and digested with DNase I in the presence of MnCl2 (12), creating random fragments of about 50 bp. The fragments were passed through a Micro Bio-Spin 6 chromatography column (Bio-Rad) to remove residual Mn2+, and the purified fragments were added to the reassembly PCR mixture at 20 ng/μl final concentration. The reassembly reaction contained the TG-15 gene fragments, 0.2 mM each dNTP, 1× Pfu buffer, and 2.5 units Pfu Turbo DNA polymerase (Stratagene) in 50 μl and was processed by using a PCR program for reassembly as described (9, 10). The product of this reaction was used as template in an amplification reaction by using the primers SS-amp S: 5′-GATATATCCCCATGGCTGTTTCTGATTCGCAGAATTCAC-3′ and SS-amp AS: 5′-GACTTAAGCTCGAGTAGTCTACTACTAAGGTGAACCTTAG-3′. The PCR mixture contained primers, 1-μl reassembly reaction as template, 0.2 mM each dNTP, 1 × cloned Pfu buffer, and 2.5 units Pfu Turbo DNA polymerase and was processed by using a PCR program for amplification as described (9, 10). The PCR product was purified by QIAquick PCR purification kit, digested with NcoI and SacI, and purified after agarose gel electrophoresis using a Qiagen QIAquick gel extraction kit. The PCR fragment was then ligated into pML-10 vector and transformed into Escherichia coli strain AC70R1-504 by electroporation.
Selection and Screening.
E. coli AC70R1–504 transformed with first-generation shuffled TG-15 were grown in Kornberg's 2% glucose-enriched liquid medium with 50 μg/ml kanamycin (Kan50) for 6 h and then mixed into osmotically equilibrated Percoll (Amersham Pharmacia) buoyant density gradient medium. The gradient mixture was centrifuged at 17,000 × g for 30 min at 4°C. AC70 cells banding low in the gradient (buoyant density ≈1.13) were harvested and amplified by growth overnight, and plasmid DNA was prepared from the overnight cell pellet. The first-generation coding sequences were removed by NcoI/SacI digestion and used as template for another iteration of the DNA shuffling procedure described above. The second-generation Devo clones were dilution plated on Kornberg/ Kan50 1% glucose-enriched plates and screened by iodine vapor staining (13). Darkly staining colonies were selected, streaked onto LB/Kan50 plates, and inoculated into 1 ml of LB/Kan50 liquid medium in 2-ml wells of 24-well microtiter plates. The microtiter plate cultures were pooled after overnight growth with shaking at 37°C, and plasmid DNA prepared from the pooled culture pellet was used as template in a PCR amplification. The PCR mixture contained second-generation plasmid DNA as template, primers Devo-amp S, Devo-amp AS, and PCR mixture A components as described above. The product of this PCR was used as template for another iteration of the DNA-shuffling procedure described above. The third-generation Devo clones were dilution plated on Kornberg/ Kan50-enriched plates containing 1.0% or 0.1% glucose and screened by iodine vapor staining (13). Darkly staining colonies on 0.1% glucose plates were selected, streaked onto LB/Kan50 plates, and inoculated into 3 ml of LB/Kan50 liquid medium. Plasmid DNA was prepared from the 3-ml overnight cultures and sequenced.
Devo Mutant Expression, Purification, and Activity Assays.
Expression of Devo mutant AGPase was done in the E. coli host strain BL21 (lon−, ompT−) by using described parameters (11). All purification steps were carried out at 4°C unless otherwise stated. Assay A was used to measure enzyme activity during partial purification procedures. Lysis buffer preparation, BL21 cell culture processing, and sonication were done as described (11). Partial purification was achieved by a single anion exchange chromatographic step by using POROS 20 HQ media as detailed (11). PAGE followed by Coomassie staining and immunodetection with anti-potato SS immune serum (11) was used to verify the integrity of the mutant enzymes.
In assay A, the pyrophosphorolysis of ADP-glucose was measured by following the formation of [32P]ATP from 32inorganic pyrophosphate (PPi) (14, 15). The reaction mixture contained 80 mM Hepes, pH 7.5/5 mM DTT/5 mM MgCl2/10 mM NaF/1 mM ADP-glucose/10 mM 3-PGA/0.4 mg/ml BSA/3.0 × 106 cpm/ml 32PPi/1.5 mM NaPPi and enzyme sample in a final volume of 0.25 ml. The reaction was initiated by addition of enzyme and incubated at 37°C for 10 min.
In assay B, kinetic parameters were determined by the synthesis (forward) assay. Synthesis of ADP-glucose was measured by following the formation of [14C]ADP-glucose from ATP and [14C]glucose 1-P (16). The reaction mixture contained 100 mM Hepes, pH 7.5/3 mM DTT/5 mM MgCl2/0.3 units of inorganic pyrophosphatase/1.5 mM ATP/5 mM 3-PGA/0.4 mg/ml BSA/0.1 μCi/reaction [14C]glucose 1-P/0.5 mM glucose 1-P and enzyme sample in a final volume of 0.1 ml. Reactions were initiated by addition of enzyme and incubated for 10 min at 37°C, then terminated by boiling for 2 min. Km, Ka, and Ki values, corresponding to the concentration giving 50% maximal activity, activation, and inhibition, respectively, are the mean of at least two determinations and were calculated by using the ENZYMEKINETICS software program (Trinity Software). Protein concentrations were determined by the Bradford standard assay method (17) with BSA as standard.
Results and Discussion
The higher plant AGPase is a heterotetramer composed of two large subunits and two SS. The latter subunit is capable of forming a homotetrameric enzyme with near-normal catalytic properties but having defective allosteric regulatory properties (see SS data, Table 1). Previous work in our lab resulted in the isolation of a mutant homotetrameric AGPase SS (TG-15) (11) that exhibited regulatory properties much more similar to the wild-type (WT) condition but at the expense of a reduction in ATP binding affinity (Table 1). Accordingly, our goal in this work was to use directed molecular evolution to generate homotetrameric AGPase of plant origin that had catalytic and allosteric regulatory properties like those exhibited by cyanobacterial and bacterial enzymes. The “Devo” mutants isolated would represent devolution of the higher plant AGPase gene and a return to the homotetrameric state from which the higher plant AGPase originated. Additionally, we also expected to generate homotetrameric AGPases that had a range of kinetic activation sensitivities including those with increased sensitivity to activation and/or increased resistance to inhibition by the effector molecules 3-PGA and Pi, respectively. These mutations would allow us to further define the amino acids that mediate effector binding as well as produce a set of molecular tools that could be used in metabolic engineering attempts to gauge the role of AGPase in photosynthetic efficiency, carbon partitioning between source and sink tissues, and carbon flow into starch.
Table 1.
Kinetic parameters of Devo mutants and selected ADP-glucose pyrophosphorylases
| rWT* | SS† | TG-15‡ | Devo mutant lines
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 201 | 202§ | 206 | 301 | 303§ | 316 | 330§ | 339 | 350 | 355§ | ||||
| Effectors | |||||||||||||
| 3-PGA (uM) | 100 | 2400 | 140 | 126 | 37 | 67 | 114 | 90 | 96 | 12 | 21 | 97 | 57 |
| Pi inhibition (mM)¶ | 0.07–0.17 | 0.064 | 0.4 | 0.4 | 0.4 | 0.4 | 2.0 | 2.0 | 1.0 | 7.5 | 0.4 | 0.2 | 2.3 |
| 3-PGA/Pi ratio | 0.6–1.4 | 37.500 | 0.350 | 0.315 | 0.093 | 0.168 | 0.057 | 0.045 | 0.096 | 0.002 | 0.053 | 0.485 | 0.025 |
| Substrates | |||||||||||||
| ATP (mM) | 0.3 | 0.2 | 1.2 | 0.4 | 0.6 | 0.5 | 0.4 | 0.2 | 0.8 | 0.5 | 0.5 | 0.4 | 0.8 |
| Glucose-1-P (mM) | 0.25 | 0.03 | 0.25 | 0.24 | 0.29 | 0.24 | 0.11 | 0.21 | 0.09 | 0.20 | 0.18 | 0.21 | 0.22 |
| Mg2+ (mM) | 2.5 | 2.2 | 1.4 | 2.1 | 2.0 | 1.9 | 2.1 | 1.8 | 3.0 | 1.4 | 1.8 | 2.5 | 1.8 |
DNA Shuffling Mutagenic Analysis.
DNA shuffling was carried out as described (9, 10) with variations as detailed in Experimental Protocol. As prior work has identified the amino and carboxyl termini of AGPase as important for assembly and allosteric regulation, Devo-amp primers were designed to amplify the SS AGPase coding region from adjacent pML-10 vector sequence homology, thereby allowing any mutations in the termini to be retained after amplification.
Three rounds of gene shuffling, beginning with a Taq polymerase PCR-amplified TG-15 gene as the template, resulted in the isolation of clones with enhanced AGPase activity as evaluated by glycogen accumulation revealed through a positive iodine-staining phenotype on Kornberg's 0.1% glucose plates. Sequencing of 30 selected clones revealed 35 mutations resulting in amino acid changes and 39 silent mutations. The mutation rate calculated from 40,770 bases sequenced was 0.18%, which corresponds to 2.4 nucleotide changes per 1,359 base pair coding sequences. This rate of change allows approximately one amino acid change per coding sequence and correlated well with the observed changes. Of the 30 Devo mutants selected based on rapidity and intensity of iodine staining, seven contained single amino acid changes and three were combinations of the single change mutants. Additional amino acid combinatorial mutants were discarded when single change mutants and redundancy indicated that a specific amino acid change was responsible for the observed phenotype.
Selection and Screening.
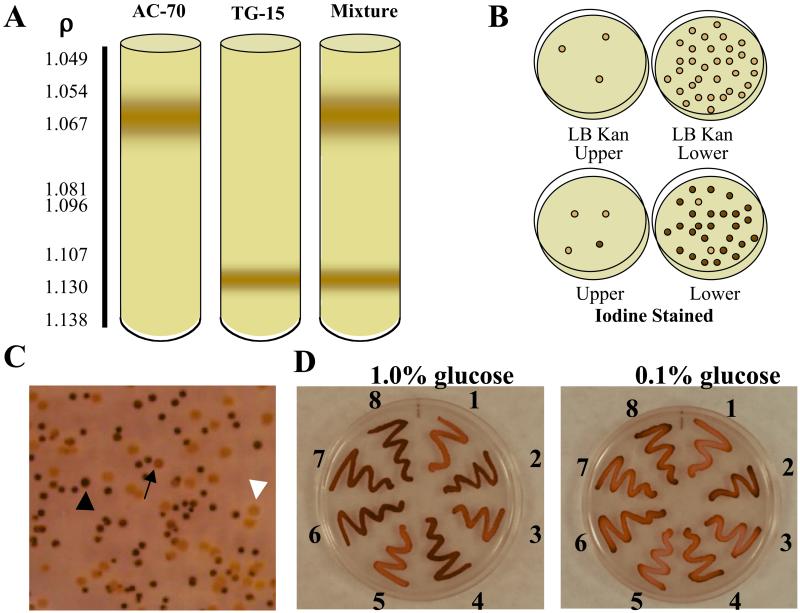
A selection method based on buoyant density differences in bacteria that contain inclusions was developed (18–20). The buoyant density of E. coli can also be shifted by osmotic shock and depends on the osmolarity of the culture medium (21, 22). To allow buoyant density differences achieved by differential accumulation of glycogen to be observable in a Percoll density gradient, the Percoll medium osmolarity was adjusted to be equal to that of the Kornberg's 2% glucose/Kan50-enriched liquid medium in which the first-generation transformants were grown. Trials using E. coli AC70R1–504 as a glycogen negative control and this strain containing pTG-15 as a glycogen positive control resulted in a clearly defined differential cellular banding pattern (Fig. 1A). Sampling of both the upper and lower bands for kanamycin resistance and glycogen accumulation revealed the upper band to consist almost entirely of AC70 cells, whereas the lower band was almost entirely AC70/pTG-15 (Fig. 1B). This technique had great utility in separating glycogen-accumulating cells from those unable to do so, but did not exhibit the sensitivity necessary to separate low-level glycogen-accumulating clones from those with greater glycogen accumulation as tested with AC-70/pTG-15 (low-level accumulation) and AC-70/Up-Reg1 (high-level accumulation, ref. 23) (data not shown). Buoyant density gradient separation proved to be a valuable initial selection step after the first round of shuffling, allowing the elimination of nonfunctional clones from inclusion in the second round while allowing a large population of positive clones to be carried forward without the need for long and laborious screening procedures.
Figure 1.
Selection and screening methodologies. (A) Percoll density gradient separation of glycogen-positive clones from cells unable to accumulate glycogen. (B) Cells were harvested from the upper and lower bands in A and tested for kanamycin resistance (B Upper) and glycogen accumulation by iodine staining (B Lower). (C) Representative iodine staining patterns exhibited by third-generation Devo bacterial colonies grown on 0.1% glucose: dark staining colony (black arrowhead), lightly staining colony (arrow), and negative staining colony (white arrowhead). (D) Iodine staining of selected third-generation Devo mutants on glucose-enriched media. Devo clones were streaked onto plates containing decreasing amounts of glucose and stained by exposure to iodine vapors as described in Experimental Protocols. 1, Devo 350; 2, Devo 339; 3, Devo 316; 4, Devo 301; 5, Devo 355; 6, Devo 303; 7, Devo 330; 8, heterotetrameric UpReg-1 AGPase (23), which has enhanced affinity for activator and is more resistant to inhibition than the WT enzyme. Cells expressing the WT heterotetrameric AGPase iodine stain very lightly in media containing 0.1% glucose.
Increasing the stringency of selection in subsequent shuffling iterations was accomplished by screening for glycogen accumulation as revealed by iodine staining on decreasing concentrations of glucose in Kornberg's/Kan50-enriched plates over decreasing time. The second-generation Devo clones were screened on 1.0% glucose-enriched media, and colonies that stained darkly within 30 s and hence had higher glycogen levels than those expressing the recombinant wild-type were selected (results not shown, but see Fig. 1C for various iodine-staining patterns observed). Over 5,700 colonies were screened, and 216 were selected to produce the third generation template genes. The third-generation Devo clones were screened on 0.1% glucose-enriched media, and colonies that stained darkly in 15 s were selected (Fig. 1C). Over 12,000 colonies were screened, 200 were subcultured to verify the rapid staining phenotype, and 30 were selected for sequence analysis. These mutants fell into 10 different classes (Fig. 1D), seven contained single amino acid changes and three were combinations of the single change mutants in addition to the two-point mutations present in TG-15. Representative mutants from each class were selected for further study.
Kinetic Parameters of Devo Mutants.
The substrate (magnesium, ATP, and glucose-1 phosphate) Km, activator (3-phosphoglycerate) Ka, and inhibitor (inorganic phosphate) Ki were determined for each of the 10 Devo mutants isolated (Table 1). The Km values for magnesium or glucose-1 phosphate for all 10 Devo mutants did not vary significantly from that of TG-15 or the recombinant WT heterotetrameric potato AGPase. The previously determined Km for ATP of TG-15 was four times that of WT AGPase, thus defining the only kinetic parameter that varied significantly between the homotetrameric mutant and the WT enzyme. All 10 of the Devo mutants displayed lower Km values for ATP than did TG-15, with one combinatorial mutant (Devo 303) having a slightly lower Km than WT AGPase. The Km values for magnesium, ATP, and glucose-1 phosphate for the Devo mutants are not significantly different from that of WT AGPase, suggesting that enhanced binding of substrate does not account for the improved iodine-staining phenotype exhibited in the colony screening.
Improvements in AGPase activity have been observed in previous mutagenic studies (23, 24). Greene et al. (23) reported AGPase variants that showed increased affinity for the activator 3-PGA and reduced sensitivity to the inhibitor Pi (class I), whereas Giroux et al. (24) isolated a mutant that had decreased sensitivity to Pi (class II). Individual Devo mutants isolated in this study exhibit characteristics of both phenotypes described above (Table 2) as well as two other classes that showed either changes in sensitivity to only 3-PGA (class III) or exhibited no significant changes from TG-15 (class IV). Devo 330 and Devo 355 have 3-PGA Ka values at least 2-fold lower than TG-15, with the combinatorial mutant Devo 330 having the lowest Ka at 12 μM, representing a 10-fold improvement in binding affinity from TG-15. In addition to enhanced affinity for 3-PGA, both mutants have increased resistance to Pi inhibition. A second class of allosteric regulatory mutants (Devo 301, Devo 303, Devo 316, and Devo 350) has significant increases in Pi resistance while having little or no effect on 3-PGA sensitivity (Table 2). A third class, represented by Devo 202, Devo 206, and Devo 339, has increased 3-PGA sensitivity without any change in Pi effector binding.
Table 2.
Fold changes in KM for ATP, A0.5 for 3-PGA activation, and I0.5 for Pi inhibition for the various Devo mutants as compared to TG-15
| Class | Strain | Mutations | ATP
|
3-PGA
|
Pi
|
|---|---|---|---|---|---|
| fold ↓ in KM | fold ↓ in A0.5 | fold ↓ in I0.5 | |||
| I | Devo 330 | Y315C, L378S | 2.4 | 11.7 | 18.8 |
| Devo 355 | K295E, Y315C | 1.5 | 2.5 | 5.8 | |
| II | Devo 301 | T426I | 3.0 | 1.2 | 5.0 |
| Devo 303 | K295E, L378S | 6.0 | 1.6 | 5.0 | |
| Devo 316 | L378S | 1.5 | 1.5 | 2.5 | |
| Devo 350 | K295E | 3.0 | 1.4 | 2.0 | |
| III | Devo 202 | S2P, K95R | 2.0 | 3.8 | 1.0 |
| Devo 206 | K341M | 2.4 | 2.1 | 1.0 | |
| Devo 339 | Y315C | 2.4 | 6.7 | 1.0 | |
| IV | Devo 201 | K377E | 3.0 | 1.1 | 1.0 |
The phenotypes of the three combinatorial mutants, Devo 303, Devo 355, and Devo 330, mirror the phenotypes of the individual mutations. Devo 330 and Devo 355 share a common Y315C mutation that confers increased 3-PGA sensitivity together with a class II mutation, K295E or L378S, which imparts increased Pi resistance. Devo 303 contains two class II mutations, and its enhanced degree of Pi resistance is an additive effect of the two single mutations with no changes in 3-PGA sensitivity.
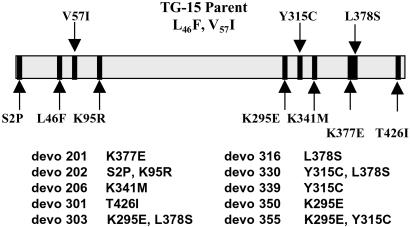
Improvements in activator binding are mediated by alteration of varied and distant amino acids, yet there is an observable pattern of mutation in both the amino and carboxyl termini in the improved Devo variants (Fig. 2). Although the majority of the improved enzymes have lysine mutations, Devo 339, with a tyrosine to cysteine change, exhibits an interesting phenotype. This enzyme requires very low levels of 3-PGA for activation (Ka of 21 μM) and has significant activity in the absence of activator, leading to the low-fold activation seen in Table 3. Combinatorial mutants of single mutations that mediate increased resistance to phosphate inhibition exhibit both additive (Devo 303, Devo 355) and synergistic effects (Devo 330). The combination of the Y317C and L380S mutations form Devo 330, which has a Ka for 3-PGA of 12 μM and is insensitive to phosphate inhibition at concentrations exceeding 7.5 mM. Increased resistance to phosphate inhibition is exhibited by half of the Devo mutants, with Devo 330 having the lowest 3-PGA/Pi ratio at 0.001; others (Table 1) showed 5 to 10-fold improvements over TG-15.
Figure 2.
Relative locations of the TG-15 and Devo mutations in the WT AGPase small subunit coding sequence. All Devo mutants retained both of the TG-15 mutations (L46F, V57I).
Table 3.
Relative metabolite activation of Devo mutants and selected ADP-glucose pyrophosphorylases
| AGPase | Fold activation with metabolic effectors at 2 mM each
|
|||||
|---|---|---|---|---|---|---|
| 3-PGA | 2-PGA | PEP | Glu-6-P | Fru-6-P | Fru-1,6-bisP | |
| Devo 201 | 30.7 | 4.0 | 18.2 | 6.5 | 22.6 | 1.4 |
| Devo 202 | 5.5 | 6.8 | 8.2 | 1.2 | 3.9 | 10.7 |
| Devo 206 | 14.9 | 5.9 | 11.9 | 3.9 | 10.8 | 5.7 |
| Devo 301 | 20.6 | 2.9 | 10.5 | 3.8 | 15.2 | 1.6 |
| Devo 303 | 22.1 | 2.5 | 11.9 | 2.9 | 15.4 | 1.9 |
| Devo 316 | 4.6 | 9.2 | 9.1 | 2.1 | 3.4 | 15.0 |
| Devo 330 | 9.6 | 3.6 | 6.6 | 4.4 | 7.9 | 3.2 |
| Devo 339 | 5.4 | 2.9 | 3.4 | 2.0 | 3.7 | 2.5 |
| Devo 350 | 19.4 | 4.7 | 13.2 | 5.0 | 15.0 | 0.9 |
| Devo 335 | 12.6 | 1.8 | 4.2 | 2.4 | 11.4 | 1.7 |
| TG-15* | 24.7 | 1.0 | 8.7 | 3.3 | 16.2 | 5.7 |
| rWT potato† | 36.0 | 1.7 | 4.0 | ND | ND | 3.3 |
| native potato‡ | 32.7 | ND | 2.6 | ND | ND | 3.3 |
| TL-46§ | 25.0 | 2.9 | 9.8 | 3.0 | 3.0 | 3.9 |
Data generated with the synthesis (forward) assay.
Homotetrameric SS AGPase (11).
Recombinant heterotetrameric AGPase.
WT AGPase purified from potato tissue.
Homotetrameric Arabidopsis SS AGPase.
Sensitivity and Selectivity of Devo Mutants to Metabolite Activators.
The blue–green algae and higher type AGPases prefer 3-PGA as their activator with little sensitivity to other phosphate esters. The homotetrameric SS enzyme TL-46 retains this specificity for 3-PGA, although phosphoenolpyruvate (PEP) is much more active as an activator compared with the WT heterotetrameric enzyme (Table 3). The SS enzyme TG-15, which contains the double mutation L46F and V57I, retains the 3-PGA and PEP specificity of the unmutated SS enzyme, but also shows significant activation by Fru-6-P.
Six of the 10 Devo mutants (Devo 201, 206, 301, 303, 350, 355) have retained 3-PGA as the main activator, whereas two are almost equally activated by 3-PGA and Fru-6-P (Devo 330 and Devo 339), and two have changed activator preference to Fru-1,6-bisP (Devo 202 and Devo 316). Activation by PEP increases parallel to that of Fru-6-P and Fru-1,6-bisP, although to a lesser degree. The fold activation of Devo 202, Devo 330, and Devo 339 is low for all metabolites because of an increased level of activity in the absence of activator, whereas Devo 206 and Devo 316 exhibit meaningful fold activation by 3-PGA and Fru-1,6-bisP, respectively, while having significant activity in the absence of activator.
The AGPases from bacteria, blue–green algae, and higher plants can be divided into seven groups based on differences in activator specificity (1). Six groups prefer pyruvate, Fru-6-P, Fru-1,6-bisP, 3-PGA, pyridoxal phosphate, or NADPH as an effector, with one group showing no activation by any known metabolite. The activation of Devo mutants by various metabolites indicates that there is considerable plasticity at the activator binding site, which allows minor sequence changes to alter effector sensitivity and selectivity (Table 3). This is best illustrated by Devo 201 and Devo 316, which contain mutations that are located adjacent to one another (Fig. 2). The K377E mutation of Devo 201 increases the specificity for PEP while maintaining the responses to 3-PGA and Fru-6-P. A mutation in the adjacent residue, L378S, as found in Devo 316, results in a complete change in effector specificity. Fru-1,6-bisP is the preferred activator followed by PEP. The normal activator 3-PGA shows less than one-third the activity of Fru-1,6-bisP. These results demonstrate that even though the various prokaryotic and higher plant AGPases show only about 20–25% identity in their primary sequences, the differences in effector specificity by these enzymes is likely because of a few key residue differences.
AGPase Subunit Role and Physiological Function.
The large subunit and SS of the heterotetrameric higher plant AGPase presumably arose through gene duplication events followed by divergence to the variety of sequences found today. The physiological function of the various AGPase isoforms, whose sequences and kinetic activities vary, has yet to be completely defined. Phylogenetic analysis of AGPase sequence data clearly divides AGPase sequences into leaf, stem, root, and endosperm forms of the enzyme (2). The 3-PGA/Pi ratio determines the activity level of allosterically regulated AGPase. Photosynthetic tissue has a pool of triose phosphates that accumulate in the chloroplast during the light cycle. Conversely, in sink tissue the 3-PGA pool is generated as an intermediate in the respiratory process of glycolysis and may be quite low at times whereas concentrations of Pi would peak during starch accumulation. These facts indicate that in sink tissue, Pi inhibition may dominate over 3-PGA activation for regulation of AGPase, and phosphate may be the key physiological regulator of AGPase activity (24). A mutant AGPase that exhibits increased sensitivity to 3-PGA activation as well as decreased sensitivity to Pi inhibition may allow for increased storage of carbon in starch reserves. The Devo mutants generated here exhibit both types of kinetic behaviors to various degrees, thus providing the tools to allow further study of these processes in vivo.
Prior investigation of the physiological role of AGPase in controlling carbon flux into storage starch had failed to show that an increased sensitivity to activator can cause an increase in starch accumulation. Previous work illustrates that lowering Pi levels (25), having an AGPase that is constitutively activated (26) or insensitive to Pi inhibition (24), can result in increased starch production. Although there is an obvious benefit of AGPase sensitivity to 3-PGA during the diurnal cycle of photosynthetic tissue, it may simply be an evolutionary carryover in sink tissue AGPase. In fact, Pi may be the main physiological regulator of reserve carbon accumulation. Experimental evidence indicates that barley (27) and wheat (28) endosperm AGPase is insensitive to 3-PGA activation, yet the enzymes display negative regulation by Pi. In maize (29) and barley (30), the major endosperm AGPase is localized in the cytosol. The extra-plastidial location of AGPase may indicate that carbon flux regulation occurs differently from that observed for the chloroplast AGPase of photosynthetic tissue. Maize endosperm AGPase exhibits significant sensitivity to Fru-6-P activation (31), which is not observed for either chloroplast or tuber AGPase. Because the principal route of cytosolic sucrose degradation occurs through the action of sucrose synthase, Fru-6-P is a good indicator of sucrose availability in the cytosol of sink tissue and would be a logical regulator of carbon flux into storage. The generation of Devo mutants with increased sensitivity to Fru-6-P activation (including Devo 355, which has a Km for Fru-6-P of 13 μM) (Table 3) would allow the opportunity to investigate the physiological role of Fru-6-P in maize AGPase regulation and carbon flux into starch.
The amino acid changes that mediate the kinetic effects outlined above are varied and diverse. The mutagenesis of key lysine residues has been shown to affect the allosteric behavior of AGPase (23, 32), and our study supports the role of lysine residues in determining AGPase activity (Devo 201, K379E; Devo 202, S4P, K97R; Devo 206, K340M; Devo 350, K297E; Devo 303, K297E, L380S; and Devo 355, K297E, Y317C) (Fig. 2). The role of charged residues in the binding of charged effectors is intuitive, yet the location of key residues in AGPase has been slow to be elucidated because of a lack of three-dimensional structural data. Mutagenesis studies, such as the present work, add valuable information to a growing understanding of the evolution of the allosteric behavior of AGPase. Structural data may allow further understanding and ultimately contribute to efforts in metabolic engineering of AGPase and starch accumulation in crops, although at present the directed molecular evolution of complex enzymatic traits offers the best alternative.
Acknowledgments
We thank Michael Kahn for helpful discussion. This study, which falls under the auspices of the NC-142 project, was supported by Department of Energy Grant DE-FG03-96ER20216. P.R.S. was supported in part by a National Institutes of Health Protein Biotechnology Traineeship. P.R.S. and I.H.K. are recipients of the Loyal Davis Fellowship.
Abbreviations
- PEP
phosphoenolpyruvate
- WT
wild type
- SS
small subunit
- Fru-6-P
fructose-6-phosphate
- Fru-1,6-bisP2
fructose-1,6-bisphosphate
- 3-PGA
3-phosphoglyceric acid
References
- 1.Preiss J. Annu Rev Microbiol. 1984;38:419–458. doi: 10.1146/annurev.mi.38.100184.002223. [DOI] [PubMed] [Google Scholar]
- 2.Smith-White B J, Preiss J. J Mol Evol. 1992;34:449–464. doi: 10.1007/BF00162999. [DOI] [PubMed] [Google Scholar]
- 3.Nakata P A, Greene T W, Anderson J M, Smith-White B J, Okita T W, Preiss J. Plant Mol Biol. 1991;17:1089–1093. doi: 10.1007/BF00037149. [DOI] [PubMed] [Google Scholar]
- 4.Ballicora M A, Laughlin M J, Fu Y, Okita T W, Barry G F, Preiss J. Plant Physiol. 1995;109:245–251. doi: 10.1104/pp.109.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laughlin M J, Chantler S E, Okita T W. Plant J. 1998;14:159–168. doi: 10.1046/j.1365-313x.1998.00102.x. [DOI] [PubMed] [Google Scholar]
- 6.Kuchner O, Arnold F H. Trends Biotechnol. 1997;15:523–530. doi: 10.1016/S0167-7799(97)01138-4. [DOI] [PubMed] [Google Scholar]
- 7.Petrounia I P, Arnold F H. Curr Opin Biotechnol. 2000;11:325–330. doi: 10.1016/s0958-1669(00)00107-5. [DOI] [PubMed] [Google Scholar]
- 8.Arnold F H, Giver L, Gershenson A, Zhao H, Miyazaki K. Ann NY Acad Sci. 1999;70:400–403. doi: 10.1111/j.1749-6632.1999.tb08913.x. [DOI] [PubMed] [Google Scholar]
- 9.Stemmer W P. Proc Natl Acad Sci USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stemmer W P. Nature (London) 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 11.Salamone P R, Greene T W, Kavakli I H, Okita T W. FEBS Lett. 2000;482:113–118. doi: 10.1016/s0014-5793(00)01985-2. [DOI] [PubMed] [Google Scholar]
- 12.Lorimer I A, Pastan I. Nucleic Acids Res. 1995;23:3067–3068. doi: 10.1093/nar/23.15.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Govons S, Vinopal R, Ingraham J, Preiss J. J Bacteriol. 1969;97:970–972. doi: 10.1128/jb.97.2.970-972.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morell M K, Bloom M, Knowles V, Preiss J. Plant Physiol. 1987;85:182–187. doi: 10.1104/pp.85.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okita T W, Nakata P A, Anderson J M, Sowokinos J, Morell M, Preiss J. Plant Physiol. 1990;93:785–790. doi: 10.1104/pp.93.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh H P, Preiss J. J Biol Chem. 1966;241:4491–4504. [PubMed] [Google Scholar]
- 17.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 18.Hwang S O. Biotechnol Techniques. 1996;10:157–160. [Google Scholar]
- 19.Mas J, Pedros-Alio C, Guerrero R. J Bacteriol. 1985;164:749–756. doi: 10.1128/jb.164.2.749-756.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mas J, Pedros-Alio C, Guerrero R. FEMS Microbiol Lett. 1989;57:231–236. doi: 10.1111/j.1574-6968.1989.tb03305.x. [DOI] [PubMed] [Google Scholar]
- 21.Baldwin W W, Myer R, Powell N, Anderson E, Koch A. Arch Microbiol. 1995;164:155–157. doi: 10.1007/s002030050248. [DOI] [PubMed] [Google Scholar]
- 22.Baldwin W W, Sheu M J, Bankston P W, Woldringh C. J Bacteriol. 1988;170:452–455. doi: 10.1128/jb.170.1.452-455.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greene T W, Kavakli I H, Kahn M L, Okita T W. Proc Natl Acad Sci USA. 1998;95:10322–10327. doi: 10.1073/pnas.95.17.10322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giroux M J, Shaw J, Barry G, Cobb B G, Greene T, Okita T, Hannah L C. Proc Natl Acad Sci USA. 1996;93:5824–5829. doi: 10.1073/pnas.93.12.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hnilo J, Okita T W. Plant Cell Physiol. 1989;30:1007–1010. [Google Scholar]
- 26.Stark D M, Timmerman K P, Barry G F, Preiss J, Kishore G M. Science. 1992;258:287–292. doi: 10.1126/science.258.5080.287. [DOI] [PubMed] [Google Scholar]
- 27.Kleczkowski L A, Villand P, Luthi E, Olsen O-A, Preiss J. Plant Physiol. 1993;101:179–186. doi: 10.1104/pp.101.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olive M, Ellis R J, Schuch W W. Plant Mol Biol. 1989;12:525–538. doi: 10.1007/BF00036967. [DOI] [PubMed] [Google Scholar]
- 29.Denyer K, Dunlap F, Thorbjornsen T, Keeling P, Smith A M. Plant Physiol. 1996;112:779–785. doi: 10.1104/pp.112.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorbjornsen T, Villand P, Denyer K, Olsen O-A, Smith A M. Plant J. 1996;10:243–250. [Google Scholar]
- 31.Plaxton W C, Preiss J. Plant Physiol. 1987;83:105–112. doi: 10.1104/pp.83.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ballicora M A, Fu Y, Nesbitt N M, Preiss J. Plant Physiol. 1998;118:265–274. doi: 10.1104/pp.118.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]