Abstract
Background
The role of centrioles in mitotic spindle function remains unclear. One approach to investigate mitotic centriole function is to ask whether mutation of centriole-associated proteins can cause genomic instability.
Results
We addressed the role of the centriole-associated EF-hand protein centrin in genomic stability using a Chlamydomonas reinhardtii centrin mutant that forms acentriolar bipolar spindles and lacks the centrin-based rhizoplast structures that join centrioles to the nucleus. Using a genetic assay for loss of heterozygosity, we found that this centrin mutant showed increased genomic instability compared to wild-type cells, and we determined that the increase in genomic instability was due to a 100-fold increase in chromosome loss rates compared to wild type. Live cell imaging reveals an increased rate in cell death during G1 in haploid cells that is consistent with an elevated rate of chromosome loss, and analysis of cell death versus centriole copy number argues against a role for multipolar spindles in this process.
Conclusion
The increased chromosome loss rates observed in a centrin mutant that forms acentriolar spindles suggests a role for centrin protein, and possibly centrioles, in mitotic fidelity.
Background
Centrioles are cylindrical structures located within the core of the centrosome. Although the localization of centrioles within the centrosome together with their precise duplication prior to mitosis has suggested a role in bipolar spindle assembly or function, the actual role of centrioles in cell division remains unclear and controversial. Cells from which centrioles and centrosomes are ablated can still form bipolar spindles via a centrosome-independent self-organization process, but the effectiveness of such spindles in terms of chromosome segregation has not been carefully measured. Based on the circumstantial evidence that tumor cells which display genomic instability also frequently show aberrations in centriole structure or copy number [1,2], it has been proposed that centrioles may participate in the maintenance of genomic stability. If centrioles play a role in chromosome segregation, then mutations in genes encoding centriolar proteins would be expected to result in genomic instability.
The EF-hand protein centrin [3] is one of the few identified centriolar proteins. Centrin is found in all eukaryotes, and is localized to centrioles in vertebrate cells [4], basal bodies in green algae [5], and spindle pole bodies (SPBs) in yeast [6,7]. Centrin is functionally required for centriole duplication [8,9] and inheritance [10]. RNAi experiments in mammalian and algal cells have indicated that reduction of centrin can result in cell division arrest [9] and cytokinesis failure [11]. However, it is unclear if these defects are indirectly caused by abnormal centriole structure or copy number, or if instead they represent pleiotropic effects due to the direct participation of centrin in the cell division process. Centrin also appears to be involved in many other cellular processes, including flagellar excision [12], nuclear mRNA export [13], cell membrane integrity [14] and homologous recombination [15]. Consistent with these multiple roles, centrin localization is not restricted to centrioles [4], and centrin has been identified as a component of nuclear pores and flagellar dynein complexes [16,17]. Centrin is thus a versatile molecule that has been adapted to perform multiple diverse cellular functions. Given the fact that centrin localizes to centrioles, which are potentially involved in genome maintenance, together with the fact that centrin is functionally involved in centriole duplication and segregation, an obvious question is whether centrin plays a role in genomic stability. In this report we address this question using a centrin mutation in the green alga Chlamydomonas reinhardtii.
Results
Genomic instability in the centrin mutant vfl2-1
We tested whether centrin plays a role in genomic instability using a genetic assay for loss of heterozygosity in Chlamydomonas reinhardtii. Chlamydomonas (Chlamydomonas refers to Chlamydomonas reinhardti throughout) is a unicellular green alga with genetics similar to yeast, but which, unlike yeast, has centrioles (known as basal bodies) that are virtually identical to centrioles of vertebrate cells. In Chlamydomonas, centrin polymerizes into long fibers that link the centrioles/basal bodies to the nucleus. These fibers are called the nucleus basal body connectors (NBBCs) or rhizoplasts [18,19]. The vfl2-1 mutant has a point mutation in the centrin gene [10], and these mutants produce a reduced quantity of centrin protein that is unable to polymerize into a rhizoplast. The vfl2 mutation appears to result in a defect in centriole segregation, because mutant cells have a variable number of centrioles (between zero and six per cell). Despite their centriole number variation, these mutants are viable and form seemingly normal bipolar mitotic spindles. It has been argued on this basis that centrin-based rhizoplasts do not play a role in mitosis [19]. Importantly, the centrioles in a vfl2 mutant cell are not located at the spindle poles [19], consistent with ultrastructural studies showing that, in green algae, centrioles are joined to the spindle poles by centrin-based fibers derived from the rhizoplast [20]. The high viability of vfl2 mutants thus allows us to measure genomic instability using genetic assays because populations of mutant cells can be grown for a sufficient number of generations to allow rare chromosome loss events to be detected.
We measured genomic instability in vfl2 mutants using a simple genetic assay for loss of heterozygosity (LOH) previously developed by Dutcher and coworkers [21]. This assay uses the act2 mutation, which confers recessive resistance to cycloheximide [22]. The act2 heterozygous diploids are sensitive to cycloheximide, while homozygous or hemizygous act2 diploids are cycloheximide-resistant. When a diploid culture is plated on media containing cycloheximide, only cells that have lost the wild-type ACT2 allele can grow. By measuring the frequency with which such cells arise, the rate of chromosome loss can be calculated (see Methods). As illustrated in Fig. 1, using 48 independently constructed diploid lines each for wild type and vfl2, we found that the LOH rate per cell division increased from 1.5 × 10-3 ± 1.6 × 10-4 SEM (n = 48) in wild type, to 8.8 × 10-3 ± 5.7 × 10-4 SEM (n = 48) in vfl2 mutants. This difference is statistically significant (P < 0.00005 based on one-tailed Welch's approximate t-test, t = 12.2). The increase in loss of heterozygosity in the vfl2 centrin mutant relative to wild-type cells indicates that centrin plays a role in genomic stability.
Figure 1.
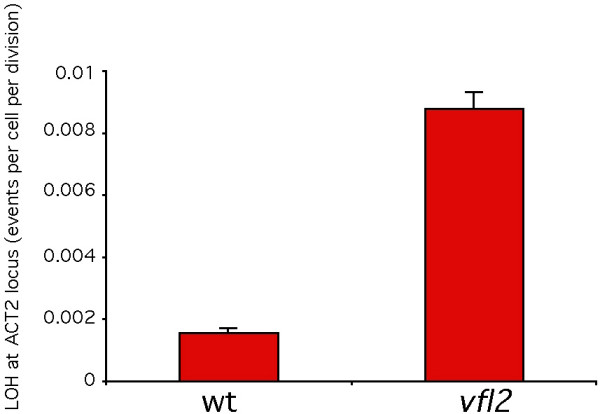
Genomic instability in the Chlamydomonas centrin mutant vfl2. Graph indicates frequency of loss of heterozygosity (LOH) at ACT2 test locus according to measurements based on the Luria-Delbrück fluctuation test. Error bars correspond to standard error of the mean.
Genomic instability occurs via chromosome loss
In principle the LOH seen in vfl2 mutants could occur either by increased recombination or increased chromosome loss (Fig. 2). The latter would be consistent with a role for centrin or centrioles in mitotic spindle function, while an effect on recombination would suggest a spindle-independent role in DNA maintenance or chromosome dynamics. It is critical to distinguish these pathways because a role for centrin in DNA recombination and repair has been documented in plants [15]. Given the close phylogenetic relation between green algae like Chlamydomonas and green plants, the involvement of centrin in recombination in plants raises the real possibility that the loss of heterozygosity observed in vfl2 might have nothing to do with mitotic spindle function and might instead be driven by increased recombination.
Figure 2.

Pathways of ACT2 LOH. LOH due to mitotic recombination, including crossover and gene conversion events, will have no effect on the mating type locus MT on the other arm of the chromosome; hence cells will retain both the MT+ and MT- alleles. This is also true for spontaneous mutation of the ACT2 gene. In contrast, chromosome loss will result in simultaneous LOH at both ACT2 and MT.
In order to determine whether genomic instability in the vfl2 mutant is caused by increased recombination or chromosome loss, we distinguished between the two pathways by analyzing the mating type (MT) locus, which is located on the opposite arm of the same chromosome from ACT2 (Fig. 2). Diploid cells are heterozygous at MT and contain both the MT+ and MT- alleles. Because the ACT2 and MT loci are on opposite arms of the same chromosome, and recombination only causes LOH on a single arm distal to the crossover site, it can be inferred that only chromosome loss, and not mitotic recombination, could lead to the simultaneous loss of both the wild-type ACT2 gene and the MT+ allele (Fig. 2).
We therefore tested lines that had undergone LOH at the ACT2 locus for chromosome loss versus recombination, by analyzing the MT locus using PCR primers specific for the MT- and MT+ alleles [23]. As illustrated in Fig. 3, prior to selection on cycloheximide, all wild-type and vfl2/vfl2 diploid lines were indeed diploid, as judged by the fact that they were heterozygous at the MT locus and contained both MT+ and MT- alleles as expected for a diploid. In wild type, of 96 independently constructed diploid lines selected on cycloheximide, only two lines showed concomitant loss of the MT+ allele and, therefore, chromosome loss. In contrast, of 96 cycloheximide resistant vfl2 lines, we found that 27 had also lost the MT+ allele, loss of which indicated that the lines had undergone chromosome loss. These chromosome loss events apparently represent the loss of individual chromosomes rather than an overall return to haploidy, as the cells were able to grow on media lacking arginine and thus seemed to have retained the two homologs of the chromosome containing the arg2/arg7 mutations used to select for diploidy (see Methods). The difference in the proportion of LOH events due to chromosome loss (2/96 for wild type vs. 27/96 for vfl2) was highly significant (P < 0.00001 based on Fisher's Exact Test). Multiplying the LOH rate by the fraction of LOH events due to chromosome loss, we found that the total rate of chromosome loss per cell division increased almost 100-fold, from 3.2 × 10-5 in wild type to 2.4 × 10-3 in vfl2 mutants.
Figure 3.

An increased proportion of LOH events is due to chromosome loss in vfl2 cells compared to wild-type cells. PCR analysis of MT locus before and after selection for LOH at ACT2 locus based on resistance to cycloheximide (chx) as described in Methods. Loss of MT+ specific band suggests that chromosome loss accompanied LOH at ACT2. Each panel shows 24 out of a total of 96 samples analyzed for each case.
Cell death during G1 in vfl2 mutants
The fact that vfl2 mutants show increased chromosome loss in the genetic assay described above leads to the prediction that haploid vfl2 cells should frequently produce inviable progeny cells. This prediction follows from the fact that all Chlamydomonas chromosomes contain essential genes, so a single chromosome loss event for any of the 17 chromosomes should produce an inviable progeny cell when haploid cell division are analyzed. (Note: chromosome loss did not lead to death in the experiments described above because those assays were conducted using diploid cells. Thus, chromosome loss led only to monosomy rather than nullisomy.) Consistent with this prediction, analysis of cell viability using the vital stain phenosaffranin indicated that vfl2 cultures contain a higher fraction of inviable cells than wild type (data not shown).
In order to test this prediction more quantitatively, we examined the outcomes of individual cell division events. Synchronized cultures of haploid Chlamydomonas cells were embedded in agarose pads under a coverslip and imaged during G1 (as judged by time relative to the light-dark cycle and by the presence of flagella on most cells) by phase contrast microscopy as previously described [24]. Viable cells were identified and their position marked, and then their progeny were examined during the first G1 phase after division. Only cells that divided successfully to give two or four progeny were scored (note that sometimes, depending on cell size at the commitment point, Chlamydomonas cells can undergo two or three rapid division cycles during the dark phase of the circadian cycle). As a test of the methodology, we compared outcomes of division for wild-type cells and fla10-1 mutant cells grown at 21°C. The fla10-1 mutation occurs in a kinesin involved in flagellar assembly, and it has been reported that fla10-1 mutants show an increased rate of mitotic chromosome loss when grown at the permissive temperature [21]. If chromosome loss leads to increased production of inviable progeny, then this should be detectable in our analysis. Indeed, as shown in Fig. 4, fla10-1 cells showed a significant increase in the fraction of divisions producing inviable progeny.
Figure 4.

Production of inviable progeny cells during division in haploid Chlamydomonas cells. Graph shows fraction of cell divisions in which one or both progeny cells are inviable based on morphology. (Green) Wild type; n = 97. (Red) The fla10 mutant, previously shown to have elevated rate of chromosome loss; n = 18. Production of inviable progeny likely reflects spontaneous missegregation of chromosomes resulting in nullisomic progeny that die due to lack of essential genes. (Blue) The vfl2 mutant; n = 312. As predicted from chromosome loss measurements, this mutant shows an elevated rate of inviable progeny. (Grey) Outcomes for vfl2 mutant cells with less than two, two, or greater than two centrioles, as judged by flagellar number (numbers of cell divisions scored: less than two centrioles, n = 105; two centrioles, n = 136; greater than two centrioles, n = 71). Cells with supernumerary centrioles do not show an elevated rate of dead progeny production compared to cells with two or fewer centrioles.
We then applied this method to vfl2 mutant cells. As indicated in Fig. 4, we found that vfl2 showed a similar level of inviable progeny production as was observed in fla10 and significantly higher than in wild type. The cell death was observed during the first G1 after division and therefore did not result from a prolonged arrest or from a failure during subsequent S phase or mitosis. Dead cells were often roughly similar in size to their live siblings, suggesting that they were initially able to grow but eventually became inviable as they grew. In most cases the viable sibling cells retained flagella, indicating the culture had not yet reached the first S phase (at which time flagella are resorbed in Chlamydomonas preparatory to division), confirming that death occurred during G1. This cell death during G1 is consistent with increased chromosome loss during mitosis, which should result in death during growth as essential gene products become depleted. We cannot, however, rule out the possibility that the cell death results from other defects in cell division.
In order to test whether supernumerary centrioles might cause the elevated cell death we observed, we asked whether the cell division defects in vfl2 correlated with the number of centrioles present. As previously shown, the fact that all centrioles are active as basal bodies in vfl2 mutants allows the number of centrioles to be inferred from the number of flagella [24]. We therefore examined the outcome of cell divisions of cells having either the correct centriole copy number (n = 2), too few centrioles (n<2) or supernumerary centrioles (n>2). As indicated in Fig. 4, the fractions of inviable progeny were statistically similar for all three classes. We especially note that the cells with supernumerary centrioles did not show an increase in cell death after division.
Discussion
These results demonstrate that the vfl2-1 point mutation [10] in centrin leads to increased genomic instability via an increase in chromosome loss, suggesting that centrin plays a role in mitotic chromosome segregation. Formally, these results suggest that the gene encoding centrin could act as a "housekeeper" type tumor suppressor gene, which when mutated would predispose a cell to tumorigenesis indirectly by increasing genomic instability.
The mechanism by which the vfl2 mutation leads to increased chromosome loss is unknown. The chromosome missegregation seen in the vfl2 centrin mutant is reminiscent of the Ipl (increase in ploidy) phenotype seen in yeast mutants defective in the yeast centrin homolog CDC31 [25], which is presumably due to a failure to form a mitotic spindle. By analogy with cdc31 mutants, it is then possible that the chromosome loss in vfl2 results from defective spindles. However, unlike in yeast where the spindle pole body is absolutely necessary to form a mitotic spindle, in higher eukaryotes it has been shown that bipolar spindles can self-assemble in the absence of centrioles or centrosomes [26]. Thus it was not obvious a priori that a defect in a centriole-associated protein (such as centrin) should have any effect on mitosis. Indeed, in the specific case of the Chlamydomonas vfl2 mutant, it has previously been reported that even though the vfl2 mutation causes centrioles to detach from the spindle poles, the acentriolar spindles are nonetheless still bipolar [19]. On this basis it was concluded by Wright and coworkers [19] that centrin does not play a role in mitosis. It might therefore seem surprising that we find an increase in chromosome loss in the vfl2 mutant. However, while the ability of cells lacking centrioles or centrosomes to form bipolar spindles is well documented, and these spindles can be seen to undergo an apparently normal anaphase chromosome separation [27], the question of whether such acentriolar spindles might have an altered fidelity of chromosome segregation compared to normal spindles has never been directly addressed. We speculate that the chromosome missegregation we observe in the vfl2 mutant reflects a decreased effectiveness of acentriolar spindles that self-assemble in this mutant. This would suggest a mitotic fidelity function for centrioles, which requires not merely the presence of centrioles in the cell, but also their specific localization at the spindle pole.
Alternatively, the chromosome loss we observe could be a byproduct of multipolar spindles. A small fraction (less than 1%) of vfl2 cells have three or more centrioles that could in theory give rise to tripolar or tetrapolar spindles. We suspect this is not the case because only bipolar spindles have been observed in these mutants [19]. Moreover, since centrioles are not located at spindle poles in this mutant, it is unlikely that the number of centrioles would have any effect on the number of spindle poles. The lack of centrioles at the poles also rules out configurations such as those seen during mitosis in multinucleate PtK1 cells [28], when multiple centriole pairs line up on either side of a single metaphase plate to form a masked multipolar spindle. Furthermore, to the extent that production of inviable cells during mitosis reflects chromosome loss, the data of Fig. 4 argue against multipolar spindles playing a dominant role in chromosome loss since cells containing supernumerary centrioles do not show any increase in the rate of production of inviable progeny cells.
It is also possible that the chromosome loss in vfl2 mutants arises through a spindle-independent mechanism. Because centrin may be involved in nuclear transport [13,17], it is possible that the vfl2 mutation could cause defects in chromosome organization due to a nuclear trafficking defect, leading indirectly to chromosome loss. We also note that disruption of centrin by RNAi in Chlamydomonas has been reported to cause cytokinesis defects leading to binucleate cells [11]. However such a cytokinesis defect cannot explain our results because the LOH assay we employed selects for loss of ACT2, rather than the presence of additional copies. Detailed cell-biological analysis of centrin mutant cells during division will be necessary to understand the basis of the chromosome loss we have observed.
Conclusion
We have found that a mutation in the centriole-associated protein centrin, which causes affected cells to form acentriolar spindles and lack centrin-based fibers, produces an increase in genomic instability caused by an increase in the rate of chromosome loss. These results indicate that either centrioles or the centrin protein itself are involved in increasing the fidelity of mitotic chromosome segregation.
Methods
Heterozygous act2 diploid cells were constructed by crossing haploid strains of genotypes act2 arg7 mt- × ACT2 arg2 mt+. The arg2 and arg7 mutations are present in the same gene but complement each other in trans, so that cells require both alleles in order to grow in media lacking arginine. The ACT2 and ARG2/ARG7 loci are on different chromosomes, allowing us to distinguish loss of an individual chromosome containing ACT2 from a general return to haploidy (based on the ability of cells to grow on media lacking arginine).
We inoculated act2 heterozygous diploid cultures in non-selective media at an average density of 1 cell per culture, and, after growth, plated them on media containing cycloheximide. We then calculated the rate of loss of heterozygosity (LOH) at ACT2 from the number of resistant cells and the number of cell divisions following the initial inoculation (determined from viable cell counts obtained by serial dilution) using the fluctuation test method of Luria and Delbrück [29].
Authors' contributions
IZ conducted PCR-based assays for chromosome loss. WFM conceived and designed the experiments, constructed strains, conducted fluctuation tests to measure LOH rate, and conducted analyses of live cells during division. Both authors contributed to the writing of the manuscript, and both authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Jessica Feldman, Lani Keller, Elisa Kannegaard, Kim Wemmer and Joel Rosenbaum for careful reading of the manuscript, and Mike Adams, Susan Dutcher, Hongmin Qin and Lotte Pedersen for helpful discussions about methodology. This work was supported by National Science Foundation grant MCB0416310 (WFM), a Hellman Family Award, startup funds from the Sandler Family Supporting Foundation and a University of California, San Franscisco Research Evaluation and Allocation Committee (REAC) grant.
Contributor Information
Ivan Zamora, Email: dsjiz@itsa.ucsf.edu.
Wallace F Marshall, Email: wmarshall@biochem.ucsf.edu.
References
- Brinkley BR, Goepfert TM. Supernumerary centrosomes and cancer: Boveri's hypothesis resurrected. Cell Motil Cytoskel. 1998;41:281–8. doi: 10.1002/(SICI)1097-0169(1998)41:4<281::AID-CM1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Duensing S, Munger K. Centrosome abnormalities, genomic instability, and carcinogenic progression. Biochim Biophys Acta. 2001;1471:M81–8. doi: 10.1016/s0304-419x(00)00025-1. [DOI] [PubMed] [Google Scholar]
- Schiebel E, Bornens M. In search of a function for centrins. Trends Cell Bio. 1995;5:197–201. doi: 10.1016/S0962-8924(00)88999-0. [DOI] [PubMed] [Google Scholar]
- Paoletti A, Moudjou M., Paintrand M, Salisbury JL, Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J Cell Sci. 1996;109:3089–102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- Salisbury JL, Baron AT, Sanders MA. The centrin-based cytoskeleton of Chlamydomonas reinhardtii :distribution in interphase and mitotic cells. J Cell Biol. 1988;107:635–41. doi: 10.1083/jcb.107.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Courtney I, Fackler U, Matzner M, Schiebel E. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J Cell Biol. 1993;123:405–16. doi: 10.1083/jcb.123.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti A, Bordes N, Haddad R, Schwartz CL, Chang F, Bornens M. Fission yeast cdc31p is a component of the half-bridge and controls SPB duplication. Mol Biol Cell. 2003;14:2793–808. doi: 10.1091/mbc.E02-10-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middendorp S, Kuntziger T, Abraham Y, Holmes S, Bordes N, Paintrand M, Paoletti A, Bornens M. A role for centrin 3 in centrosome reproduction. J Cell Biol. 2000;148:405–16. doi: 10.1083/jcb.148.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury JL, Suino KM, Busby R, Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol. 2002;12:1287–92. doi: 10.1016/S0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- Taillon BE, Adler SA, Suhan JP, Jarvik JW. Mutational analysis of centrin: an EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas. J Cell Biol. 1992;119:1613–24. doi: 10.1083/jcb.119.6.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblentz B, Schoppmeier J, Grunow A, Lechtreck KF. Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation, and maturation. J Cell Sci. 2003;116:2635–46. doi: 10.1242/jcs.00497. [DOI] [PubMed] [Google Scholar]
- Sanders MA, Salisbury JL. Centrin plays an essential role in microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J Cell Biol. 1994;124:795–805. doi: 10.1083/jcb.124.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Rodriguez-Navarro S, Pereira G, Racz E, Schiebel E, Hurt E. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat Cell Biol. 2004;6:840–848. doi: 10.1038/ncb1163. [DOI] [PubMed] [Google Scholar]
- Ivanovska I, Rose MD. Fine structure analysis of the yeast centrin, Cdc31p, identifies residues specific for cell morphology and spindle pole body duplication. Genetics. 2000;157:503–518. doi: 10.1093/genetics/157.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J, Ramos C, Fritsch O, Hohn B. CENTRIN2 modulates homologous recombination and nucleotide excision repair in Arabidopsis. Plant Cell. 2004;16:1633–43. doi: 10.1105/tpc.021378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDizet M, Piperno G. The light chain p28 associates with a subset of inner dynein arm heavy chains in Chlamydomonas axonemes. Mol Biol Cell. 1995;6:697–711. doi: 10.1091/mbc.6.6.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–51. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater JM. Morphology and division of Chlamydomonas with reference to the phylogeny of the flagellate neuromotor system. Univ Calif Pub Zool. 1929;33:125–68. [Google Scholar]
- Wright RL, Adler SA, Spanier JG, Jarvik JW. Nucleus-basal body connector in Chlamydomonas : evidence for a role in basal body segregation and against essential roles in mitosis or in determining cell polarity. Cell Motil Cytoskel. 1989;14:516. doi: 10.1002/cm.970140409. [DOI] [PubMed] [Google Scholar]
- Lechtreck KF, Grunow A. Evidence for a direct role of nascent basal bodies during spindle pole initiation in the green alga Spermatozopsis similis. Protist. 1999;150:163–81. doi: 10.1016/S1434-4610(99)70019-2. [DOI] [PubMed] [Google Scholar]
- Miller MS, Esparza JM, Lippa AM, Lux FG, 3rd, Cole DG, Dutcher SK. A genetic analysis of mutations a the FLA10 locus of Chlamydomonas reinhardtii. Mol Biol Cell. 2005.
- Smyth RD, Martinek GW, Ebersold WT. Linkage of six genes in Chlamydomonas reinhardtii and the construction of linkage test strains. J Bacteriol. 1975;124:1615–7. doi: 10.1128/jb.124.3.1615-1617.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora I, Feldman JL, Marshall WF. PCR-based assay for mating type and diploidy in Chlamydomonas. BioTechniques. 2004;37:1–3. doi: 10.2144/04374BM01. [DOI] [PubMed] [Google Scholar]
- Marshall WF, Vucica Y, Rosenbaum JL. Kinetics and regulation of de novo centriole assembly. Implications for the mechanism of centriole duplication. Curr Biol. 2001;11:308–17. doi: 10.1016/S0960-9822(01)00094-X. [DOI] [PubMed] [Google Scholar]
- Chan CS, Botstein D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 1993;135:677–91. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Salmon E. Pathways of spindle assembly. Curr Opin Cell Biol. 1997;9:37–43. doi: 10.1016/S0955-0674(97)80149-4. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/S0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Sluder G, Thompson EA, Miller FJ, Hayes J, Rieder CL. The checkpoint control for anaphase onset does not monitor excess numbers of spindle poles or bipolar spindle symmetry. J Cell Sci. 1997;110:421–429. doi: 10.1242/jcs.110.4.421. [DOI] [PubMed] [Google Scholar]
- Luria SE, Delbruck M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]


