Abstract
Genetic map length and gene number in eukaryotes vary considerably less than genome size, giving rise to the hypothesis that recombination is restricted to genes. The complex genome of maize contains a large fraction of repetitive DNA, composed principally of retrotransposons arranged in clusters. Here, we assess directly the contribution of retrotransposon clusters and genes to genetic length. We first measured recombination across adjacent homozygous genetic intervals on either side of the bronze (bz) locus. We then isolated and characterized two bacterial artificial chromosome clones containing those intervals. Recombination was almost 2 orders of magnitude higher in the distal side, which is gene-dense and lacks retrotransposons, than in the proximal side, which is gene-poor and contains a large cluster of methylated retrotransposons. We conclude that the repetitive retrotransposon DNA in maize, which constitutes the bulk of the genome, most likely contributes little if any to genetic length.
Although the amount of nuclear DNA in eukaryotes varies greatly, the total length of their genetic maps does not. This can be considered the genetic version of the C-value paradox, which states that the size of eukaryotic genomes bears little relationship to organismic complexity or number of genes (1). To account for the discrepancy between the sizes of genomes and genetic maps, Thuriaux hypothesized that meiotic recombination may be largely restricted to genes (2). Maize, with its large amount of repetitive DNA (3), is a particularly suitable organism for testing this hypothesis. One prediction of the hypothesis is that recombination within genes, expressed as the ratio of genetic to physical length (cM/kb), should be much higher than the genome's average (4). Data from intragenic recombination experiments support that prediction (5–9). Recombination within the bronze (bz) locus, for example, is at least 100 times higher than the maize genome's average (5). A second prediction of the hypothesis is that repetitive DNA should not contribute significantly to genetic length, i.e., recombination across repetitive DNA should be much lower than across genic DNA. Here we analyze how this prediction holds around the bz locus, one of the regions of the maize genome where recombination has been studied in greatest detail (5, 10).
Approximately 80% of the 2,500-megabase maize nuclear genome (11) is present in more than 100 copies (12). As in other plants with large genomes, much of this repetitive DNA consists of different families of retrotransposons (13–15). In maize, at least 50% of the repetitive DNA is made up of retrotransposon blocks dispersed throughout the gene-containing regions of the genome (16). We recently isolated a 240-kb bacterial artificial chromosome (BAC) contig centered around Bz-McC, one of the bz alleles used in the earlier intragenic recombination studies, and established that the bz gene is located in an unusually gene-dense region flanked at either end by retrotransposon blocks (17, 18). Thus, the bz genomic domain constitutes an excellent experimental system in which to assess directly the contribution of adjacent genic and retrotransposon regions to genetic length.
To determine the ratio between genetic and physical distance in the gene- and retrotransposon-rich regions flanking the bz gene, we have measured the physical lengths of defined genetic intervals in the vicinity of the bz locus. These intervals are marked by Ac (Activator) elements that transposed from a common bz progenitor allele to nearby sites. Because they are completely homozygous, potential effects of intervening sequence heterozygosity on recombination are eliminated. To correlate the ratio of genetic to physical length with the nature of the DNA in those intervals, we have characterized the DNA by sequencing and methylation analysis.
Materials and Methods
Genetic Stocks.
All the alleles used in this study were in the common genetic background of the inbred W22. The aleurone phenotypes conditioned by the various alleles in the presence of all the complementary factors for anthocyanin pigmentation are given in parentheses: Bz-McC (purple), the normal progenitor allele of the bz-m2(Ac) mutation; bz-m2(Ac) (purple spots on a bronze background), an allele arising from the insertion of the 4.6-kb Ac element at position 755–762 in the second exon of Bz-McC (refs. 19 and 20, GenBank accession number AF355378); bz-s derivatives (bronze), stable mutant derivatives of bz-m2(Ac) arisen from independent excisions of Ac (ref. 21; the mutations carry excision footprints at position 755–762 and a transposed Ac element in the vicinity of bz); bz-m2(D1) (bronze in the absence of Ac; spotted, in its presence), derivative 1 from bz-m2(Ac), harboring a 3.3-kb internally deleted Ds element at the same position as Ac in bz-m2(Ac) (19, 22); and bz-R (bronze), the bz reference allele, associated with a 340-bp deletion that extends from within the single intron to the second exon of bz and includes the Ac insertion site in bz-m2(Ac) (20, 23). The mutations sh (shrunken endosperm) and wx (waxy endosperm) were used as markers flanking bz. They map, respectively, ≈3 cM distal and 25 cM proximal to bz in 9S.
Genetic Methods.
Recombination between bz and transposed Ac elements was measured in test crosses of the type Sh (Ac bz-s) Wx/sh bz-m2(D1) wx to sh bz-R wx. Putative recombinants were selected as single spotted seed (bz-m) in ears otherwise containing only bz seeds. The selections were classified for outside markers and back-crossed to the male parent to verify their heritability and determine the segregation of bz-m and bz seeds in the progeny. The 95% confidence limits of the map distance were calculated according to Stevens (24).
DNA Sequencing and Assembly.
The isolation of a NotI BAC clone containing the 5′ half of the Bz-McC allele and 125 kb proximal to it has been described (17). The BAC clone was sequenced by the shotgun sequence strategy (www.genome.ou.edu/proto.html) with some modifications. BAC DNAs were purified by double equilibrium density gradient centrifugation in CsCl-ethidium bromide gradients and sheared to 3–6-kb fragments with hydroshearing (Genemachines, San Carlos, CA). The resulting fragments were used to generate the shotgun library. DNA minipreps were prepared from randomly picked clones. Sequencing reactions were performed by using the ABI PRISM BigDye terminator cycle sequencing ready reaction kit (Perkin–Elmer Applied Biosystems, Foster City, CA) and analyzed on ABI377 sequencing gels. The 10-fold redundant sequence data were assembled with the PHRED/PHRAP software (25). Contigs were extended and joined by custom specific primer walking to close the gaps. The reliability of the sequence was confirmed by the location of expected restriction sites.
Sequence Analysis.
The final sequence was divided into 5-kb fragments that served as queries to search the GenBank databases with the various BLAST programs (26). Programs from the LASERGENE package (DNAstar, Madison, WI) were used for sequence comparisons and alignments.
DNA Methylation Analysis.
Bz-McC genomic DNA (10 μg) was digested overnight with 50 units of different C-methylation-sensitive restriction endonucleases (identified in Fig. 2). BAC DNA (0.5 μg) was digested to completion with 5–10 units of the same methylation-sensitive restriction endonucleases for 2–3 h. The digested genomic and BAC DNAs were resolved in the same agarose gel; genomic DNAs were loaded on one side of the gel and BAC DNAs on the other. After electrophoresis, the gel was blotted to a Hybond+ nylon membrane (Amersham Pharmacia), and the membrane was cut in half to separate the two sides containing genomic and BAC DNA. The DNA blots were hybridized separately with random primer labeled P32 probes. The conditions for hybridization, high-stringency washing, and exposure to x-ray film were standard.
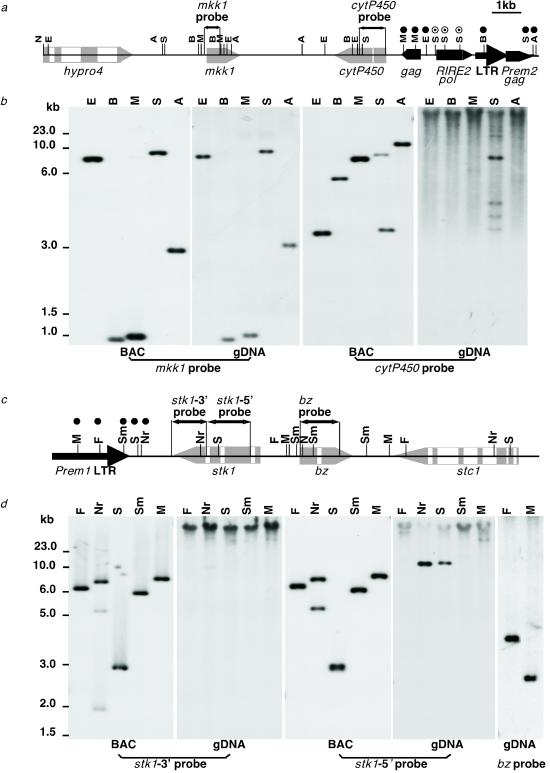
Figure 2.
Methylation analysis of restriction sites in genes and retrotransposons. (a and c) Restriction maps of the gene-retrotransposon junctions at the proximal (a) and distal (c) end of the 94-kb retrotransposon block in Fig. 1. The location of the probes used in the Southern blots is indicated above each map. The following C-methylation-sensitive restriction endonucleases were used: A, AatII; B, BssHII; E, EagI; F, FspI; M, MluI; N, NotI; Nr, NruI, S, SalI, and Sm, SmaI. Shown in the map on either side of each probe are the nearest restriction sites that were predicted from the DNA sequence and were confirmed to be completely, partially (⊙), or not (●) cut in the genomic DNA blots shown below. (b and d) Southern blots of BAC DNA and genomic DNA (gDNA) digested with the enzymes shown above each blot and hybridized to either the mkk1 (b Left) or cytP450 (b Right) probes from the proximal end of the BAC and the stk1-3′ (d Left), stk1-5′ (d Center), or bz (d Right) probes from the distal end of the BAC (17). The unmethylated BAC DNA serves as control. Restriction sites for all enzymes used in the analysis of the proximal end are present within the BAC; some sites used in the analysis of the distal end are missing from this BAC DNA, which terminates in the NotI site within bz, but are present in the adjacent stc1 gene shown in Fig. 1. The bz probe reveals the methylation status of the FspI and MluI restriction sites distal to stk-5′.
Results
Measurement of Genetic Distances Between bz and Adjacent Genes.
As genetic markers closely linked to bz, we have used Ac elements that transposed proximally or distally from the mutable allele bz-m2(Ac), leaving behind a bz-s stable bz allele (21). bz-m2(Ac) harbors an Ac insertion in the second exon of Bz-McC, and most transpositions generate a footprint that does not restore gene function (19, 20). Recombination between bz and the transposed Ac elements was measured in test crosses of the type Sh (Ac bz-s) Wx/sh bz-m2(D1) wx to sh bz-R wx (Table 1). This marker arrangement enables us to use the convenient seed-pigmentation phenotype conditioned by the bz locus to screen for rare recombinants between bz and a nearby site. The bz-m2(D1) reporter allele was derived from bz-m2(Ac) by an internal deletion in Ac that produced a Ds element (22, 27). Because the bz-s alleles carrying closely linked transposed Ac elements originated from the same progenitor, the region around the bz locus is identical in the two homologues. Therefore, the effect of sequence heterozygosity on recombination is eliminated, allowing us to focus on the nature of the intervening DNA segment itself. The flanking markers sh and wx lie outside of the interval where recombination is being measured and serve mainly to determine the proximal-distal location of Ac relative to bz.
Table 1.
Recombination between the bz locus and nearby Ac elements
| Ac | No. of recombinants | Ac location (relative to bz) | Population |
bz-Ac distance
|
|
|---|---|---|---|---|---|
| cM | 95% C.L. | ||||
| Ac6067 | 13 | Distal | 8,210 | 0.32 | 0.16–0.53 |
| Ac6058 | 19 | Distal | 15,200 | 0.25 | 0.15–0.39 |
| Ac6087 | 10 | Distal | 8,237 | 0.24 | 0.12–0.45 |
| Ac2094 | 11 | Proximal | 35,370 | 0.06 | 0.03–0.11 |
Results are from test crosses of the type Sh (Ac bz-s) Wx/sh bz-m2(D1) wx to sh bz-R wx. The data extend to those presented in an earlier paper (21), which also explains in more detail the basis of the mapping.
The vast majority of seed from the above crosses are bronze in color. Rare cosegregation of Ac and the Ds reporter allele bz-m2(D1) in the homologous chromosome results in a bronze seed with purple spots (bz-m). These seeds can arise from recombination between Ac and bz-m2(D1) or from transpositions of Ac to an unlinked site. bz-m exceptions are selected, scored for flanking markers, and back-crossed to the bz-R parent. Recombinants will segregate 1 bz-m: 1 bz, whereas transpositions of Ac to an unlinked site will segregate 1 bz-m: 3 bz. The location of Ac relative to bz is determined by examining the number of recombinants in the two bz-m classes bearing reciprocal recombinant arrangements of outside markers. Recombinants occur predominantly or exclusively in one of the two classes: Sh wx, if Ac is located distal to bz, or sh Wx, if Ac is located proximal to bz. The genetic distance between bz and Ac, in cM, is calculated as the number of recombinants × 2/population. The Ac insertions Ac6067, Ac6058, and Ac6087 map 0.32, 0.25, and 0.24 cM distal to bz, respectively, whereas Ac2094 maps 0.06 cM proximal to bz.
The DNA adjacent to the four transposed Ac elements (tac sites) has been isolated and sequenced. Each Ac has inserted in a gene. Ac6087 and Ac6067 are inserted in stc1, a gene encoding a sesquiterpene cyclase that is inducible by the insect elicitor volicitin (28). Ac6058 is inserted in a gene that is expressed abundantly in developing endosperms; it has no homology to sequences in the databases and has been provisionally designated tac6058 (18). Ac2094 is inserted in mkk1, a gene encoding a mitogen-activated protein kinase that is preferentially expressed in the developing ear (ref. 29 and B. Shen and H.K.D., unpublished observations). Therefore, the map distances in Table 1 represent intergenic distances between bz and a nearby gene, or more precisely, between the insertion sites of Ds in bz-m2(D1) and Ac in a closely linked gene. These distances are considerably higher than the 0.03-cM intragenic distance measured previously between two Ds insertion sites in bz separated by 0.6 kb, an interval representing about half the length of the bz gene (10). This result suggests that much of the recombination contributing to the distances in Table 1 is occurring outside of bz.
Characterization of the Proximal Genetic Interval.
To obtain the physical distances corresponding to those genetic distances, we have sequenced a 128-kb stretch of continuous DNA from the Bz-McC BAC contig (Fig. 1) and placed the tac sites precisely in the contig by direct sequence comparison. We had previously determined that the bz-tac2094 intergenic distance measured between 25 and 120 kb based on the sizes of fragments that hybridized to only one or both probes (17, 29). By DNA sequencing, we have now established that the exact distance separating the Ds insertion site in bz and the Ac2094 insertion site in mkk1 is 108 kb. Therefore, the ratio of genetic to physical distance in the mkk1-bz interval is 6 × 10−4 cM/kb. This is very close to the average ratio for the maize genome (7–8 × 10−4 cM/kb) calculated from a total genetic map that ranges from 1,727 (30) to 2,049 cM (B. Burr, ftp://ftp.bio.bnl.gov/pub/maize/chrom.map) and a genome size of 2.5 × 106 kb (11).
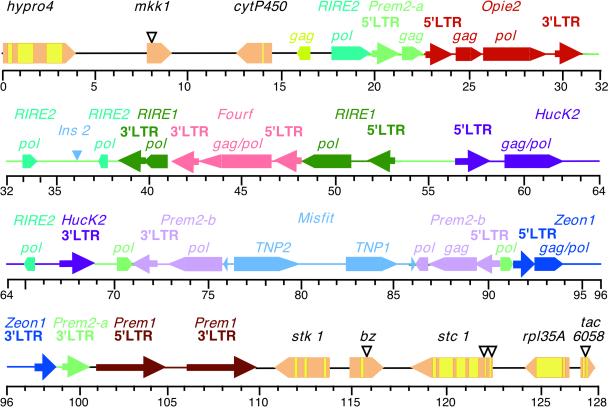
Figure 1.
Organization of genomic DNA around the bz locus in maize. The proximal end is at 0 kb, and the distal end is at 128 kb. The sequence (GenBank accession number AF391808) is a composite from two adjacent BAC clones centered around a NotI site in the Bz-McC allele (17). The NotI ends of the proximal BAC are at positions 0 and 115 kb. The part of the distal BAC shown here extends from 115 to 128 kb. Genes are shown as pentagons pointing in the direction of transcription. Gene names are in black, exons in bronze, and introns in yellow. Two gene islands at the proximal and distal end of the sequence are interrupted by a 94-kb cluster of mostly nested retrotransposons and two DNA transposons. Each transposon is in a different color. To facilitate the identification of interrupted retrotransposons, the name, LTRs and encoded proteins of the same element are in the same color. The location of the markers (inverted triangles) used in the various mapping experiments are: Ac2094, 8.0 kb; bz-m2(D1), 115.8 kb; Ac6067, 122.3 kb; Ac6087, 122.5 kb, and Ac6058, 127.4 kb.
Most of the DNA separating bz and the proximal gene mkk1 is composed of a 94-kb retrotransposon cluster arranged mainly in the same type of nested configuration described originally in the Adh1 region (16, 31) and found subsequently in the zein region (32). This very large retrotransposon block makes up 89% of the mkk1-bz interval. The main component of the block is an 81-kb retrotransposon nest, which has a highly fragmented Prem2 element at its base (Prem2-a in Fig. 1). Inserted into Prem2-a from proximal to distal end are: a full-size but probably nonfunctional Opie2 retroelement (16); a RIRE1 element (33) interrupted by an intact Fourf element (31); an incomplete Huck2 element (31); a second Prem2 element (Prem2-b) interrupted by a 10-kb member of the CACTA family of DNA transposons that we have called Misfit and an incomplete Zeon1 element (34). The 10,630-bp Misfit transposon terminates in a 19-bp imperfect inverted repeat having four mismatches and, similar to other CACTA elements, encodes TNP1- and TNP2-like proteins and is flanked by a 3-bp target site duplication. The TNP2-like reading frame contains frameshift mutations that create premature termination of translation.
As expected of this insertion hierarchy, the long terminal repeats (LTRs) of the Prem2-a element at the base are the most widely divergent (35). The sequence similarity index of the LTRs for the seven transposons in the cluster are as follows: Zeon1, 100%; Fourf, 98.2%; RIRE1, 97.1%; Prem1, 94.6%; Prem2b, 91.6%; Opie2, 91.2%; Huck2, 85.8%; and Prem2a, 63.8%. Besides Misfit, the 94-kb retrotransposon cluster contains only one other DNA or class II element, Ins2, which is related to the Ac-Ds family (20) and is inserted between Opie2 and RIRE1. This arrangement appears to differ from that of a 183-kb chromosomal knob in Arabidopsis, in which several DNA transposons are interspersed with the retrotransposons in the heterochromatin (36). In addition, fragments of pol genes homologous to RIRE2 retroelements from rice (37) are found scattered inside and immediately proximal to the 81-kb retrotransposon nest. Immediately distal to it are the unusually long 5′ and 3′ LTRs of a Prem1 element. Flanking the 94-kb retrotransposon cluster and separating it from mkk1 and bz are cytP450 and stk1, two genes encoding proteins with homology to a cytochrome P450 and a serine-threonine kinase, respectively.
Retrotransposons have been reported to be hypermethylated in maize and Arabidopsis (36, 38, 39). In agreement with those observations, all four NotI sites located within the retrotransposon block in the 115-kb NotI bz BAC clone are methylated in genomic DNA (17). To analyze further the methylation status of the retrotransposon cluster between mkk1 and bz, we digested genomic and BAC DNA with several methylation-sensitive enzymes and hybridized the resulting digests with single-copy probes located next to the proximal and distal ends of the cluster (Fig. 2). In all, 137 sites were assayed by 18 different methylation-sensitive enzymes. The methylation-sensitive restriction sites located within the retrotransposon block are mostly undigested, whereas those located in the genes outside of the block are digested completely. Therefore, the DNA in the large retrotransposon block proximal to the bz locus appears to be heavily methylated.
Characterization of the Distal Genetic Interval.
In contrast to the proximal side, there are no retrotransposon sequences between bz and the distal genes stc1 and tac6058. The physical distances from the Ds insertion site in bz to the Ac insertion sites in those genes are as follows: to Ac6067, 6.5 kb; to Ac6087, 6.7 kb; and to Ac6058, 11.6 kb. Therefore, the ratios of genetic to physical distances in the respective intervals are 0.049, 0.035, and 0.022 cM/kb. These ratios are 40–80 times higher than the genome's average and the corresponding ratio in the mkk1-bz proximal interval. However, they are only slightly lower than the bz intragenic recombination rates, which can vary between 0.05 and 0.2 cM/kb depending on the heteroalleles used (10). Furthermore, the bz-Ac intergenic distances were obtained from heterozygotes at two insertions, a condition that has been shown to reduce recombination (10), and thus they may represent low-end estimates. The DNA in the region immediately distal to bz is not methylated and is present in single copy in the maize genome (18). Eight genes have been identified in the 25 kb of DNA that separate bz from the nearest retrotransposon cluster on the distal side: stc1 is the first and tac6058 is the third of those genes. Thus, recombination distal to bz, a region composed principally of genic DNA, also is much higher than the genome's average. These data strongly suggest that recombination in maize is confined to the low-copy DNA component in which most genes reside.
Discussion
The highly recombinogenic nature of maize genes has been documented at a number of loci (40). This property of maize genes supports the hypothesis advanced by Thuriaux years ago that meiotic recombination in eukaryotes takes place mostly in genes (2). A corollary of that hypothesis is that recombination in repetitive DNA should be much lower than in genic DNA. Consequently, most of the genome in organisms with extensive repetitive DNA such as maize would be recombinationally inert. Because the majority of the repetitive DNA in maize is made up of retrotransposon clusters (16), recombination in regions containing such clusters should be much lower than in adjacent regions containing genes. We have examined here how well that prediction holds in the bz region of the maize genome.
The disproportionately high intragenic recombination rate of maize genes was reported first at bz, a gene where the ratio of genetic to physical distance is more than 100 times higher than the genome's average (5, 41). In the present work, we analyzed the relationship between genetic and physical distance in the regions immediately proximal and distal to bz. The genetic distance between bz and adjacent genes was measured by using as markers Ac elements that had transposed from the bz-m2(Ac) allele to very closely linked sites. Physical distances were measured by placing these Ac elements in one of the two Bz-McC BAC clones. Finally, the nature of the intervening region was characterized by DNA sequencing and methylation analysis.
There are several reasons why these Ac markers are useful for our study. (i) The dominant trans-acting effect of Ac on a Ds reporter facilitates the measurement of genetic distance between bz and any point nearby by taking advantage of bz's readily scorable seed phenotype. (ii) Ac inserts preferentially into genes (18, 28, 42), thus enabling measurements of intergenic distances. (iii) Ac insertions can be localized easily in a physical map. (iv) The transposed Acs are contained in the same chromosomal region as Bz-McC, the progenitor allele of bz-m2(Ac), for which we have isolated a 240-kb BAC contig (17). (v) Perhaps most importantly they enable the measurement of recombination between two points in the chromosome separated by identical DNA segments such that the effect of any potential sequence heterozygosity is eliminated.
We recently reported that bz lies in an unusually gene-rich region of the genome (18). Ten genes are contained in a 32-kb stretch of DNA that is uninterrupted by retrotransposons. The Ac elements that transposed to sites located distal to bz landed in two of these genes: stc1, encoding a sesquiterpene cyclase induced by insect herbivory, and tac6058, a gene of unknown function that is expressed abundantly in the endosperm. The ratios of genetic to physical distance in the distal intervals approach the values reported for bz. Thus, recombination in the distal interval, which is devoid of retrotransposons and composed principally of genic DNA, also is much higher than the genome's average.
The Ac element that transposed proximally landed in mkk1, a gene encoding a mitogen-activated protein kinase kinase that is preferentially expressed in the ear. The ratio of genetic to physical distance in the mkk1-bz proximal interval is much lower than in the distal intervals and is close to the average value for the maize genome. This interval is made up principally of a 94-kb retrotransposon cluster with a similar type of nested organization to that first described at the Adh region (16). In addition, this cluster is heavily methylated. There is evidence that retrotransposons in maize and other plants are hypermethylated in general (38, 39, 43). Most likely, they also are recombinationally inert. In barley, intrachromosomal recombination between LTRs has been postulated to explain the excess of solo LTRs relative to intact BARE retrotransposons (44), but that may be a property of recently transposed, and presumably hypomethylated, elements (45). Given their dispersed distribution in the genome (16), ectopic recombination between retrotransposons often would have serious deleterious consequences. Suppression of recombination in repetitive DNA, which acts to stabilize the genome by preventing ectopic exchanges, seems to be a common feature of eukaryotic genomes. In Drosophila (46) and Arabidopsis (47), the rate of meiotic recombination also is reduced in transposon-rich chromosomal regions. The only multicellular eukaryote in which this correlation has not been observed is Caenorhabditis elegans (48), which also is unusual in that most exchanges occur in gene-poor regions (49).
Recombination rates within genes are known to vary with location in the chromosome. For example, genes close to the centromere recombine at very low frequencies (50). In addition, our own data suggest that the position of a gene in a gene-dense region may affect recombination. If the frequency of recombination within the two genes flanking the retrotransposon cluster proximal to bz (cytP450 and stk1) was as high as within bz, the mkk1-bz distance should have been higher. Possibly, genes next to retrotransposon clusters are less recombinogenic than other genes in a gene-rich region. A 94-kb methylated retrotransposon cluster most likely behaves as a heterochromatic island in maize, as it does in Arabidopsis (36). The more condensed chromatin state of the retrotransposon cluster may interfere with access of the recombination machinery to the adjacent euchromatic regions. Thus, recombination rates in gene clusters such as the one in which bz resides may show gradients in the transition zones between genes and retrotransposons. If so, the gradients would not extend into neighboring genes, because recombination between heteroalleles derived from the same progenitor occurs uniformly across the bz gene (10).
Our experimental set-up enabled us to examine recombination between genes separated by a completely homozygous retrotransposon block. There is increasing evidence that the make-up of retrotransposons upstream and downstream of genes differs among inbred lines (ref. 35 and H.F. and H.K.D., unpublished observations). Thus, in most maize mapping experiments, the intergenic retrotransposon blocks will be structurally heterogeneous. Yet, because recombination is highly suppressed in this large component of the genome, retrotransposon block heterozygosity per se should not contribute significantly to the observed variability in estimates of total map length in maize (30).
Acknowledgments
We thank Kim McKim, Joachim Messing, and members of the Dooner lab, especially Matt Cowperthwaite, for comments on the manuscript. This research was supported by National Science Foundation Grant MCB 99-04646.
Abbreviations
- BAC
bacterial artificial chromosome
- LTR
long terminal repeat
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF391808).
References
- 1.Thomas C A. Annu Rev Genet. 1971;5:237–256. doi: 10.1146/annurev.ge.05.120171.001321. [DOI] [PubMed] [Google Scholar]
- 2.Thuriaux P. Nature (London) 1977;268:460–462. doi: 10.1038/268460a0. [DOI] [PubMed] [Google Scholar]
- 3.Hake S, Walbot V. Chromosoma. 1980;79:251–270. [Google Scholar]
- 4.Puchta H, Hohn B. Trends Plant Sci. 1996;1:340–348. doi: 10.1016/S1360-1385(03)00004-9. [DOI] [PubMed] [Google Scholar]
- 5.Dooner H K. Genetics. 1986;113:1021–1036. doi: 10.1093/genetics/113.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown J, Sundaressan V. Theor Appl Genet. 1991;81:185–188. doi: 10.1007/BF00215721. [DOI] [PubMed] [Google Scholar]
- 7.Civardi L, Xia Y, Edwards K J, Schnable P S, Nikolau B J. Proc Natl Acad Sci USA. 1994;91:8268–8272. doi: 10.1073/pnas.91.17.8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eggleston W B, Alleman M, Kermicle J L. Genetics. 1995;141:347–360. doi: 10.1093/genetics/141.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patterson G I, Kubo K M, Shroyer T, Chandler V L. Genetics. 1995;140:1389–1406. doi: 10.1093/genetics/140.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dooner H K, Martinez-Ferez I M. Plant Cell. 1997;9:1633–1646. doi: 10.1105/tpc.9.9.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arumuganathan K, Earle E D. Plant Mol Bio Rep. 1991;9:208–218. [Google Scholar]
- 12.Flavell R B, Bennett M D, Smith J B, Smith D B. Biochem Genet. 1974;12:257–269. doi: 10.1007/BF00485947. [DOI] [PubMed] [Google Scholar]
- 13.Grandbastien M A. Trends Genet. 1992;8:103–108. doi: 10.1016/0168-9525(92)90198-d. [DOI] [PubMed] [Google Scholar]
- 14.Bennetzen J L. Trends Microbiol. 1996;4:347–353. doi: 10.1016/0966-842x(96)10042-1. [DOI] [PubMed] [Google Scholar]
- 15.Kumar A, Bennetzen J. Annu Rev Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- 16.SanMiguel P, Tikhonov A, Jin Y K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 17.Fu H, Dooner H K. Genome Res. 2000;10:866–873. doi: 10.1101/gr.10.6.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu H, Park W, Yan X, Zheng Z, Shen B, Dooner H K. Proc Natl Acad Sci USA. 2001;98:8903–8908. doi: 10.1073/pnas.141221898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClintock B. Carnegie Inst Washington, Year Book. 1955;54:245–255. [PubMed] [Google Scholar]
- 20.Ralston E J, English J, Dooner H K. Genetics. 1988;119:185–197. doi: 10.1093/genetics/119.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dooner H K, Belachew A. Genetics. 1989;122:447–457. doi: 10.1093/genetics/122.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dooner H K, English J, Ralston E, Weck E. Science. 1986;234:210–211. doi: 10.1126/science.234.4773.210. [DOI] [PubMed] [Google Scholar]
- 23.Rhoades M M. Am Nat. 1952;86:105–108. [Google Scholar]
- 24.Stevens W L. J Genet. 1942;43:301–306. [Google Scholar]
- 25.Green P. DOE Human Genome Program Contractor-Grantee Workshop V. Washington, DC: U.S. Dept. of Energy; 1996. p. 157. [Google Scholar]
- 26.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClintock B. Carnegie Inst Washington, Year Book. 1962;61:448–461. [PubMed] [Google Scholar]
- 28.Shen B, Zheng Z, Dooner H K. Proc Natl Acad Sci USA. 2000;97:14807–14812. doi: 10.1073/pnas.240284097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ralston E J, English J, Dooner H K. Proc Natl Acad Sci USA. 1989;86:9451–9455. doi: 10.1073/pnas.86.23.9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis G L, McMullen M D, Baysdorfer C, Musket T, Grant D, Staebell M, Xu G, Polacco M, Koster L, Melia-Hancock S, et al. Genetics. 1999;152:1137–1172. doi: 10.1093/genetics/152.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tikhonov A P, SanMiguel P J, Nakajima Y, Gorenstein N M, Bennetzen J L, Avramova Z. Proc Natl Acad Sci USA. 1999;96:7409–7414. doi: 10.1073/pnas.96.13.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Llaca V, Messing J. Plant J. 1998;15:211–220. doi: 10.1046/j.1365-313x.1998.00200.x. [DOI] [PubMed] [Google Scholar]
- 33.Noma K, Nakajima R, Ohtsubo H, Ohtsubo E. Genes Genet Syst. 1997;72:131–140. doi: 10.1266/ggs.72.131. [DOI] [PubMed] [Google Scholar]
- 34.Hu W, Das O P, Messing J. Mol Gen Genet. 1995;248:471–480. doi: 10.1007/BF02191647. [DOI] [PubMed] [Google Scholar]
- 35.SanMiguel P, Gaut B S, Tikhonov A, Nakajima Y, Bennetzen J L. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 36.Arabidopsis Sequencing Consortium. Cell. 2000;100:377–386. [PubMed] [Google Scholar]
- 37.Ohtsubo H, Kumekawa N, Ohtsubo E. Genes Genet Syst. 1999;74:83–91. doi: 10.1266/ggs.74.83. [DOI] [PubMed] [Google Scholar]
- 38.Bennetzen J L, Schrick K, Springer P S, Brown W E, SanMiguel P. Genome. 1994;37:565–576. doi: 10.1139/g94-081. [DOI] [PubMed] [Google Scholar]
- 39.Rabinowicz P D, Schutz K, Dedhia N, Yordan C, Parnell L D, Stein L, McCombie W R, Martienssen R A. Nat Genet. 1999;23:305–308. doi: 10.1038/15479. [DOI] [PubMed] [Google Scholar]
- 40.Schnable P S, Hsia A P, Nikolau B J. Curr Opin Plant Biol. 1998;1:123–129. doi: 10.1016/s1369-5266(98)80013-7. [DOI] [PubMed] [Google Scholar]
- 41.Dooner H K, Weck E, Adams S, Ralston E, Favreau M, English J. Mol Gen Genet. 1985;200:240–246. [Google Scholar]
- 42.Cowperthwaite, M., Park, W., Xu, Z., Yan, X., Maurais, S. & Dooner, H. K. (2002) Plant Cell14, in press. [DOI] [PMC free article] [PubMed]
- 43.Hirochika H, Okamoto H, Kakutani T. Plant Cell. 2000;12:357–369. doi: 10.1105/tpc.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vicient C M, Jaaskelainen M J, Kalendar R, Schulman A H. Plant Physiol. 2001;125:1283–1292. doi: 10.1104/pp.125.3.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman A H. Proc Natl Acad Sci USA. 2000;97:6603–6607. doi: 10.1073/pnas.110587497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charlesworth B, Sniegowski P, Stephan W. Nature (London) 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 47.Arabidopsis Genome Initiative. Nature (London) 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 48.Duret L, Marais G, Biemont C. Genetics. 2000;156:1661–1669. doi: 10.1093/genetics/156.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barnes T M, Kohara Y, Coulson A, Hekimi S. Genetics. 1995;141:159–179. doi: 10.1093/genetics/141.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Copenhaver G P, Nickel K, Kuromori T, Benito M I, Kaul S, Lin X, Bevan M, Murphy G, Harris B, Parnell L D, et al. Science. 1999;286:2468–2474. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]