Abstract
8-Oxo-7,8-dihydroguanine (8-oxoGua) is produced in cells by reactive oxygen species normally formed during cellular metabolic processes. This oxidized base can pair with both adenine and cytosine, and thus the existence of this base in messenger RNA would cause translational errors. The MutT protein of Escherichia coli degrades 8-oxoGua-containing ribonucleoside di- and triphosphates to the monophosphate, thereby preventing the misincorporation of 8-oxoGua into RNA. Here, we show that for human the MutT-related proteins, NUDT5 and MTH1 have the ability to prevent translational errors caused by oxidative damage. The increase in the production of erroneous proteins by oxidative damage is 28-fold over the wild-type cells in E.coli mutT deficient cells. By the expression of NUDT5 or MTH1 in the cells, it is reduced to 1.4- or 1.2-fold, respectively. NUDT5 and MTH1 hydrolyze 8-oxoGDP to 8-oxoGMP with Vmax/Km values of 1.3 × 10−3 and 1.7 × 10−3, respectively, values which are considerably higher than those for its normal counterpart, GDP (0.1–0.5 × 10−3). MTH1, but not NUDT5, possesses an additional activity to degrade 8-oxoGTP to the monophosphate. These results indicate that the elimination of 8-oxoGua-containing ribonucleotides from the precursor pool is important to ensure accurate protein synthesis and that both NUDT5 and MTH1 are involved in this process in human cells.
INTRODUCTION
Oxidation of the components of nucleic acids occurs through normal cellular metabolism, especially in aerobic states. More than 20 different types of oxidatively altered purine and pyrimidine bases have been detected in nucleic acids (1,2). Among them, an oxidized form of guanine base, 8-oxo-7,8-dihydroguanine (8-oxoGua), has a potential to alter genetic information, since it can pair with adenine and cytosine at almost equal efficiencies (3–5). 8-OxoGua-containing nucleotides can be incorporated into DNA as well as RNA, and would cause both replicational and transcriptional errors (6–8).
Studies of Escherichia coli mutator mutants have revealed that organisms possess mechanisms to prevent spontaneous mutations caused by misincorporation of 8-oxoGua into DNA. In this organism, MutT protein, which has a potent activity to degrade 8-oxoGua-containing deoxyribonucleoside triphosphate, is almost solely responsible for reducing the level of mutagenic nucleotides in cell (9,10). Mammalian cells also possess enzymes capable of eliminating 8-oxoGua-containing nucleotides from the DNA precursor pool. These include MTH1 (11,12), MTH2 (NUDT15) (13) and NUDT5 (14). MTH1 and MTH2 degrade preferentially 8-oxodGTP, whereas NUDT5 specifically hydrolyses 8-oxodGDP but hardly 8-oxodGTP. Despite their different substrate specificities, all of these mammalian proteins have abilities to replace the MutT function. When each of the cDNAs for these proteins was expressed in mutT-defective E.coli mutant cells, the mutator phenotype was almost completely suppressed (13–15).
The error rate of transcription is estimated to be ∼10−5 per residue (16), and the fidelity of transcription is worse in an aerobic state. It has been shown that 8-oxoGua can be incorporated into RNA by the normal action of RNA polymerase, and that the E.coli MutT protein has the ability to prevent this misincorporation (17). The MutT catalyzes the hydrolysis of both 8-oxoGDP and 8-oxoGTP to the monophosphate, thereby cleaning up the nucleotide pool to ensure accurate transcription (18). 8-OxoGua can be incorporated into RNA by mammalian RNA polymerase II (19). For mammalian cells, particularly those which rarely undergo cell division, the conservation of RNA seems to be important. In fact, a relatively large amount of 8-oxoGua was formed in the RNA in the neuronal tissues of patients suffering from some neurodegenerative diseases, such as Parkinson's disease (20,21), Alzheimer's disease (21) and Down's syndrome (22). However, the mechanisms for cleaning up the ribonucleotide pools of mammalian cells remain unclear. The actions of the mammalian MutT-related proteins on 8-oxoGua-containing ribonucleotides have not been reported, except for MTH1, which has been shown to carry a weak hydrolytic activity for 8-oxoGTP (19). Therefore, we initiated a survey of human enzymes that act on 8-oxoGua-containing ribonucleotides. Here, we report that among the three human MutT-related proteins, NUDT5 and MTH1, but not MTH2, are capable of eliminating oxidized ribonucleotides from the RNA precursor pool.
MATERIALS AND METHODS
Bacterial strains and plasmids
E.coli 101 (wild type) and 101T (mutT-deficient) carry an amber mutation in codon 461 of the lacZ gene, where the A:T to C:G transversion mutation is reversed phenotypically to produce the β-galactosidase protein (17). Plasmid pTT100 is a derivative of pTrc99A (Amersham Pharmacia) lacking the lacIq gene (15). pTT100::hMTH1 and pTT100::hNUDT5 were constructed as described previously (14,15). The human MTH2 cDNA in the I.M.A.G.E. clone 4472716 (purchased from Kurabo, gene bank accession no. 12761620) was PCR amplified and cloned into the KpnI/HindIII site of pTT100 using two primers, 5′-CGCTGGTACCATGACGGCCAGCGCACAGCC-3′ and 5′-GGAAAGCTTAAGAGACTGCAAATAAACTG-3′, to generate pTT100::hMTH2.
Measurement of β-galactosidase activity
5-Bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal, Wako) was dissolved in dimethylformamide (100 mg/ml) and added to Luria–Bertani medium (10 g of bacto-tryptone, 5 g of bacto-yeast extract and 10 g of NaCl per liter) to give a final concentration of 0.5 mg/ml. Quantitative assays for β-galactosidase activity were performed by measuring the UV absorbance at 420 nm, which represents the hydrolysis of o-nitrophenyl-β-d-galactoside (ONPG, Sigma) to o-nitrophenol. Three independent clones from each transformant were used, and at least four experiments were performed with each clone.
Hydrolysis of 8-oxoGua-containing ribonucleotides
8-OxoGTP, 8-oxoGDP and 8-oxoGMP were prepared as described previously (23). The reaction mixture for the hydrolysis assay contained 20 mM Tris–HCl, pH 8.0, 80 μg/ml BSA, 8 mM MgCl2, 40 mM NaCl, 5 mM DTT, 2% glycerol and the protein sample to be examined. The reaction was run at 30°C for 10–30 min and was terminated by adding SDS to a final concentration of 0.1%. The samples were fractionated by high-performance liquid chromatography (HPLC) with a TSK-GEL DEAE-2SW column (Tosoh) in an isocratic flow of 0.1 M sodium phosphate (pH 6.0)–40% acetonitrile at a flow rate of 0.9 ml/min. Nucleotides were quantified by measuring the area of UV absorbance, using a Whatmann HPLC detection system and the Millennium program. The relative velocity for hydrolysis was determined in time-course experiments. The substrate concentrations for NUDT5 ranged from 1 to 50 μM for 8-oxoGDP, 20 to 500 μM for GDP and 50 to 2000 μM for 8-oxoGTP and GTP. For MTH1, they ranged from 5 to 400 μM for 8-oxoGDP, 0.1 to 3 mM for GDP, 40 to 2000 μM for 8-oxoGTP and 0.2 to 4 mM for GTP. Km and Vmax were obtained from Lineweaver–Burk plots of the data.
Analysis of reaction products
The [γ-32P]labeled 8-oxoGTP was prepared by the oxidation of [γ-32P]labeled GTP (MP Biomedicals, Inc.) and was purified as described previously (18). The enzyme reaction was carried out as mentioned above, and an aliquot of the reaction mixture was spotted onto a PEI-cellulose plate (MERCK) and developed in 2 M LiCl/0.2 M Na2HPO4 (1:1). Yeast pyrophosphatase was purchased from Sigma and the reaction was performed at 30°C for 30 min.
RESULTS
Prevention of transcriptional errors caused by oxidized guanine nucleotides
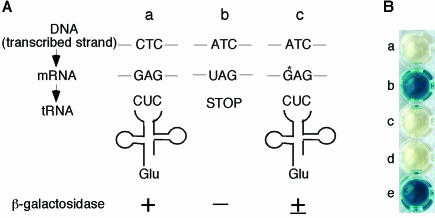
8-OxoGTP is produced by either the oxidation of GTP or the phosphorylation of 8-oxoGDP, the latter of which is formed by the oxidation of GDP (18). Once 8-oxoGTP is formed, this oxidized nucleotide would be incorporated into RNA and cause transcriptional errors (17). Since 8-oxoG is placed opposite adenine in the DNA template, this misincorporation would cause partial phenotypic suppression when an E.coli lacZ− strain with a 5′-T·A·G-3′ stop codon is used as a tester strain (Figure 1A). In the presence of 8-oxoGTP, the 3′-A·T·C-5′ trinucleotide in the transcribed strand of the mutant would be copied to 5′-8-oxoG·A·G-3′, which would then pair with 5′-C·U·C-3′ glutamic acid anticodon. This RNA transcript would encode a wild-type β-galactosidase protein, whereas the vast majority of the mRNAs encode truncated proteins. We have used this assay system to examine the abilities of human proteins to eliminate the mismatch-evoking oxidized nucleotides from the RNA precursor pool.
Figure 1.
Partial phenotypic suppression by 8-oxoGua. (A) Schematic explanation. a, Lac+ cells; b, Lac− cells with an amber mutation at codon 461 of the lacZ gene; c, the same Lac− cells with an additional mutT− mutation. 8-OxoGua (G*) can be incorporated by the RNA polymerase opposite adenine in the DNA template during transcription. The 8-oxoG·A·G could pair with the CUC anticodon of tRNA for glutamic acid, thereby allowing elongation of the β-galactosidase polypeptide chain. The cells would then have low levels of β-galactosidase activity. (B) Tests for production of β-galactosidase. E.coli 101 (wild type) or 101T (mutT-deficient) cells harboring various plasmids were grown for 12 h in the presence of 0.5 mg/ml of X-gal, and the supernatants of the cultures were placed in the wells. a, 101 (wild type); b, 101T (mutT−); c, 101T with cDNA for human NUDT5. d, 101T with cDNA for human MTH1; e, 101T with cDNA for human MTH2.
The β-galactosidase activities produced by partial phenotypic suppression are relatively low, but can be detected when cells are cultured in the presence of X-gal, as shown in Figure 1B. E.coli 101 mutT+ cells carrying an amber mutation at codon 461 in the lacZ gene yield white colonies, since they are unable to produce an active β-galactosidase protein. On the other hand, 101T cells, which carry a mutT mutation in addition to the lacZ amber mutation, produce blue colonies, probably due to the partial phenotypic suppression of the lacZ mutation caused by the misincorporation of 8-oxoGua into mRNA (17) (Table 1). When the cDNA for either NUDT5 or MTH1 was introduced into the 101T cells, the formation of blue colonies was almost completely suppressed, implying that these human proteins can replace the defective MutT function in E.coli cells. On the other hand, no suppression was induced by the expression of MTH2.
Table 1.
Partial phenotypic suppression of an amber mutation of the lacZ gene
| E.coli strain | Plasmid | β-Galactosidase activitya (Miller units) | Relative value |
|---|---|---|---|
| 101 (wild type) | pTT100 | 0.012 ± 0.002 | 1 |
| 101T (mutT−) | pTT100 | 0.34 ± 0.17 | 28 |
| 101T (mutT−) | pTT100::hNUDT5 | 0.017 ± 0.005 | 1.4 |
| 101T (mutT−) | pTT100::hMTH1 | 0.014 ± 0.002 | 1.2 |
| 101T (mutT−) | pTT100::hMTH2 | 0.41 ± 0.14 | 34 |
aAverage ± standard error of all independent experiments for each transformant.
More quantitative data were obtained by measuring the actual β-galactosidase activities in the cultures of E.coli 101T cells harboring plasmids bearing various cDNAs. An ∼30-fold increase in β-galactosidase activity was observed in the mutT-defective 101T strain as compared with the wild-type 101 strain. This increased level of enzyme activity was almost completely suppressed by the expression of the cDNA encoding either NUDT5 or MTH1 in the cells. In accordance with the results of the colony color test, the cDNA encoding MTH2 was unable to prevent the phenotypic suppression.
Actions of NUDT5 protein on oxidized guanine ribonucleotides
The human NUDT5 protein was expressed as a His-tagged protein in E.coli M15 cells and was purified to near homogeneity (14). Assays for enzyme activities were carried out using 5 μM 8-oxoGDP or 8-oxoGTP, and the products were analyzed by HPLC. As shown in Figure 2A, NUDT5 efficiently degraded 8-oxoGDP to its monophosphate form. Under the same conditions, the hydrolysis of 8-oxoGTP was hardly detected.
Figure 2.
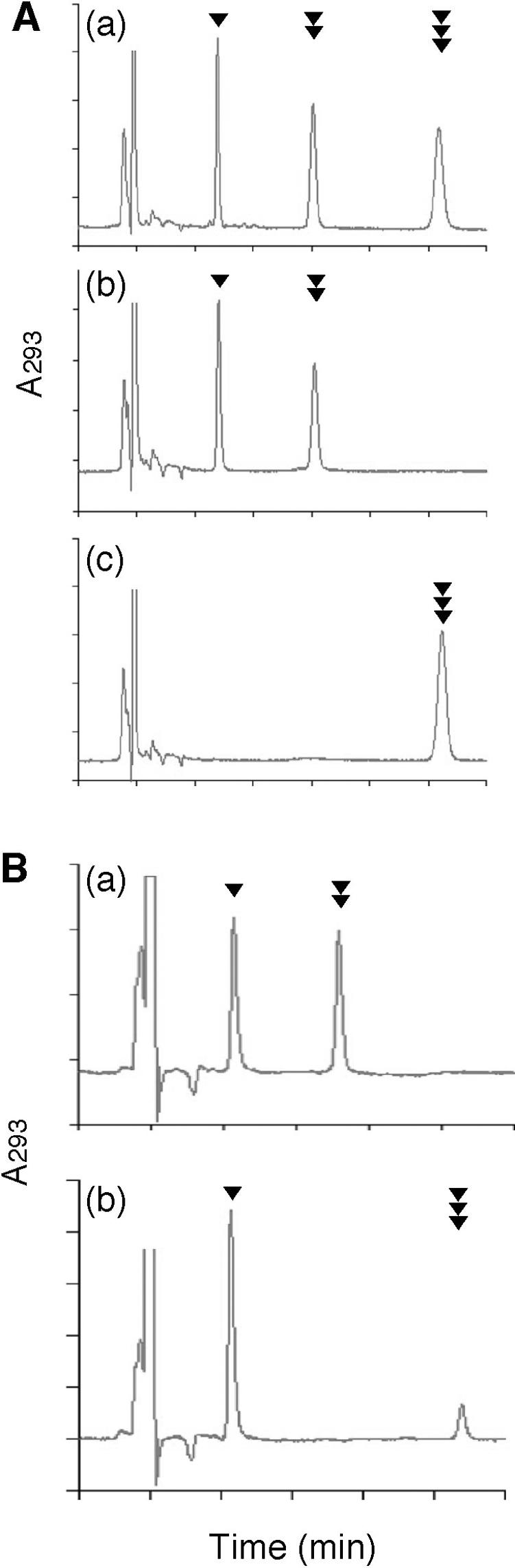
Actions of the NUDT5 and MTH1 proteins toward 8-oxoGTP and 8-oxoGDP. (A) Action of NUDT5. a, Reference nucleotides; b, treatment of 8-oxoGDP with NUDT5 protein; c, treatment of 8-oxoGTP with NUDT5 protein. (B) Action of MTH1. a, treatment of 8-oxoGDP with MTH1 protein; b, treatment of 8-oxoGTP with MTH1 protein. In these experiments, 5 μM of 8-oxoGDP or 8-oxoGTP were incubated with a purified preparation of 0.41 μM human N-terminal His-tagged NUDT5 protein at 30°C for 30 min for (A), or 10 μM 8-oxoGDP or 8-oxoGTP were incubated with a purified preparation of 0.049 μM human MTH1 protein at 30°C for 10 min for (B) under the conditions described in Materials and Methods, and the products were analyzed by HPLC. Arrowheads indicate peaks for 8-oxoGMP (single black triangle), 8-oxoGDP (double black triangles) and 8-oxoGTP (triple black triangles).
The kinetic parameters of the NUDT5 enzyme (Km and Vmax) were measured for the hydrolysis of the normal and oxidized forms of guanine ribonucleotides (Table 2). The Km for the hydrolysis of 8-oxoGDP is ten times lower than that for GDP. The Km values for both 8-oxoGTP and GTP are extremely high and the Vmax values are very low, indicating that these nucleotides are hardly degraded by NUDT5. On the basis of these results, we may conclude that 8-oxoGDP is a specific substrate for the human NUDT5 protein.
Table 2.
Substrate specificity of human NUDT5 and MTH1
| Protein | Substrate | Km (μM) | Vmax (pmol/min/ng) | Vmax/Km (×1000) |
|---|---|---|---|---|
| NUDT5 | 8-oxoGDP | 4.9 | 0.0065 | 1.3 |
| GDP | 48 | 0.023 | 0.49 | |
| 8-oxoGTP | 230 | 0.0021 | 0.009 | |
| GTP | 210 | 0.0091 | 0.04 | |
| MTH1 | 8-oxoGDP | 44 | 0.075 | 1.7 |
| GDP | 420 | 0.048 | 0.11 | |
| 8-oxoGTP | 290 | 12 | 41 | |
| GTP | 790 | 10 | 13 |
Hydrolysis of oxidized guanine ribonucleotides by the MTH1 protein
The human MTH1 protein was purified to homogeneity (24). The data presented in Figure 2B indicate that both 8-oxoGDP and 8-oxoGTP can be hydrolyzed by the MTH1 protein. The incubation of 8-oxoGDP with MTH1 effectively converted it to the monophosphate. A more rapid conversion was observed with 8-oxoGTP; almost all of the triphosphate was converted to the monophosphate, under the same conditions.
The kinetic parameters of the MTH1 protein are also given in Table 2. The Km for the hydrolysis of 8-oxoGDP is 10 times lower than that for the normal counterpart, GDP, as observed with NUDT5. However, the actual Km values of the MTH1 protein are considerably higher than those of the NUDT5 protein. This low affinity of MTH1 for the substrates appears to be compensated by its high velocity in the enzyme reaction. It vastly exceeds the values of NUDT5 for 8-oxoGDP, thus providing almost the same Vmax/Km value for 8-oxoGDP. In addition, the Km of MTH1 for 8-oxoGTP is rather high, and the high Vmax/Km value of this protein was attained by its extremely high Vmax in the 8-oxoGTP cleavage reaction. These characteristics of the MTH1 protein could imply that the protein may act on the two types of error-evoking nucleotides, 8-oxoGDP and 8-oxoGTP, under different circumstances.
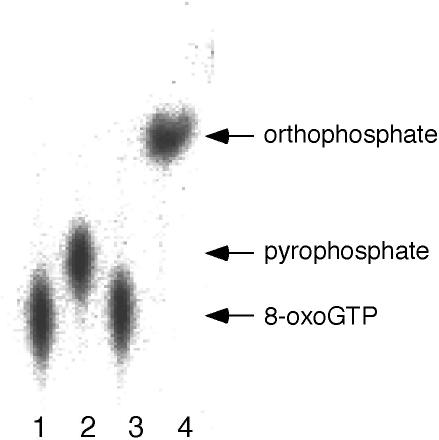
Another question regarding the action of MTH1 is whether the 8-oxoGTP is directly converted to 8-oxoGMP or degraded through the formation of 8-oxoGDP as an intermediate. To distinguish between these possibilities, [γ-32P]labeled 8-oxoGTP was treated with a purified preparation of MTH1 protein and the products were analyzed on thin-layer chromatography. As shown in Figure 3, a spot for [32P]labeled pyrophosphate appeared, which was further converted to orthophosphate by a treatment with pyrophosphatase. It appears that the MTH1 protein has the ability to cleave the phosphoanhydride bond between the α and the β phosphate of 8-oxoGua-containing nucleoside di- and triphosphates.
Figure 3.
Analysis of products cleaved by MTH1. Reaction mixtures (total of 5 μl) containing 50 pmol of [γ-32P]labeled 8-oxoGTP were incubated with or without enzymes at 30°C for 30 min and the reaction products were separated on thin-layer chromatography. Lane 1, no enzyme; lane 2, treated with 5 ng of purified MTH1 protein; lane 3, treated with 0.05 unit of yeast inorganic pyrophosphatase; lane 4, treated with both 5 ng of MTH1 protein and 0.05 U of yeast inorganic pyrophosphatase.
DISCUSSION
The basal level of spontaneous errors in RNA synthesis is estimated to be 10−5 per residues, which is much higher than the error rate of DNA replication (10−9 per residues). This means that numerous erroneous proteins are synthesized in a normal cell. Furthermore, the transcriptional fidelity would become lower when the RNA bases are modified by internal or external agents. Among such modifications, 8-oxoGua is particularly important since this modified base can pair with adenine and cytosine at almost equal efficiencies (3–5). It has been shown that the incorporation of 8-oxoGua opposite the adenine residues of DNA by RNA polymerase causes partial phenotypic suppression (17). In this case, although the majority of the proteins are normal, some of the products are abnormal; erroneous proteins may lose their functions or exhibit dominant characteristics to cause disorders of some cellular functions, which could lead to catastrophic consequences. In mammals, many differentiated cells remain in the G1/G0 state and exert their cellular functions via interactions with sophisticated networks. The dysfunction of a single cell, caused by the accumulation of proteins translated from the erroneous RNA, may be amplified with increasing age. Therefore, mechanisms to control the RNA quality would be important in facilitating the normal functions of organisms.
8-OxoGua can be formed in RNA by the direct oxidation of the base and also by the incorporation of the oxidized base into RNA (17). Once 8-oxoGua is formed in RNA, it cannot be eliminated, in contrast to the case of in DNA, in which damaged bases are excised by a specific glycosylase and repaired (25). Thus, organisms must be equipped with other mechanisms to maintain the high quality of RNA against oxidative stress. Proteins that specifically bind to oxidized RNA have been implicated in a mechanism to scavenge damaged RNA. The E.coli polynucleotide phosphorylase (PNP) protein and the humanYB1 protein function in such mechanisms (26,27). Another mechanism to prevent transcriptional errors caused by oxidative damage is the sanitization of nucleotide pools. The E.coli MutT protein eliminates 8-oxoGTP and prevents the occurrence of transcriptional errors, which are induced particularly in the aerobic state (17). Recently, it was revealed that the MutT protein has an additional activity to sanitize the nucleotide pools; it degrades 8-oxoGDP as efficiently as 8-oxoGTP (18). However, in mammalian cells it is unclear what types of enzymes are involved in this process. These situations prompted us to search for enzymes that eliminate 8-oxoGua-containing ribonucleotides from the RNA precursor pool in mammalian cells. In the present study, we identified two MutT-related proteins, MTH1 and NUDT5, as the candidates responsible for this process.
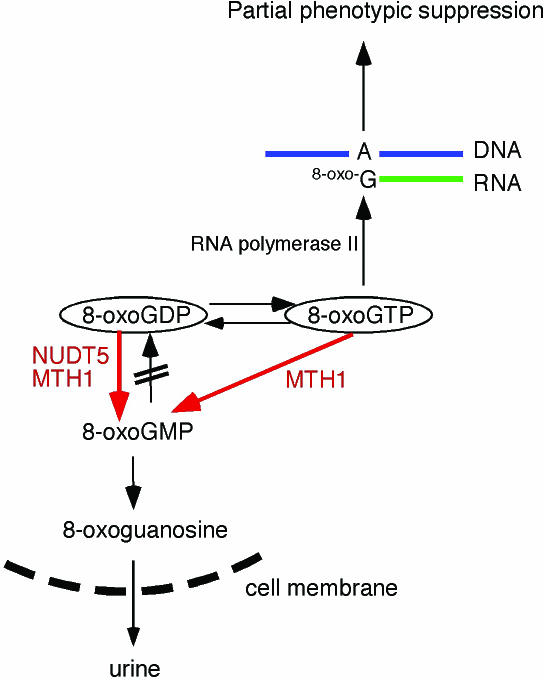
Both MTH1 and NUDT5 exhibit considerably lower Km values for 8-oxoGDP than for 8-oxoGTP. In the case of MTH1, the selective cleavage of 8-oxoGTP was also observed. Since 8-oxoGTP and 8-oxoGDP are interconvertible within the cell (19), NUDT5 and MTH1 may collaborate to prevent the misincorporation of 8-oxoGua into RNA. These situations can be seen in Figure 4. The relatively high Vmax/Km values for the hydrolysis of 8-oxoGDP by NUDT5 and MTH1 (1.3 × 10−3 and 1.7 × 10−3, respectively) support this view (Figure 1). On the other hand, the Vmax/Km of MTH1 for the hydrolysis of 8-oxoGTP is considerably high, implying that MTH1 is capable of eliminating 8-oxoGTP from the precursor pool. In contrast to MTH1, NUDT5 hardly acts on 8-oxoGTP, yet the expression of NUDT5 prevented of partial phenotypic suppression in the mutT-deficient background. Taking all of these results into account, it is conceivable that the hydrolysis of 8-oxoGDP may be sufficient for the high fidelity of RNA synthesis. To establish the biological significance of these enzymes, it is necessary to construct mouse models deficient in one or both of the activities. These studies are in progress in this laboratory.
Figure 4.
A model for the exclusion of 8-oxoGua-containing ribonucleotides from the RNA precursor pools in mammalian cells. 8-OxoGTP and 8-oxoGDP are interconvertible by the actions of nucleoside diphosphate kinase and nucleoside triphosphatase. Partial phenotypic suppression is caused by the misincorporation of 8-oxoGTP into RNA by RNA polymerase II. 8-OxoGDP is hydrolyzed to 8-oxoGMP by NUDT5 and MTH1, while 8-oxoGTP is cleaved directly to 8-oxoGMP by MTH1. 8-OxoGMP, a non-utilizable form for RNA synthesis, is further degraded to 8-oxoguanosine by nucleotidases. Nucleosides are readily transported through the cell membrane and can be excreted into the urine.
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by Biomolecular Engineering Research Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gajewski E., Rao G., Nackerdien Z., Dizdaroglu M. Modification of DNA bases in mammalian chromatin by radiation-generated free radicals. Biochemistry. 1990;29:7876–7882. doi: 10.1021/bi00486a014. [DOI] [PubMed] [Google Scholar]
- 2.Demple B., Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 3.Wood M.L., Dizdaroglu M., Gajewski E., Essigmann J.M. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 4.Moriya M., Ou C., Bodepudi V., Johnson F., Takeshita M., Grollman A.P. Site-specific mutagenesis using a gapped duplex vector: a study of translesion synthesis past 8-oxodeoxyguanosine in E. coli. Mutat. Res. 1991;254:281–288. doi: 10.1016/0921-8777(91)90067-y. [DOI] [PubMed] [Google Scholar]
- 5.Shibutani S., Takeshita M., Grollman A.P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 6.Kasai H., Crain P.F., Kuchino Y., Nishimura S., Ootsuyama A., Tanooka H. Formation of 8-hydroxyguanine moiety in cellular DNA by agents producing oxygen radicals and evidence for its repair. Carcinogenesis. 1986;7:1849–1851. doi: 10.1093/carcin/7.11.1849. [DOI] [PubMed] [Google Scholar]
- 7.Ames B.N., Gold L.S. Endogenous mutagens and the causes of aging and cancer. Mutat. Res. 1991;250:3–16. doi: 10.1016/0027-5107(91)90157-j. [DOI] [PubMed] [Google Scholar]
- 8.Sekiguchi M., Tsuzuki T. Oxidative nucleotide damage: consequences and prevention. Oncogene. 2002;21:8895–8904. doi: 10.1038/sj.onc.1206023. [DOI] [PubMed] [Google Scholar]
- 9.Yanofsky C., Cox E.C., Horn V. The unusual mutagenic specificity of E. coli mutator gene. Proc. Natl Acad. Sci. USA. 1966;55:274–281. doi: 10.1073/pnas.55.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maki H., Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 11.Mo J.Y., Maki H., Sekiguchi M. Hydrolytic elimination of a mutagenic nucleotide, 8-oxodGTP, by human 18-kilodalton protein: sanitization of nucleotide pool. Proc. Natl Acad. Sci. USA. 1992;89:11021–11025. doi: 10.1073/pnas.89.22.11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakumi K., Furuichi M., Tsuzuki T., Kakuma T., Kawabata S., Maki H., Sekiguchi M. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J. Biol. Chem. 1993;268:23524–23530. [PubMed] [Google Scholar]
- 13.Cai J.P., Ishibashi T., Takagi Y., Hayakawa H., Sekiguchi M. Mouse MTH2 protein which prevents mutations caused by 8-oxoguanine nucleotides. Biochem. Biophys. Res. Commun. 2003;305:1073–1077. doi: 10.1016/s0006-291x(03)00864-7. [DOI] [PubMed] [Google Scholar]
- 14.Ishibashi T., Hayakawa H., Sekiguchi M. A novel mechanism for preventing mutations caused by oxidation of guanine nucleotides. EMBO Rep. 2003;4:479–483. doi: 10.1038/sj.embor.embor838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furuichi M., Yoshida M.C., Oda H., Tajiri T., Nakabeppu Y., Tsuzuki T., Sekiguchi M. Genomic structure and chromosome location of the human mutT homologue gene MTH1 encoding 8-oxo-dGTPase for prevention of A:T to C:G transversion. Genomics. 1994;24:485–490. doi: 10.1006/geno.1994.1657. [DOI] [PubMed] [Google Scholar]
- 16.Ninio J. Connections between translation, transcription and replication error-rates. Biochimie. 1991;73:1517–1523. doi: 10.1016/0300-9084(91)90186-5. [DOI] [PubMed] [Google Scholar]
- 17.Taddei F., Hayakawa H., Bouton M., Cirinesi A., Matic I., Sekiguchi M., Radman M. Counteraction by MutT protein of transcriptional errors caused by oxidative damage. Science. 1997;278:128–130. doi: 10.1126/science.278.5335.128. [DOI] [PubMed] [Google Scholar]
- 18.Ito R., Hayakawa H., Sekiguchi M., Ishibashi T. Multiple enzyme activities of Escherichia coli MutT protein for sanitization of DNA and RNA precursor pools. Biochemistry. 2005;44:6670–6674. doi: 10.1021/bi047550k. [DOI] [PubMed] [Google Scholar]
- 19.Hayakawa H., Hofer A., Thelander L., Kitajima S., Cai Y., Oshiro S., Yakushiji H., Nakabeppu Y., Kuwano M., Sekiguchi M. Biochemistry. 1999;38:3610–3614. doi: 10.1021/bi982361l. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J., Perry G., Smith M.A., Robertson D., Olson S.J., Graham D.G., Montine T.J. Parkinson's disease is associated with oxidative damage to cytoplasmic DNA and RNA in substantia nigra neurons. Am. J. Pathol. 1999;154:1423–1429. doi: 10.1016/S0002-9440(10)65396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nunomura A., Perry G., Aliev G., Hirai K., Takeda A., Balraj E.K., Jones P.K., Ghanbari H., Wataya T., Shimohama S., et al. Oxidative damage is the earliest event in Alzheimer disease. J. Neuropathol. Exp. Neurol. 2001;60:759–767. doi: 10.1093/jnen/60.8.759. [DOI] [PubMed] [Google Scholar]
- 22.Nunomura A., Perry G., Pappolla M.A., Friedland R.P., Hirai K., Chiba S., Smith M.A. Neuronal oxidative stress precedes amyloid-beta deposition in Down syndrome. J. Neuropathol. Exp. Neurol. 2000;59:1011–1017. doi: 10.1093/jnen/59.11.1011. [DOI] [PubMed] [Google Scholar]
- 23.Fujikawa K., Kamiya H., Yakushiji H., Fujii Y., Nakabeppu Y., Kasai H. The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein. J. Biol. Chem. 1999;274:18201–18205. doi: 10.1074/jbc.274.26.18201. [DOI] [PubMed] [Google Scholar]
- 24.Yakushiji H., Maraboeuf F., Takahashi M., Deng Z., Kawabata S., Nakabeppu Y., Sekiguchi M. Biochemical and physicochemical characterization of normal and variant forms of human MTH1 protein with antimutagenic activity. Mutat. Res. 1997;384:181–194. doi: 10.1016/s0921-8777(97)00025-6. [DOI] [PubMed] [Google Scholar]
- 25.Michaels M.L., Cruz C., Grollman A.P., Miller J.H. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl Acad. Sci. USA. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayakawa H., Kuwano M., Sekiguchi M. Specific binding of 8-oxoguanine-containing RNA to polynucleotide phosphorylase protein. Biochemistry. 2001;40:9977–9982. doi: 10.1021/bi010595q. [DOI] [PubMed] [Google Scholar]
- 27.Hayakawa H., Uchiumi T., Fukuda T., Ashizuka M., Kohno K., Kuwano M., Sekiguchi M. Binding capacity of human YB-1 protein for RNA containing 8-oxoguanine. Biochemistry. 2002;41:12739–12744. doi: 10.1021/bi0201872. [DOI] [PubMed] [Google Scholar]