Abstract
In the unicellular green alga Chlamydomonas reinhardtii, the epigenetic silencing of transgenes occurs, as in land plants, at both the transcriptional and posttranscriptional levels. In the case of single-copy transgenes, transcriptional silencing takes place without detectable cytosine methylation of the introduced DNA. We have isolated two mutant strains, Mut-9 and Mut-11, that reactivate expression of a transcriptionally silenced single-copy transgene. These suppressors are deficient in the repression of a DNA transposon and a retrotransposon-like element. In addition, the mutants show enhanced sensitivity to DNA-damaging agents, particularly radiomimetic chemicals inducing DNA double-strand breaks. All of these phenotypes are much more prominent in a double mutant strain. These observations suggest that multiple partly redundant epigenetic mechanisms are involved in the repression of transgenes and transposons in eukaryotes, presumably as components of a system that evolved to preserve genomic stability. Our results also raise the possibility of mechanistic connections between epigenetic transcriptional silencing and DNA double-strand break repair.
Epigenetic processes, which result in heritable changes in gene expression without modifications in DNA sequence, play important roles in the control of development as well as in the cellular responses to viruses, viroids, transposable elements, and transgenes (1–5). In plants, fungi, and animals, analyses of transgene expression have revealed a wide range of epigenetic silencing processes and are providing new insights into mechanisms of gene regulation. Depending on the level at which silencing occurs, two types of phenomena have been distinguished: transcriptional gene silencing (TGS) and posttranscriptional gene silencing (PTGS) (1, 4–7). In addition, the introduction of double-stranded RNA triggers a process similar to PTGS, called RNA interference, in a variety of protozoa, invertebrate, and vertebrate species (8–10). In Chlamydomonas reinhardtii, transgenes are silenced by epigenetic phenomena similar to those in land plants (11–13).
PTGS involves sequence specific degradation of RNAs. Several genes required for RNA interference (RNAi) or related posttranscriptional processes, such as quelling, have been isolated in animal and fungal systems (4, 9, 10, 14–16). Thus far, four genes have been implicated in PTGS in Arabidopsis thaliana: SDE1/SGS2, encoding an RNA-dependent RNA polymerase; AGO1, encoding a protein similar to rabbit eIF2C; SDE3, encoding an RNA helicase; and SGS3 encoding a coiled-coil protein of unknown function (4, 7, 9, 10, 17). In Chlamydomonas, we have recently described a DEAH-box RNA helicase that functions in the posttranscriptional silencing of transgenes and transposons (13). Although the molecular mechanism(s) of RNAi/PTGS is not fully understood, recent evidence indicates that double-stranded RNA, generated by alternative pathways, is processed to 21- to 25-nt RNAs by an RNase-III-related protein (4, 7, 10, 15, 16). These small RNAs target the cleavage of homologous transcripts through an RNA-directed ribonuclease, a multisubunit complex named RNA-induced silencing complex in Drosophila (4, 7, 10, 15, 16).
TGS involves transcriptional repression. In plants, it is usually associated with cytosine methylation of promoter regions and reduced accessibility to DNase I, suggesting an altered chromatin structure (1, 5, 6, 18). Many transcriptionally silenced transgenes have complex structures, such as arrays of rearranged copies integrated at a single genomic site (5, 6). In a phenomenon resembling paramutation, some of these loci can also silence homologous sequences in trans (5, 6). DNA–DNA interactions have long been postulated to trigger this homology-dependent process (1, 18–20). However, double-stranded RNA derived from promoter regions has recently been implicated in the transcriptional inactivation of homologous sequences in ectopic positions (4, 21–23). These findings also raise the possibility of mechanistic connections between PTGS and TGS (18, 21–24).
Several genes required for TGS of transgenes have been identified in Arabidopsis. DDM1 (Decrease in DNA Methylation) encodes a chromatin-remodeling protein belonging to the SWI2/SNF2 superfamily, which affects both genomic DNA methylation and TGS (25). MOM1 (Morpheus' Molecule) encodes a nuclear protein that releases TGS without changes in transgene methylation (26). These genes also control some transposable elements (3, 27–29). Histone deacetylases and DNA methyltransferases also play a role in the epigenetic regulation of (trans)gene expression in Arabidopsis (2, 22, 24, 30, 31). In Drosophila melanogaster and Caenorhabditis elegans, the transcriptional silencing of repeated transgenes depends on Polycomb Group (PcG) genes, initially defined by their function in the repression of developmental genes (32, 33). However, the role of plant PcG homologs in TGS is currently unknown (3).
In Chlamydomonas and other volvocine algae, as in land plants, silenced multiple-copy transgenes exhibit high levels of DNA methylation (12, 34). In contrast, single-copy transgenes are subject to TGS without detectable cytosine methylation (12). The molecular mechanism(s) of TGS for simple single-copy transgenes has not been examined extensively in higher plants (5, 6, 35, 36). However, some transgenic loci in Arabidopsis remain transcriptionally silent despite a drastic reduction in DNA methylation caused by the depletion of methyltransferase 1 (5, 18, 24, 37). Further, in a recent study of silencing of a neomycin phosphotransferase transgene in Arabidopsis, single-copy transgenes did not show methylation of a diagnostic SacII promoter site that was partially or completely methylated in all examined multiple-copy lines (36). As previously proposed, these observations suggest that TGS in photosynthetic eukaryotes can also operate through a methylation-independent pathway (12).
To identify the genetic determinants of TGS for single-copy transgenes, we have isolated Chlamydomonas mutants deficient in this process. We report here the characterization of two mutant strains, Mut-9 and Mut-11, that reactivate transgenic expression. In addition, the suppressors are defective in the regulation of transposable elements. Interestingly, these Chlamydomonas mutants are also very sensitive to DNA-damaging agents causing DNA double-strand breaks (DSBs). Emerging evidence in a variety of eukaryotes suggests that repair of DSBs is associated with chromatin modifications (38, 39). We speculate that the proteins disrupted in Mut-9 and Mut-11 likely function in the formation of a distinct chromatin structure that is required for transcriptional repression and, possibly, DSB repair.
Materials and Methods
Culture Conditions, Strains, and Genetic Screen for Suppressors of Transgenic Silencing.
Unless noted otherwise, C. reinhardtii cells were grown photoheterotrophically in Tris-acetate-phosphate (TAP) medium (40) as previously described (11, 12). Strain 11-P[300] was generated by transformation of the wild-type strain CC-124 and contains a transcriptionally silenced single copy of the RbcS2∷aadA∷RbcS2 transgene (11, 12). To identify suppressors of transgene silencing, we mutagenized 11-P[300] by transformation with a mutant form of protoporphyrinogen oxidase (rs-3 gene) (41), conferring resistance to diphenyl ether herbicides. Herbicide-resistant transformants, containing the rs-3 gene integrated at random into the nuclear genome, were tested for their ability to grow in the presence of spectinomycin as an indication of reactivation of expression of the aadA transgene.
Genetic Analyses.
We isolated two spectinomycin-resistant mutant strains, Mut-9 and Mut-11. To test whether the insertional mutagen (rs-3 gene) cosegregated with reactivation of transgenic expression, Mut-9 and Mut-11 were crossed to the wild-type strain of opposite mating type, CC-125, and tetrads were dissected as previously described (40). Meiotic tetrad products of each mutant, containing exclusively the rs-3 plasmid, were then backcrossed to 11-P[300]. Tetrad products of Mut-9 and Mut-11 were also crossed to each other to generate a double mutant (Mut-9 Mut-11). Expression of the RbcS2∷aadA∷RbcS2 transgene in the tetrad progeny was evaluated by spot tests on medium containing spectinomycin. Five-microliter aliquots of appropriately diluted cells were pipetted onto the plates and incubated as previously described (12). The presence of the transgene and/or the rs-3 plasmid in the genome was examined by Southern blot analyses.
DNA and RNA Analyses.
Total cell DNA was isolated, fractionated by agarose gel electrophoresis, transferred to a nylon membrane, and hybridized as previously described (11, 42). Total cell RNA was purified with the RNeasy Plant Mini Kit (Qiagen, Chatsworth, CA), and standard techniques were used for fractionation by formaldehyde–agarose gel electrophoresis, blotting, and hybridization (11, 42). The PhosphorImager System (Molecular Dynamics) was used for quantitation of 32P radioactivity.
Transposon Mobilization Analyses.
To test the effect of the mutations on the activity of Chlamydomonas transposons, we established parallel subcultures of strains 11-P[300], Mut-9, Mut-11, and Mut-9 Mut-11. Cells were plated at low density to obtain individual colonies. Ten independent colonies from each strain were subcultured by transfer to fresh TAP plates every 2 weeks. After 3 months, we isolated total cell DNA from the subclones and evaluated the mobilization of transposable elements by Southern blot analyses. Genomic DNA was digested with restriction enzymes that cut inside each transposon (conserved site) and in a flanking chromosomal region (polymorphic site depending on the place of insertion) and probed with short DNA sequences that hybridize to the transposon termini.
Growth Rate and Cell Survival on Exposure to DNA-Damaging Agents.
Cells were grown photoheterotrophically in TAP medium under continuous light (300 μmol·m−2·s−1 photosynthetically active radiation) at 23°C. To determine growth rates, cells in middle logarithmic growth phase were inoculated into fresh TAP medium to a density of 1 × 105 cells/ml. The cells were then cultured under the same conditions and cell densities determined by measuring optical absorbance at 750 nm. For treatment with DNA-damaging agents, cells were grown to logarithmic phase and spread to a density of 500–700 cells per plate. To test for sensitivity to UV light below 280 nm (UV-C), cells spread on minimal HS medium (40) were irradiated with a Stratalinker (Stratagene). After 24 h in the dark, to prevent photoreactivation, the plates were incubated under moderate light (50 μmol·m−2·s−1 photosynthetically active radiation) at 23°C for 10–14 days before the surviving colonies were counted. For treatments with bleomycin (Zeocin; Invitrogen) and methyl methanesulfonate (Sigma), cells were spread on TAP plates containing the appropriate concentrations of each genotoxic agent and incubated as described above.
Results
Isolation of Chlamydomonas Mutants Defective in Transcriptional Transgene Silencing.
To identify genes responsible for epigenetic silencing in C. reinhardtii, we carried out random insertional mutagenesis on strain 11-P[300], which contains a transcriptionally silenced single copy of the RbcS2∷aadA∷RbcS2 transgene (12). This transgene consists of the coding sequence of the eubacterial aadA gene (conferring spectinomycin resistance) under the control of the 5′ and 3′ regulatory regions of the endogenous RbcS2 gene (encoding the small subunit of Rubisco) (11). Because Chlamydomonas is haploid, nonlethal mutations in genes required for silencing allow reactivation of expression of aadA and cell survival on media containing spectinomycin.
Cells from 11-P[300] were transformed with the rs-3 gene, which encodes a mutated form of Chlamydomonas protoporphyrinogen oxidase conferring resistance to diphenyl ether herbicides (41). Herbicide-resistant transformants were recovered and tested for their ability to grow on media containing different concentrations of spectinomycin. By using this approach, we isolated two mutant strains (Mut-9 and Mut-11) that showed reactivation of the chimeric aadA transgene (Fig. 1). In Mut-9 and Mut-11, the rs-3 gene integrated into different genomic locations, providing a molecular tag to identify either mutant or a double mutant by Southern blot analysis (Fig. 1A). Blots of total cell DNA hybridized to the pBluescript vector backbone, common to plasmids containing the aadA or rs-3 genes, showed a 4.5-kb HindIII fragment corresponding to the RbcS2∷aadA∷RbcS2 transgene and ≈13 kb and ≈20 kb segments corresponding to the rs-3 inserts in Mut-9 and Mut-11, respectively (Fig. 1A). Tetrad analyses confirmed that the introduced rs-3 marker cosegregated with reactivation of expression of the RbcS2∷aadA∷RbcS2 transgene (data not shown).
Figure 1.
Expression of the RbcS2∷aadA∷RbcS2 transgene is reactivated in the mutant strains. (A) Southern blot analysis of the wild-type untransformed strain (CC-124), the silenced parental strain (11-P[300]), the mutant strains (Mut-9 and Mut-11), and a double mutant strain (Mut-9 Mut-11). Total cell DNA was digested with HindIII and hybridized to the pBluescript vector backbone, which is common to the plasmids containing the chimeric aadA transgene or the tagging rs-3 gene. The fragments corresponding to the transgene (aadA) or the insertional mutagen (rs-3) are indicated. (B) Northern blot analysis of the strains described above. Total cell RNA was isolated from each strain, separated under denaturing conditions, and hybridized to the aadA coding sequence (Upper). The same blot was reprobed with the coding sequence of RbcS2 as a control for equal loading of the lanes (Lower). The faint transcript seen above RbcS2 corresponds to the RbcS1 gene (11). (C) Growth and survival on TAP medium or on TAP medium containing spectinomycin (TAP + SPEC) of the indicated strains. Five-fold serial dilutions of cells, starting with 1 × 105 cells on the left, were spotted on each plate and incubated for 15 days (12).
Expression of the RbcS2∷aadA∷RbcS2 Transgene.
We examined the expression of the chimeric aadA transgene by Northern blot analysis and by cell survival on medium containing 100 μg/ml of spectinomycin. Hybridization to the aadA coding sequence was observed in Mut-9, Mut-11, and in a double mutant, Mut-9 Mut-11, but was undetectable in the silenced strain 11-P[300] and in the untransformed wild-type strain CC-124 (Fig. 1B). As previously reported (11), the RbcS2∷aadA∷RbcS2 transcripts showed several discrete bands superimposed on a smear of hybridizing RNA, presumably because of improper mRNA processing. As a control for equal loading of the lanes, the same blot was rehybridized with a probe specific for RbcS2 (Fig. 1B). Consistent with the steady-state levels of RbcS2∷aadA∷RbcS2 transcripts, Mut-9, Mut-11, and Mut-9 Mut-11 were able to grow in the presence of spectinomycin, whereas 11-P[300] and CC-124 could not survive (Fig. 1C). In addition, the double mutant showed greater aadA RNA levels (Fig. 1B) as well as greater resistance to spectinomycin (Fig. 1C) than either of the individual mutants. These results indicate that integration of the rs-3 marker disrupted two genes, designated Mut9 and Mut11, required for epigenetic silencing of the RbcS2∷aadA∷RbcS2 transgene.
Reactivation of Transposable Elements.
We also analyzed whether the mutations affected mobilization of Chlamydomonas transposons. Transposable elements are grouped into two major classes depending on their mode of transposition. Class I elements transpose via an RNA intermediate and include retrotransposons and other retroelements, such as Chlamydomonas TOC1 (43). In contrast, Class II elements transpose via a DNA intermediate by a “cut-and-paste” mechanism and include Chlamydomonas Gulliver (44).
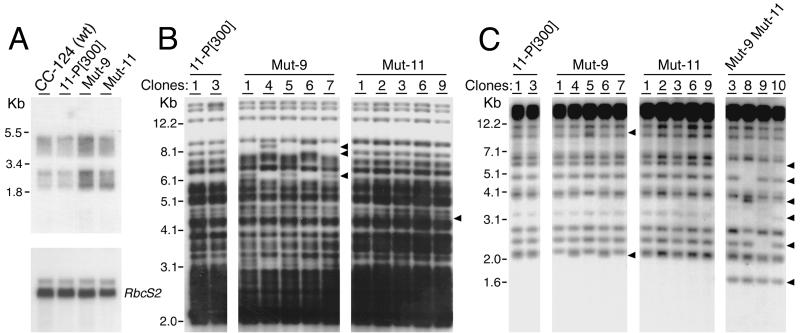
As previously reported (13, 43), the majority of TOC1 transcripts are nonpolyadenylated and heterogeneous in size, which produces a smeary signal on Northern blots of total RNA (Fig. 2A). The steady-state level of TOC1 RNA is about 2.5-fold higher in Mut-9 compared with the parental strain 11-P[300] (Fig. 2A). TOC1 transcripts are also somewhat elevated in Mut-11. Accordingly, the transposition frequency of TOC1 is significantly enhanced in Mut-9 but only slightly affected in Mut-11. Southern blot analyses of 10 parallel subcultures of 11-P[300], Mut-9, and Mut-11 revealed additional TOC1 copies in the genome of many Mut-9 subclones (Fig. 2B and data not shown). In contrast, the Mut-11 subcultures displayed very few changes in the copies of TOC1 (Fig. 2B, Mut-11 subclones 3 and 6), and the 11-P[300] subcultures showed no detectable transposition.
Figure 2.
Reactivation of a retroelement, TOC1, and a DNA transposon, Gulliver, in the mutant strains. Abbreviations are as in the legend to Fig. 1. (A) Northern blot of total RNA probed sequentially for TOC1 (Upper) to examine transcript levels and for RbcS2 (Lower) to test for equal loading of the lanes. (B) Southern blot analysis of TOC1 transposition. Genomic DNA from parallel subcultures (Clones) of the indicated strains was digested with HincII and probed for TOC1. The arrowheads indicate new fragments in the subclones of Mut-9 and Mut-11. (C) Southern blot analysis of Gulliver transposition. Total cell DNA from parallel subcultures (Clones) of the indicated strains was digested with HindIII and probed for Gulliver. The arrowheads indicate missing or new fragments in the subclones of Mut-9 and Mut-9 Mut-11. Although only two subclones are shown for 11-P[300], we did not detect mobilization of either TOC1 or Gulliver in 10 parallel subcultures grown under the same conditions as the mutant strains.
We also examined the mobilization of a Class II transposable element, Gulliver (Fig. 2C). Total cell DNA from 10 parallel subcultures of each strain was digested with HindIII and hybridized with a terminal repeat sequence of Gulliver. Whereas 11-P[300] and Mut-11 showed no changes in the banding pattern of Gulliver, a few subcultures of Mut-9 displayed differences indicative of transposon mobilization (Fig. 2C, Mut-9 subclones 1, 5, and 7). However, the changes were most dramatic in the double mutant Mut-9 Mut-11, where many subcultures showed missing fragments (indicating excision from the genome) as well as new fragments (indicating integration into other genomic locations) (Fig. 2C). These observations suggest that Mut9 and Mut11, in addition to their role in the epigenetic silencing of transgenes, participate in the suppression of transposable elements.
Photoheterotrophic Cell Growth.
Because Mut-9 and Mut-11 were deficient in both transgene and transposon silencing, we tested for defects in other biological processes that might indicate additional roles of the mutated gene products on global gene regulation. To determine growth rates, cells pregrown to logarithmic phase were inoculated at low density into fresh medium and cultured under the same conditions. Cell densities were measured at fixed intervals. The growth rate of Mut-9 was similar to that of the wild-type strain CC-124 under standard photoheterotrophic conditions (Fig. 3). In contrast, Mut-11 and the double mutant grew at a slower rate. In the exponential phase of growth, all mutants had doubling times similar to that of the wild-type strain (Fig. 3). However, Mut-11 and the double mutant showed a much longer lag phase. Thus, Mut-11 seems to be defective in the initial survival and/or adaptation to grow at low density in new medium, suggesting that Mut11 might regulate a physiological adaptive response(s).
Figure 3.
Photoheterotrophic growth of the mutant and wild-type strains. Abbreviations are as in the legend to Fig. 1. Each time point represents the mean (± standard error) of six replicates (three independent experiments). Where the error bars are not visible, they are smaller than the symbols. The exponential phase of the growth curve was used to calculate doubling times. Even though all strains show similar doubling times, Mut-11 and Mut-9 Mut-11 took considerably longer to reach exponential growth (represented by a linear increase in optical density in the semilogarithmic scale).
Sensitivity to DNA-Damaging Agents.
Because of the possible connections between DNA repair and chromosomal mechanisms of epigenetic regulation (38, 39, 45–50), we also examined the response of the mutants to several genotoxic agents. Mut-9 and Mut-11 were particularly sensitive to chemical agents that induce DSBs (51), such as methyl methanesulfonate or bleomycin (Fig. 4 A and C). The dose resulting in 30% cell survival (Fig. 4, horizontal dashed lines) was significantly lower for each mutant compared with the wild-type strain CC-124. Moreover, the double mutant was much more sensitive to these treatments than each of the single mutants. In contrast, Mut-11 was as resistant as the wild-type strain to UV-C irradiation (<280 nm) (Fig. 4B), a treatment that mainly causes formation of cyclobutane pyrimidine dimers (51). Similarly, Mut-9 and Mut-9 Mut-11 showed only a moderate defect in survival on exposure to low doses of UV-C light, although they were clearly sensitive at higher doses. Mut-9 also displayed a greater than 10-fold reduction in the frequency of transformation with exogenous DNA, when compared with the parental strain 11-P[300] (see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). These observations are consistent with a deficiency in the integration of transforming DNA into the nuclear genome, presumably because of defective DSB repair.
Figure 4.
Effect of DNA-damaging agents on the survival of the mutant and wild-type strains. Each graph point represents the mean (± standard error) of nine replicates (three independent experiments). Where the error bars are not visible, they are smaller than the symbols. The dashed horizontal lines indicate 30% cell survival. Symbols: , wild-type CC-124; ●, Mut-9; Δ, Mut-11; ♦, Mut-9 Mut-11. (A) Survival of the mutants and wild-type C. reinhardtii grown on TAP medium containing increasing concentrations of bleomycin. (B) Survival of the mutants and wild-type C. reinhardtii exposed to increasing doses of UV-C irradiation under nonphotoreactivating conditions. (C) Survival of the mutants and wild-type C. reinhardtii grown on TAP medium containing increasing concentrations of methyl methanesulfonate (MMS).
Discussion
Epigenetic Silencing of Transgenes.
In C. reinhardtii, nuclear run-on assays with isolated nuclei and Northern blot analyses have revealed that transgene inactivation occurs at both transcriptional and posttranscriptional levels (12, 13). We describe here the characterization of two Chlamydomonas mutants, Mut-9 and Mut-11, defective in the epigenetic silencing of transgenes. The strain used to isolate these suppressors, 11-P[300], contains a single copy of the RbcS2∷aadA∷RbcS2 transgene that is silenced at the transcriptional level without detectable cytosine methylation (12). Mut-9 and Mut-11 reactivate expression of this chimeric aadA transgene, as shown by Northern blot analyses and by the ability of the mutant cells to survive on spectinomycin-containing medium. Moreover, a double mutant (Mut-9 Mut-11) exhibited more pronounced transgene reactivation than each of the single mutants. Nuclear run-on assays confirmed that the RbcS2∷aadA∷RbcS2 transgene becomes transcriptionally active in the mutant backgrounds (data not shown). Thus, Mut9 and Mut11 are required for the transcriptional silencing of transgenes in Chlamydomonas.
Epigenetic Silencing of Transposons.
Transposable elements are widespread constituents of all eukaryotic genomes (52). Epigenetic processes, particularly DNA methylation, have been implicated in regulating the activity of plant transposable elements (53). In Arabidopsis, Robertson's Mutator transposons and members of the CACTA superfamily are controlled by the SWI2/SNF2 chromatin-remodeling gene DDM1 (28, 29). DDM1 also has a slight effect on endogenous retrotransposon mobilization (27, 54). A chromomethylase (encoded by CMT3), which is required for maintenance of CpXpG methylation, has also been shown to participate in the silencing of retrotransposons (30). Moreover, a truncated Athila (a putative retrotransposon) transcript was induced in several Arabidopsis mutants defective in TGS (27).
The effect(s) of posttranscriptional gene silencing mechanisms on transposon mobilization in plants has not been reported. However, in C. elegans and D. melanogaster, transposon and/or retrotransposon mobilization is regulated by RNA interference/PTGS processes (8, 9, 55, 56). Similarly, we have previously reported that a Chlamydomonas mutant defective in PTGS shows enhanced transpositional activity of both TOC1 and Gulliver (13). We have now found that mutations affecting TGS also enable mobilization of transposable elements in Chlamydomonas. The steady-state RNA level of the TOC1 retrotransposon, as well as its transposition frequency, is enhanced in Mut-9 compared with the parental strain 11-P[300]. The transpositional activity of the DNA element Gulliver is also slightly increased in Mut-9. Mut-11 shows some mobilization of TOC1, but the activity of Gulliver is not affected. Interestingly, in the double mutant, Gulliver seems to transpose at a much higher frequency than in either of the single mutants. Taken together, our results suggest that Chlamydomonas transposable elements are regulated by multiple epigenetic mechanisms operating at both transcriptional and posttranscriptional levels. Furthermore, there is also redundant repression at the transcriptional level because, as discussed below, Mut9 and Mut11 appear to play a role(s) in partly different pathways. Likewise, the I factor retrotransposon in D. melanogaster appears to be controlled by various epigenetic processes (55, 57). Thus, multiple epigenetic mechanisms might operate as a defense system against the massive expansion of transposable elements in eukaryotes.
Molecular Mechanism(s) of Epigenetic Transcriptional Silencing.
Molecular characterization of the suppressor strains provided insights into the silencing mechanism(s). Because of deletions caused by integration of the rs-3 plasmid, the mutations in Mut-9 and Mut-11 result in complete loss-of-function null phenotypes (data not shown). Since a double mutant shows greater transgenic reactivation and greater transposon mobilization than either of the single mutants, epistatic analysis suggests that Mut9 and Mut11 function in (at least) partly distinct pathways of transcriptional repression. This explanation is also supported by the differences in the phenotypes of Mut-9 and Mut-11.
Phenotypic characterization of the parental strain 11-P[300] suggested the involvement of chromatin domains in transcriptional silencing of unmethylated transgenes (12). Consistent with this interpretation, the Mut9 and Mut11 gene products (Mut9p and Mut11p) might function in the formation of a repressive chromatin structure that leads to the transcriptional inactivation of transgenes and transposons. Mut11 encodes a WD40-repeat containing protein (GenBank accession no. AF443204) with homology to the C-terminal domain of Saccharomyces cerevisiae Tup1p, a global transcriptional repressor (58–60). Yeast Tup1p interacts with many proteins, including components of the basal transcription machinery, histones, and histone deacetylases, and has been suggested to play an architectural role in organizing repressive chromatin domains (58–60). By analogy, Mut11p may also have a structural function and/or interfere directly with transcription factors. Mut9 encodes a novel serine/threonine protein kinase (GenBank accession no. AF443205). In D. melanogaster, phosphorylation of Heterochromatin Protein 1 is correlated with heterochromatin assembly and silencing (61). Perhaps in similar fashion, Mut9p may modulate chromatin structure by phosphorylation of one or more of its components.
Transcriptional Silencing, DNA Repair, and Cell Growth.
Although Mut-9 and Mut-11 were isolated on the basis of their ability to reactivate expression of the aadA transgene, they also show enhanced sensitivity to DNA-damaging agents, particularly radiomimetic chemicals inducing DSBs. In addition, Mut-9 displays a greater than 10-fold reduction in the frequency of transformation with foreign DNA, presumably because of a deficiency in the integration of transforming DNA into the nuclear genome. As discussed below, these results are consistent with a role of Mut9 and Mut11 in the repair of DSBs. It should be noted, however, that increased sensitivity to DNA-damaging agents could also result from overall defects in genome stability and cell survival. However, this explanation seems less likely because the mutants are only moderately sensitive to UV-C irradiation, and Mut-9 is not affected in cellular growth.
DSBs are a common form of DNA damage in proliferating cells, and their repair is a fundamental mechanism of genome protection (62). Observations in a variety of eukaryotic organisms suggest that the repair of DSBs is associated with chromatin modifications (39). After exposure to ionizing radiation, a member of the histone H2A family becomes rapidly phosphorylated in domains around the damaged sites (38, 39). In S. cerevisiae, DSBs cause the relocalization of SIR (Silent Information Regulator) proteins from telomeres, where they are responsible for epigenetic silencing, to the site of damage (45, 46). The ATM (Ataxia Telangiectasia Mutated) and ATR (ATM and Rad3 Related) protein kinases, which have been implicated in the response of mammalian cells to multiple forms of DNA damage, are present in complexes with histone deacetylases and/or chromatin remodeling factors (47, 48). The ATM-associated deacetylase activity increases on cellular exposure to ionizing radiation (47). Moreover, factors involved in DNA repair, DNA replication, and chromatin assembly, such as CAF-1 (Chromatin Assembly Factor 1) and PCNA (Proliferating Cell Nuclear Antigen), have been implicated in a marking system for the inheritance of epigenetic states (49, 50). Even though these processes have not been examined in detail in plants, it is intriguing that mutagenesis treatments, such as exposure to ethyl methanesulfonate or irradiation, occasionally lead to the formation of silenced epi-alleles in genes regulating plant development (3, 63). The latter observations and our results suggest that the connections between DNA repair and epigenetic chromosomal mechanisms may also extend to the plant kingdom.
In Chlamydomonas, Mut9p and Mut11p might play a role in establishing the proper chromatin environment for DNA repair. Because Mut9p is a protein kinase, it might also participate in the signaling response to DNA damage. Indeed, many proteins directly involved in DSB repair or cell-cycle checkpoints are regulated by phosphorylation (62, 64). Another explanation for the mutant phenotypes is that Mut9p and Mut11p might control the expression of genes required for DNA repair, perhaps indirectly, as reported in yeast for SIR regulation of the nonhomologous end-joining repair pathway through mating type factors (65).
Besides sensitivity to DNA-damaging agents, Mut-11 also shows defects in growth when cells are inoculated into fresh medium at low density. S. cerevisiae Tup1p is required for the repression of multiple families of genes, including those responsive to different physiological conditions such as osmotic stress and hypoxia (58, 60). By analogy, Mut11p might participate in the regulation of genes involved in a physiological adaptive response(s).
In summary, Chlamydomonas Mut-9 and Mut-11 are defective in the transcriptional silencing of transgenes, the repression of transposable elements, the tolerance of DNA damage (particularly DSBs), and, in the case of Mut-11, cell growth. The simplest explanation for these pleiotropic phenotypes is that Mut9p and Mut11p are involved in the formation of a distinct chromatin structure that is required, directly or indirectly, for repression of transgenes and transposons, for controlling endogenous gene expression, and possibly for repairing DNA damage.
Supplementary Material
Acknowledgments
We thank J. Kovar and D. Weeks for kindly donating plasmid pJK7 and various lab members for critical reading of the manuscript. This work was supported by funds to H.C. from the National Science Foundation (MCB-9808473) and from the Nebraska Research Initiative.
Abbreviations
- TGS
transcriptional gene silencing
- PTGS
posttranscriptional gene silencing
- DSBs
DNA double-strand breaks
- TAP
Tris-acetate-phosphate
- UV-C
UV light <280 nm
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wolffe A P, Matzke M A. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 2.Finnegan E J, Peacock W J, Dennis E S. Curr Opin Genet Dev. 2000;10:217–223. doi: 10.1016/s0959-437x(00)00061-7. [DOI] [PubMed] [Google Scholar]
- 3.Habu Y, Kakutani T, Paszkowski J. Curr Opin Genet Dev. 2001;11:215–220. doi: 10.1016/s0959-437x(00)00182-9. [DOI] [PubMed] [Google Scholar]
- 4.Matzke M, Matzke A J M, Kooter J M. Science. 2001;293:1080–1083. doi: 10.1126/science.1063051. [DOI] [PubMed] [Google Scholar]
- 5.Vaucheret H, Fagard M. Trends Genet. 2001;17:29–35. doi: 10.1016/s0168-9525(00)02166-1. [DOI] [PubMed] [Google Scholar]
- 6.Chandler V L, Vaucheret H. Plant Physiol. 2001;125:145–148. doi: 10.1104/pp.125.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vance V, Vaucheret H. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- 8.Fire A. Trends Genet. 1999;15:358–363. doi: 10.1016/s0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- 9.Plasterk R H A, Ketting R F. Curr Opin Genet Dev. 2000;10:562–567. doi: 10.1016/s0959-437x(00)00128-3. [DOI] [PubMed] [Google Scholar]
- 10.Sharp P A. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- 11.Cerutti H, Johnson A M, Gillham N W, Boynton J E. Genetics. 1997;145:97–110. doi: 10.1093/genetics/145.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerutti H, Johnson A M, Gillham N W, Boynton J E. Plant Cell. 1997;9:925–945. doi: 10.1105/tpc.9.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu-Scharf D, Jeong B-r, Zhang C, Cerutti H. Science. 2000;290:1159–1162. doi: 10.1126/science.290.5494.1159. [DOI] [PubMed] [Google Scholar]
- 14.Cogoni C, Macino G. Curr Opin Genet Dev. 2000;10:638–643. doi: 10.1016/s0959-437x(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein E, Caudy A A, Hammond S M, Hannon G J. Nature (London) 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 16.Knight S W, Bass B L. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalmay T, Horsefield R, Braunstein T H, Baulcombe D C. EMBO J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paszkowski J, Whithman S A. Curr Opin Plant Biol. 2001;4:123–129. doi: 10.1016/s1369-5266(00)00147-3. [DOI] [PubMed] [Google Scholar]
- 19.Luff B, Pawlowski L, Bender J. Mol Cell. 1999;3:505–511. doi: 10.1016/s1097-2765(00)80478-5. [DOI] [PubMed] [Google Scholar]
- 20.Matzke M, Mette M F, Jakowitsch J, Kanno T, Moscone E A, van der Winden J, Matzke A J. Genetics. 2001;158:451–461. doi: 10.1093/genetics/158.1.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mette M F, Matzke A J M, Matzke M A. Curr Biol. 2001;11:1119–1123. doi: 10.1016/s0960-9822(01)00315-3. [DOI] [PubMed] [Google Scholar]
- 22.Jones L, Ratcliff F, Baulcombe D C. Curr Biol. 2001;11:747–757. doi: 10.1016/s0960-9822(01)00226-3. [DOI] [PubMed] [Google Scholar]
- 23.Sijen T, Vijn I, Rebocho A, van Blokland R, Roelofs D, Mol J N, Kooter J M. Curr Biol. 2001;11:436–440. doi: 10.1016/s0960-9822(01)00116-6. [DOI] [PubMed] [Google Scholar]
- 24.Morel J, Mourrain P, Beclin C, Vaucheret H. Curr Biol. 2000;10:1591–1594. doi: 10.1016/s0960-9822(00)00862-9. [DOI] [PubMed] [Google Scholar]
- 25.Jeddeloh J A, Stokes T L, Richards E J. Nat Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- 26.Amedeo P, Habu Y, Afsar K, Mittelsten Scheid O, Paszkowski J. Nature (London) 2000;405:203–206. doi: 10.1038/35012108. [DOI] [PubMed] [Google Scholar]
- 27.Steimer A, Amedeo P, Afsar K, Fransz P, Mittelsten Scheid O, Paszkowski J. Plant Cell. 2000;12:1165–1178. doi: 10.1105/tpc.12.7.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singer T, Yordan C, Martienssen R A. Genes Dev. 2001;15:591–602. doi: 10.1101/gad.193701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T. Nature (London) 2001;411:212–214. doi: 10.1038/35075612. [DOI] [PubMed] [Google Scholar]
- 30.Lindroth A M, Cao X, Jackson J P, Zilberman D, McCallum C M, Henikoff S, Jacobsen S E. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 31.Tian L, Chen Z J. Proc Natl Acad Sci USA. 2001;98:200–205. doi: 10.1073/pnas.011347998. . (First Published December 26, 2000; 10.1073/pnas.011347998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birchler J A, Pal Bhadra M, Bhadra U. Curr Opin Genet Dev. 2000;10:211–216. doi: 10.1016/s0959-437x(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 33.Hsieh J, Fire A. Annu Rev Genet. 2000;34:187–204. doi: 10.1146/annurev.genet.34.1.187. [DOI] [PubMed] [Google Scholar]
- 34.Babinger P, Kobl I, Mages W, Schmitt R. Nucleic Acids Res. 2001;15:1261–1271. doi: 10.1093/nar/29.6.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer P, Heidmann I, Niedenhof I. Plant J. 1993;4:89–100. doi: 10.1046/j.1365-313x.1993.04010089.x. [DOI] [PubMed] [Google Scholar]
- 36.Meza T J, Kamfjord D, Håkelien A-M, Evans I, Godager L H, Mandal A, Jakobsen K S, Aalen R B. Transgenic Res. 2001;10:53–67. doi: 10.1023/a:1008903026579. [DOI] [PubMed] [Google Scholar]
- 37.Mittelsten Scheid O, Afsar K, Paszkowski J. Proc Natl Acad Sci USA. 1998;95:632–637. doi: 10.1073/pnas.95.2.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paull T T, Rogakou E P, Yamazaki V, Kirchgessner C U, Gellert M, Bonner W M. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 39.Modesti M, Kanaar R. Curr Biol. 2001;11:R229–R232. doi: 10.1016/s0960-9822(01)00112-9. [DOI] [PubMed] [Google Scholar]
- 40.Harris E H. The Chlamydomonas Sourcebook. San Diego: Academic; 1989. [Google Scholar]
- 41.Randolph-Anderson B L, Sato R, Johnson A M, Harris E H, Hauser C R, Oeda K, Ishige F, Nishio S, Gillham N W, Boynton J E. Plant Mol Biol. 1998;38:839–859. doi: 10.1023/a:1006085026294. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 43.Day A, Rochaix J-D. J Mol Biol. 1991;218:273–291. doi: 10.1016/0022-2836(91)90712-f. [DOI] [PubMed] [Google Scholar]
- 44.Ferris P J. Genetics. 1989;122:363–377. doi: 10.1093/genetics/122.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mills K, Sinclair D, Guarente L. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- 46.Martin S, Laroche T, Suka N, Grunstein M, Gasser S M. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- 47.Kim G D, Choi Y H, Dimtchev A, Jeong S J, Dritschilo A, Jung M. J Biol Chem. 1999;274:31127–31130. doi: 10.1074/jbc.274.44.31127. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt D R, Schreiber S L. Biochemistry. 1999;38:14711–14717. doi: 10.1021/bi991614n. [DOI] [PubMed] [Google Scholar]
- 49.Ridgway P, Almouzni G. J Cell Sci. 2000;113:2647–2658. doi: 10.1242/jcs.113.15.2647. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Z, Shibahara K, Stillman B. Nature (London) 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- 51.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 52.SanMiguel P, Gaut B S, Tikhonov A, Nakajima Y, Bennetzen J L. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 53.Fedoroff N V. Genes Cell. 1999;4:11–19. doi: 10.1046/j.1365-2443.1999.00233.x. [DOI] [PubMed] [Google Scholar]
- 54.Hirochika H, Okamoto H, Kakutani T. Plant Cell. 2000;12:357–369. doi: 10.1105/tpc.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jensen S, Gassama M-P, Heidmann T. Nat Genet. 1999;21:209–212. doi: 10.1038/5997. [DOI] [PubMed] [Google Scholar]
- 56.Aravin A A, Naumova N M, Tulin A V, Vagin V V, Rozovsky Y M, Gvozdev V A. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 57.Chaboissier M-C, Bucheton A, Finnegan D J. Proc Natl Acad Sci USA. 1998;95:11781–11785. doi: 10.1073/pnas.95.20.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ducker C E, Simpson R T. EMBO J. 2000;19:400–409. doi: 10.1093/emboj/19.3.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fleming A B, Pennings S. EMBO J. 2001;18:5219–5231. doi: 10.1093/emboj/20.18.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith R L, Johnson A D. Trends Biochem Sci. 2000;25:325–330. doi: 10.1016/s0968-0004(00)01592-9. [DOI] [PubMed] [Google Scholar]
- 61.Zhao T, Heyduk T, Eissenberg J C. J Biol Chem. 2001;276:9512–9518. doi: 10.1074/jbc.M010098200. [DOI] [PubMed] [Google Scholar]
- 62.Zhou B-B S, Elledge S J. Nature (London) 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 63.Jacobsen S E, Meyerowitz E M. Science. 1997;277:1100–1103. doi: 10.1126/science.277.5329.1100. [DOI] [PubMed] [Google Scholar]
- 64.Karran P. Curr Opin Genet Dev. 2000;10:144–150. doi: 10.1016/s0959-437x(00)00069-1. [DOI] [PubMed] [Google Scholar]
- 65.Kegel A, Sjöstrand J O O, Åström S U. Curr Biol. 2001;11:1611–1617. doi: 10.1016/s0960-9822(01)00488-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.