Abstract
Targeted mutagenesis directed by oligonucleotides (ONs) is a promising method for manipulating the genome in higher eukaryotes. In this study, we have compared gene editing by different ONs on two new target sequences, the eBFP and the rd1 mutant photoreceptor βPDE cDNAs, which were integrated as single copy transgenes at the same genomic site in 293T cells. Interestingly, antisense ONs were superior to sense ONs for one target only, showing that target sequence can by itself impart strand-bias in gene editing. The most efficient ONs were short 25 nt ONs with flanking locked nucleic acids (LNAs), a chemistry that had only been tested for targeted nucleotide mutagenesis in yeast, and 25 nt ONs with phosphorothioate linkages. We showed that LNA-modified ONs mediate dose-dependent target modification and analyzed the importance of LNA position and content. Importantly, when using ONs with flanking LNAs, targeted gene modification was stably transmitted during cell division, which allowed reliable cloning of modified cells, a feature essential for further applications in functional genomics and gene therapy. Finally, we showed that ONs with flanking LNAs aimed at correcting the rd1 stop mutation could promote survival of photoreceptors in retinas of rd1 mutant mice, suggesting that they are also active in vivo.
INTRODUCTION
Targeted mutagenesis is a major method of genetics which, in practice, still remains restricted to the mouse model organism and a few specific cell lines in other higher eukaryotes (1). Furthermore, it relies on homologous recombination between the targeted gene and large DNA molecules requiring the construction of complex plasmid, BAC or adeno-associated viral vectors. Therefore, strategies using simpler vectors for targeted mutagenesis, such as oligonucleotides (ONs) or small DNA fragments, constitute attractive alternatives to manipulate the genome for both experimental and therapeutic purposes. ON-mediated targeted mutagenesis has been used to correct point mutations in cultured cells and in animal models of human disease (2–4). Several types of ONs have been shown to be active, including small RNA/DNA ONs, single-stranded and branched ONs (3–5). Triplex-forming ONs have also been used to induce sequence-specific deletions (6). However, the critical experimental parameters and the mechanisms involved in these methods are not well known. Indeed, the high mutagenesis rates originally reported when using small RNA/DNA ONs could often not be reproduced on different targets or on identical targets using seemingly identical experimental conditions (3,7,8). Some authors have suggested that mutagenesis rates could have been overestimated in some studies because of PCR artifacts (3,9). Differences in the sequences targeted as well as in the assays used to measure mutagenesis rates make it difficult to compare the activity of the vectors described in the various studies.
In this study, we have compared the targeting efficiency of several types of ONs previously shown to mediate targeted mutagenesis on both episomal and integrated single copy chromosomal targets. ONs with flanking LNAs, which contain a methylene bridge connecting the 2′-oxygen of ribose with the 4′-carbon (10), had been previously shown to direct targeted nucleotide modification in yeast (11), but their activity had not been reported in higher eukaryotes so far. Here, we show that short specific ONs with flanking LNAs induced the desired mutation at the DNA level and, importantly, that modified cell clones could be isolated and grown for each of the sequences targeted, the eBFP and the rd1 mutant photoreceptor cDNAs. As previously shown for phosphorothioate ONs (C. Andrieu-Soler et al., manuscript submitted), specific ONs with flanking LNAs could increase the number of photoreceptors in vivo in the retina of rd1 mice (12). Taken together, our results suggest that short ONs with flanking LNAs can be powerful reagents for targeted mutagenesis in cultured cells and in animal models of human disease.
MATERIALS AND METHODS
Oligonucleotides
ONs were synthesized and purified by high-pressure liquid chromatography by Proligo (Paris, France) with the exception of the branched-PS-eGFP ON, which was synthesized by Eurogentech. ONs in distilled water were quantified by absorbance at 260 nm.
Plasmids
The pFRT/lacZeo vector contains a FRT site and a LacZ–Zeocin fusion gene (Invitrogen, Cergy Pontoise, France). pOG44 vector (Invitrogen) constitutively expresses the Flp recombinase (13). peBFP and peGFP* were constructed by inserting eBFP cDNA and eGFP cDNA, respectively, without the translation initiation codon into BamH1/Xho1 sites of the pcDNA5/FRT/TO plasmid (Invitrogen). prd1-eGFP* and pPDE-eGFP* were constructed by inserting a 153 bp fragment containing βPDE sequence spanning the rd1 mutation or its wild-type counterpart, respectively, in HindIII/BamH1 sites of the peGFP* (inserting a shorter fragment of rd1 sequence in peGFP* or the 153 bp fragment in a peGFP containing the translation initiation codon led to background green fluorescence).
Cells cultures and establishment of stable cell lines
All media, fetal bovine serum (FBS) and Penicillin–Streptomycin were from Gibco BRL. 293T cells were maintained in DMEM supplemented with 5% FBS (optionally supplemented with Zeocin or 50 μg/ml hygromycin). Following electroporation of linearized pFRT/LacZeo plasmid, 293T cell clones having integrated pFRT/LacZeo were selected on Zeocin. The clones containing a single FRT site were identified by Southern blot analysis. One of these clones was then co-transfected with pOG44 and peBFP. Clones that had integrated the peBFP plasmid at the FRT site via Flp recombinase-mediated DNA recombination were selected with hygromycin. Southern blot analysis was used to confirm that these stable clones contained a single copy of the eBFP cDNA. The same protocol was used to obtain 293T cells with an integrated single copy of the rd1/eGFP* fusion construction.
In vitro analysis of gene conversion by episomal transfection
One day before transfection, 3 × 105 293T cells were plated per well for a 4 cm2 12-well plate. For one well, 2.5 μl of Plus Reagent (Invitrogen) were added to a mixture of 66 pmol of ON and 0.5 μg of plasmid diluted in 62.5 μl of Opti-MEM 1. After 15 min of incubation at room temperature, 2 μl of Lipofectamine (Invitrogen) diluted in 62.5 μl of Opti-MEM 1 were added. After 15 min of incubation at room temperature, the mixture was added to the cells plated in 750 μl of appropriated DMEM growth medium with 5% FBS without antibiotic. After 3 h at 37°C, 750 μl of DMEM growth medium with 15% FBS was added. The percentage of episomal eBFP conversion induced by different ONs (see Figure 2) was quantified by flow cytometry (see below) by dividing the percentage of green fluorescent protein (GFP)-positive cells per the percentage of BFP-positive cells. Each experiment was repeated at least three times.
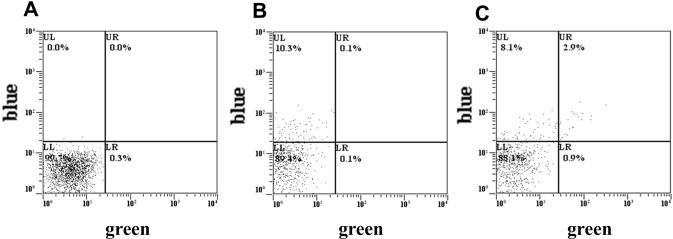
Figure 2.
Conversion of episomal eBFP to GFP. Representative dotplots of 293T cells 3 days after co-transfection with (A) pLacZ and 45-PS-eGFPas ON, (B) peBFP and 25-PS-PDEas ON and (C) peBFP and 45-PS-eGFPas ON as detailed in Materials and Methods. The control ON, 25-PS-PDEas, was of sequence complementary to βPDE. The control plasmid, pLacZ, was an expression vector for β-galactosidase. Blue fluorescent only cells are in the upper left quadrant (UL) and green and blue fluorescent cells, corresponding to converted cells, in the upper right (UR).
In vitro analysis of gene conversion by ON transfection in stable cell lines
Optimal percentage of conversion was obtained with Lipofectamine 2000 using the following protocol. One day before transfection, 2.2 × 105 293T cells were plated per well for a 2 cm2 24-well plate. For one well, 2.5 μl of Lipofectamine 2000 were diluted in 62.5 μl of Opti-MEM 1 medium (Gibco BRL). After 5 min at room temperature, Lipofectamine 2000 solution was mixed to 132 pmol of ON pre-diluted in 62.5 μl of Opti-MEM 1 and incubated for 20 min at room temperature. The mixture was added to the cells plated in 375 μl of appropriated growth medium with 5% FBS without antibiotic. The resulting ON concentration was therefore of 264 nM. After 1 h at 37°C, the medium was replaced by growth medium with 5% FBS with hygromycin. The total number of green fluorescent cells (that were not red fluorescent cells) contained in each well was counted under a 10× objective on a DM-IRB fluorescence microscope (Leica, Rueil Malmaison, France) with a mobile stage. The percentage of integrated eBFP conversion induced by different ONs (see Figure 2) was quantified by reporting this number by the total number of cells. In cells containing rd1-GFP construct, conversion of the rd1 stop codon mutation lead to the expression of GFP but fluorescence levels were too low for visual counting. The percentage of GFP-positive cells was therefore quantified by flow cytometry (as described below). Each experiment was repeated at least three times.
Flow cytometry
Transfected 293T cells were analyzed on an EPICS Altra flow cytometer (Beckman Coulter). The 488 nm argon-ion laser was run at 250 mW of power, and the second argon laser was tuned to 325 nm UV and operated at 50 mW. The green fluorescence was collected after a 525–25 nm band pass filter. For analysis, the blue emission from HO342 was filtered through a 424/44 band pass filter and a minimum of 10 000 events was collected in a gate of viable cells. Analysis of the data was performed with expo32 MultiCOMP software (Beckman Coulter). After cell sorting, green-positive cells were seeded in a 96-well plate and clones were expanded on DMEM growth medium with 5% FBS and 50 μg/ml hygromycin. One 96-well plate allowed growing between 4 and 10 clones of green cells when starting from cells transfected with LNA-modified ONs (n = 6 plates), whereas no clones could be expanded from cells transfected with phosphorothioate-modified ONs (n = 6 plates). These data suggest that fraction cell survival is ∼4–10% for LNA ON-treated cells and <0.1% (1/600) for phosphorothioate-treated cells.
DNA sequence analyses
Genomic DNA was extracted from expanded GFP-positive clones by isopropanol precipitation following lysis by proteinase K digestion. Corrected eBFP cDNA was amplified by PCR using CMV-F (cytomegalovirus-F) (5′-CGCAAATGGGCGGTAGGCGTG-3′) and BGH-R (5′-TAGAAGGCACAGTCGAGG-3′) primers and sequenced by MWG-Biotech (Germany).
Analysis of gene correction in rd1 mutant mice
C3H/HeN mice homozygous for the nonsense mutation (amino acid position 347) in the βPDE gene (Janvier, Le Genest, France) were used. Mice were maintained in clear plastic cages and subjected to a standard light:dark cycle for 12 h. Experiments were conducted in accordance with the ARVO Statement for the Use of Animals in Ophthalmologic and Vision Research and the institutional guidelines regarding animal experimentation in Ophthalmic and Vision Research. At days PN4, 6 and 8, rd1 mice received saline iontophoresis coupled to phosphate-buffered saline (PBS) or ONs intravitreal injections (further details are reported elsewhere, C. Andrieu-Soler et al., manuscript submitted). For tissue harvest, mice were sacrificed by a lethal dose of pentobarbital (Ceva Santé Animale, Libourne, France) injected intraperitoneally. Eyes of PN28 untreated, PBS- or ON-treated (six eyes for each condition) rd1 mice were enucleated, quickly frozen in Tissue-Tek OCT-compound (Bayer Diagnostics) and sectioned (10 μm). For each eye, five sections that included optic nerve were stained with hematoxylin and eosin. For each section, the number of nuclei in the outer nuclear layer (ONL) was counted at 400 μm from each edge of the optic nerve on a 400 μm length (n = 10 values for each eye). Results were expressed as means ± SD and compared using the non-parametric Mann–Whitney test. P < 0.05 was considered as significant.
RESULTS
eBFP cDNA as a target for sequence-specific mutation
Blue fluorescent coding sequence was originally obtained by screening GFP mutants for altered fluorescence. It was found to depart from GFP cDNA by only two point mutations, a C to T substitution resulting in a Y66H change in the fluorophore and a second substitution resulting in a Y145F change in a region considered to have no role in fluorescence specificity (14). We therefore tested whether ONs of eGFP sequence spanning codon 66 could convert eBFP cDNA into a new GFP cDNA (coined nGFP, Figure 1) leading to the expression of a GFP in two contexts: episomal or integrated as a single copy transgene in cultured cells. nGFP differs from eGFP at codon145, making it possible to distinguish bona fide conversion from potential contamination by eGFP cDNA (Figure 1). Importantly, in this model, stable transmission of the targeted gene modification can be examined by selection and expansion of green fluorescent cells.
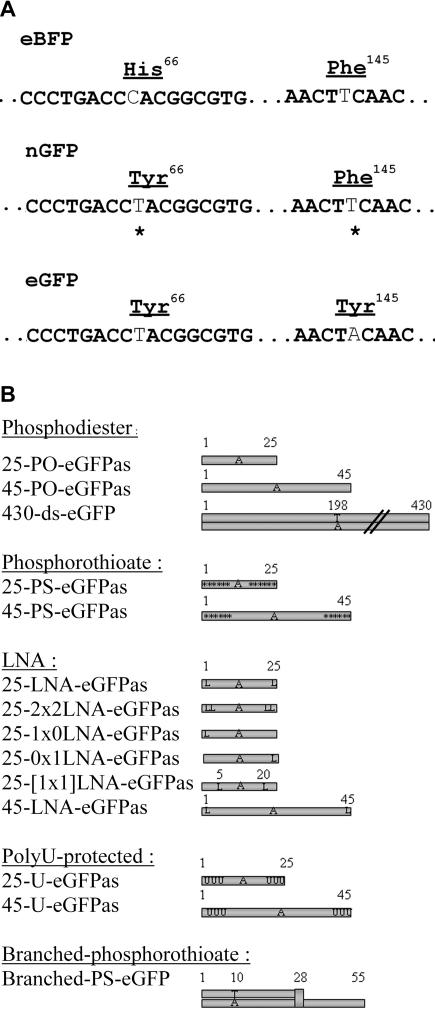
Figure 1.
Features of assay for measuring gene editing. (A) Sequence of eBFP and eBFP converted into a new GFP (nGFP) coding Tyr66 and Phe145. eGFP sequence is also indicated to show the difference from eBFP and nGFP at codon 145. (B) Schematic representation of ONs tested for targeted mutagenesis. Nomenclature: as, antisense; s, sense; ds, double-stranded; PO, phosphodiester; PS, six phosphorothioate linkages starting from 5′ and 3′ ends; asterisk, phosphorothioate linkage; L, LNA nucleotide; LNA, LNA nucleotide at 5′ and 3′ end position; 1x0-LNA, LNA nucleotide at 5′ end only; 0x1-LNA, LNA at 3′ end only; [1x1]-LNA, LNA nucleotides at internal nucleotide positions 5 and 20; U, three 2′-O-methyl uracyl RNA base starting from 5′ and 3′ ends; branched-PS-eGFP, ON with specific structure consisting of double-stranded DNA with 5′ single-stranded DNA tail coupled together by a baseless deoxyribose branching monomer described as B7 ON in (5). 430-ds-eGFP: 1–430 bp of eGFP sequence. Sequences were as follows: 25-eGFPas, 5′-ACTGCACGCCGTAGGTCAGGGTGGT-3′; 25-eGFP-s, 5′-ACCACCCTGACCTACGGCGTGCAGT-3′; 45-eGFPas, 5′-CGGCTGAAGCACTGCACGCCGTAGGTCAGGGTGGTCACGAGGGTG-3′; 45-eGFPs, 5′-CACCCTCGTGACCACCCTGACCTACGGCGTGCAGTGCTTCAGCCG-3′; 25-PDEas, 5′-ACTTTCTGCTACGTAGGTTGGAAGG-3′; 25-PDEs, 5′-CCTTCCAACCTACGTAGCAGAAAGT-3′.
As described in Figure 1, single-stranded ONs of sequence identical to either sense or antisense strands of eGFP cDNA were synthesized according to several structures previously reported to direct sequence-specific gene modification: standard phosphodiester chemistry (25-PO-eGFPs, 25-PO-eGFPas and 45-PO-eGFPas), which was shown to be active in some reports (15), modified with phosphorothioate linkages between the six bases starting from 5′ and 3′ ends (25-PS-eGFPs, 25-PS-eGFPas and 45-PS-eGFPas) (16), with 2′Omethyl-uracyl-RNA at the 3 positions from the 5′ and 3′ ends (25-U-eGFPs, 25-U-eGFPas and 45-U-eGFPas) (17) or with LNAs at the 5′ and 3′ ends (25-LNA-eGFPs, 25-LNA-eGFPas, 45-LNA-eGFPas). A branched ON, consisting of double-stranded DNA with a 5′ single-stranded DNA tail coupled together by a baseless deoxyribose branching monomer, following upon the original design reported by Olsen et al. (5), was also tested (branched-PS-eGFP) as well as a double-stranded fragment, consisting in a PCR product of 430 bp amplified from eGFP cDNA (18) (430-ds-eGFP).
Relative conversion of episomal eBFP sequence
As expected from previous studies, all the ONs encoding eGFP described in Figure 1 allowed detecting green fluorescent cells after being co-transfected with peBFP, whereas a control ON encoding an unrelated sequence did not. We compared the relative activities of the ONs to convert eBFP cDNA into nGFP cDNA by measuring the percentage of green fluorescent cells (converted cells) relative to the number of blue fluorescent cells (transfected cells) by flow cytometry 3 days after transfection (Figure 2 and Table 1). Based on green-cell quantification, only relative conversion rates were calculated but this allows to compare the activity of different ONs. The most efficient were the branched-PS-eGFP and 430-ds-eGFP vectors, which allowed obtaining green fluorescence among ∼74% of transfected cells. The less efficient were 25-U-eGFPas and 25-PO-eGFPas, which allowed detecting only 0.4 and 4.9% green cells, respectively. All other antisense ONs tested showed a roughly equivalent efficiency ranging between 27 and 57%. It appeared that for 25 nt long ONs, the phosphorothioate and LNA modifications tested provided a 7-fold improvement over standard phosphodiester chemistry. In contrast, for 45 nt long ONs, the chemical modifications tested did not significantly improve conversion efficiency. In addition, short 25 nt ONs were superior to their longer 45 nt counterparts. We also observed that, for a given chemistry, both sense and antisense ONs mediated gene modification; however, as frequently reported in other models (3), a higher efficiency was observed with the antisense ONs.
Table 1.
Relative percentage of episomal eBFP conversion
| ON | Percentage of episomal eBFP conversion ± SD |
|---|---|
| 25-LNA-scr | 0.06 ± 0.05 |
| 25-U-eGFPas | 0.4 ± 0.1 |
| 25-LNA-eGFPs | 1.3 ± 0.4 |
| 25-PO-eGFPas | 5 ± 0.7 |
| 25-PS-eGFPs | 14 ± 2 |
| 45-PS-eGFPas | 27 ± 2 |
| 45-PO-eGFPas | 29 ± 4 |
| 25-PS-eGFPas | 36 ± 8 |
| 25-LNA-eGFPas | 37 ± 3 |
| 45-U-eGFPas | 47 ± 6 |
| 45-LNA-eGFPas | 57 ± 2 |
| 430-ds-eGFP | 74 ± 7 |
| Branched-PS-eGFP | 74 ± 5 |
Episomal eBFP conversion frequency was calculated as the number of green and blue fluorescent cells (UR) divided by the number of blue fluorescent cells (UL plus UR), which were determined by flow cytometry at 3 days after transfection in 293T cells of the indicated ONs. Transfection experiments were repeated three times.
Conversion of a single copy eBFP transgene
The same set of ONs was next tested for their ability to convert integrated eBFP cDNA into nGFP cDNA in 293T cells. For this purpose, we created cells containing a unique copy of eBFP, as demonstrated by Southern blot analysis (data not shown), by the application of Flp recombinase methodology (13). Briefly, peBFP, containing an FRT site, was co-transfected with a Flp recombinase expression vector into 293T-FRT cells, containing a single copy FRT-target site, and a cell clone containing appropriate single copy integration of eBFP at the FRT site was selected. In the resulting 293T-eBFP cell line, the number of green cells counted after transfection of ONs is therefore expected to directly reflect the number of target gene copies specifically mutated.
As shown in Figure 3, when 293T-eBFP cells were transfected with a control ON of sequence unrelated to GFP, no green fluorescent cell was detected 3 days after transfection. In contrast, green fluorescent cells could be observed following transfection with the 25-LNA-eGFPas ON. Table 2 shows the rates of conversion obtained with the different ONs tested. The branched-PS-eGFP, 25-PS-eGFPas and 25-LNA-eGFPas ONs were the most efficient ONs with efficiencies between 0.4 and 2‰. The less efficient ONs were 25-PO-eGFPas and 25-U-eGFPas, as in assays of episomal targets, together with, surprising, 430-ds-eGFP, which had proven to be the most efficient on episomal targets. Other ONs tested showed efficiencies ranging between 0.13 and 0.2‰. It appeared that for 25 nt long ONs, similar to the results obtained on episomal targets, the phosphorothioate and LNA modifications tested provided a strong improvement over standard phosphodiester chemistry (30- and 60-fold, respectively). In contrast, for 45 nt long ONs, the chemical modifications tested did not appear to significantly improve efficiency. Once more, short 25 nt ONs were superior to their longer 45 nt counterparts. Concerning ON length, we found in independent experiments that the short 25 nt ONs with phosphorothioate and LNA modifications were also superior to a 60 nt long ON end-capped with six phosphorothioate linkages (data not shown). For a given chemistry, both sense and antisense ONs were active; however, as observed for episomal targets, a higher efficiency of gene modification was observed with the antisense ONs (15- and 30-fold higher for LNA and PS chemistries, respectively).
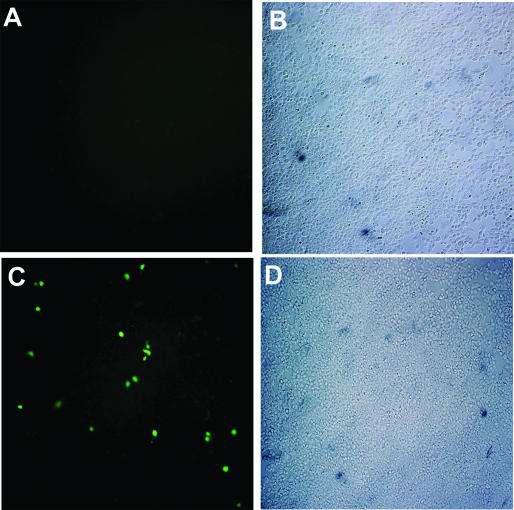
Figure 3.
Chromosomal eBFP conversion after transfection of 293T-eBFP cells with control or 25-LNA-eGFPas ONs. GFP expression was detected by fluorescence microscopy of 293T-eBFP cells 3 days after transfection with control (A) or 25-LNA-eGFPas ONs (C). Brightfield microscopy of 293T-eBFP cells transfected with control (B) or 25-LNA-eGFPas ONs (D). The control ON used was 25-LNA-PDEas of sequence complementary to photoreceptor cGMP-phosphodiesterase. BFP expression could be detected by western blot of 293T-eBFP cells but not by standard fluorescence microscopy due to its low fluorescence yield and strong photobleaching compared with GFP (14).
Table 2.
Rates of chromosomal eBFP conversion in 293T-eBFP cells
| ON | Rate of chromosomal eBFP conversion ± SD (×10−3) |
|---|---|
| 25-LNAscr | 0.007 ± 0.003 |
| 25-U-eGFPas | 0.02 ± 0.01 |
| 430-ds-eGFP | 0.04 ± 0.01 |
| 25-PO-eGFPas | 0.05 ± 0.04 |
| 25-PS-eGFPs | 0.06 ± 0.005 |
| 25-LNA-eGFPs | 0.13 ± 0.06 |
| 45-U-eGFPas | 0.16 ± 0.04 |
| 45-LNA-eGFPas | 0.17 ± 0.03 |
| 45-PS-eGFPas | 0.2 ± 0.05 |
| 45-PO-eGFPas | 0.2 ± 0.08 |
| Branched-PS-eGFP | 0.4 ± 0.08 |
| 25-PS-eGFPas | 1.7 ± 0.2 |
| 25-LNA-eGFPas | 2 ± 0.3 |
Rates of chromosomal eBFP conversion were calculated as the number of green fluorescent cells detected by fluorescence microscopy 3 days after transfection of 293T-eBFP cells with the indicated ONs divided by the total number of cells. Transfection experiments were repeated at least three times. The control ON, 25-LNA-scr, was of identical chemistry as 25-LNA-eGFPas but with scrambled sequence.
DNA sequence analysis of chromosomal eBFP cDNA converted to a GFP cDNA
To demonstrate that the green fluorescence observed resulted from conversion of eBFP to GFP coding sequence, we analyzed DNA sequence of the transgene. Single 293T-eBFP cells emitting green fluorescence after transfection by 25-LNA-eGFPas ON were isolated by flow cytometry and expanded. Confirmation of the conversion of eBFP to GFP conversion came from DNA sequence analysis of amplified genomic DNA. The analysis of four clones derived from single 293T-eBFP cells emitting green fluorescence demonstrated the specific modification of the targeted base after transfection with 25-LNA-eGFPas ON (Figure 4). No other difference from eBFP sequence was found in the entire coding sequence analyzed. In particular, codon TTC145 specific to eBFP in the region having no role in fluorescence specificity (14) was still present. This control finding excluded the possibility of any cross-contamination by eGFP cDNA during the course of our experiments.
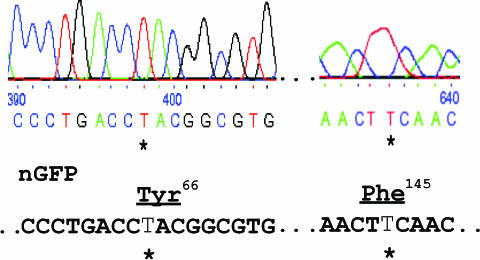
Figure 4.
DNA sequence analysis of converted clones. Sequence of chromosomal eBFP target in GFP-positive clones of 293T-eBFP cells treated with 25-LNA-eGFPas. The coding sequence was amplified by PCR from genomic DNA and directly sequenced, which confirmed that the eBFP sequence had been converted to nGFP. The asterisk indicates the position of the modified base. The sequences shown, coding respectively Tyr66 and Phe145, were from the same chromatogram.
In contrast, we were unable to expand colonies of green fluorescent cells from 293T-eBFP cultures transfected with 25-PS-eGFPas or the branched-PS-eGFP ONs, the other two ONs showing the highest efficiency of integrated transgene conversion at 3 days. Following cell sorting, we also attempted to grow cultures of pools of green fluorescent cells in order to avoid the stress due to growth of isolated cells, and we observed that the ratio of green fluorescent cells steadily decreased over passages in culture, suggesting that the growth of green fluorescent cells was severely impaired by 25-PS-eGFPas and the branched-PS-eGFP ONs and therefore precluded stable transmission of the gene modification.
Conversion of chromosomal eBFP sequence by ONs with flanking LNAs
Since 25-LNA-eGFPas proved to be one of the most efficient ONs tested in 293T cells and allowed to isolate and expand clones of cells containing the desired sequence modification, LNA chemistry was further investigated.
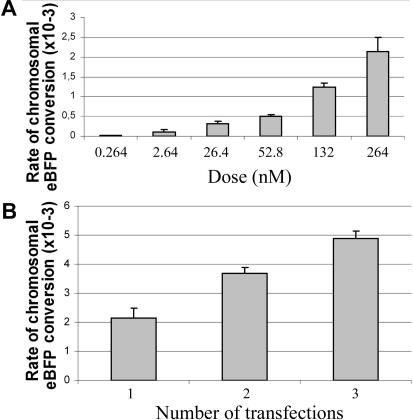
First, dose–response studies were performed. A clear dose–response was obtained when transfecting gradually higher doses of 25-LNA-eGFPas ON into 293T-eBFP cells (Figure 5A). We also examined whether repeating transfections could increase the percentage of conversion. As can be observed in Figure 5B, one, two and three transfections every 24 h led to a progressive increase in the number of converted cells detected.
Figure 5.
Dose-dependent conversion of chromosomal eBFP by 25-LNA-eGFPas ON. (A) Rates of GFP-positive cells in 293T-eBFP cells at 3 days after transfection with indicated doses of 25-LNA-eGFPas ON. The amount of DNA in the transfection mixture was kept constant by the addition of a control LNA ON of unrelated sequence. (B) Rates of GFP-positive cells in 293T-eBFP cells transfected 1, 2 or 3 times with 25-LNA-eGFPas ON (determined 3 days after the last transfection). Results are expressed as means ± SD, represented as vertical bars, corresponding to triplicates from a single transfection experiment.
We next tested ONs of different content in LNA nucleotides on integrated (Table 3) eBFP target. 25-LNA-eGFPas and 25-1x0LNA-eGFPas (containing one LNA at the 5′ end position and no LNA at 3′ end position) were the most efficient ONs. The 25-0x1LNA-eGFPas ON (containing no LNA at the 5′ end position and one LNA at 3′ end position) and the control 25-LNA-eGFP-scr ON were virtually inactive on integrated targets, suggesting that the presence of an LNA at the 5′ end position is essential to allow detectable sequence-specific modification. In addition, 25-2x2LNA-eGFPas and 25-[1x1]LNA-eGFPas ONs resulted in a slightly lower efficiency than 25-LNA-eGFPas, suggesting that increasing the number of LNAs or inserting LNAs at internal rather flanking positions does not significantly improve conversion efficiency. Increasing the number of LNAs had previously been shown to reduce gene editing in yeast (11).
Table 3.
Rates of chromosomal eBFP conversion using different types of LNA-modified ONs in 293T-eBFP cells
| ON | Rate of chromosomal eBFP conversion ± SD (×10−3) |
|---|---|
| 25-LNA-scr | 0.007 ± 0.003 |
| 25-0x1LNA-eGFPas | 0.05 ± 0.03 |
| 25-[1x1]LNA-eGFPas | 0.1 ± 0.03 |
| 25-2x2LNA-eGFPas | 0.7 ± 0.2 |
| 25-LNA-eGFPas | 2.2 ± 0.3 |
| 25-1x0LNA-eGFPas | 2.5 ± 0.08 |
Rates of chromosomal eBFP conversion were calculated as the number of green fluorescent cells detected by fluorescence microscopy 3 days after transfection of 293T-eBFP cells with the indicated ONs divided by the total number of cells. Transfection experiments were repeated three times. The control ON, 25-LNA-scr, was of identical chemistry as 25-LNA-eGFPas but with scrambled sequence.
Correction of chromosomal rd1 mutation by ONs with flanking LNAs
We next chose to compare the ability of phosphorothioate and LNA-modified ONs to correct a second unrelated sequence, the rd1 stop mutation in the photoreceptor βPDE that is responsible for retinal degeneration in the mouse rd1 mutant (12). A fragment of the βPDE cDNA containing the rd1 stop mutation was fused upstream of eGFP cDNA to produce a chimeric rd1-eGFP* sequence target for correction by ONs. Stable integration of the rd1-eGFP* sequence was performed by Flp recombinase methodology into the same 293T-FRT cell line as above and the rd1-eGFP* transgene was therefore integrated at the same genomic site than the eBFP cDNA previously studied.
In this model, correction of the rd1 mutation will result in the appearance of eGFP fluorescence. Phosphorothioate and LNA-modified ONs were designed to correct the rd1 mutation to wild-type βPDE. For each chemistry, sense (25-LNA-PDEs and 25-PS-PDEs) or antisense (25-LNA-PDEas and 25-PS-PDEas) ONs were used to target the transcribed or non-transcribed DNA strand, respectively. When 293T-rd1-eGFP* cells were transfected with control ON of unrelated sequence, no GFP-positive cells were detected at 3 days after transfection. In contrast, GFP-positive cells were observed following transfection with 25-LNA-PDEas ON (data not shown). Rates of gene correction by the different ONs were measured by flow cytometry and results are given in Table 4. Phosphorothioate ONs exhibited slightly higher rd1 correction rates than LNA ONs of the same sequence. Interestingly, even though the rd1-eGFP* target was integrated in 293T-FRT cells at the same genomic site than the eBFP cDNA, the antisense and sense ONs mediated gene correction at roughly identical levels (antisense being only 1.2- and 1.4-fold more efficient than sense sequence for PS and LNA-modifications, respectively). ONs with partial LNA modification were also tested. LNA modification of either 5′ or 3′ end was sufficient for gene editing activity; however, LNA modification at the 5′ end resulted in higher activity than at the 3′ end.
Table 4.
Rates of chromosomal rd1-eGFP* correction to βPDE-eGFP*
| ON | Rate of chromosomal rd1/eGFP* correction ± SD (×10−3) |
|---|---|
| 25-1x0LNA-PDEas | 3 ± 0.6 |
| 25-LNA-PDEs | 5 ± 3 |
| 25-0x1LNA-PDEas | 6 ± 2 |
| 25-LNA-PDEas | 7 ± 4 |
| 25-PS-PDEs | 13 ± 6 |
| 25-PS-PDEas | 16 ± 5 |
Rates of chromosomal rd1-eGFP* correction to βPDE-eGFP* were calculated as the number of green fluorescent cells divided by the total number of cells, which were determined by flow cytometry at 3 days after transfection of the indicated βPDE ONs in 293T-rd1-eGFP* cells. Transfection experiments were repeated three times.
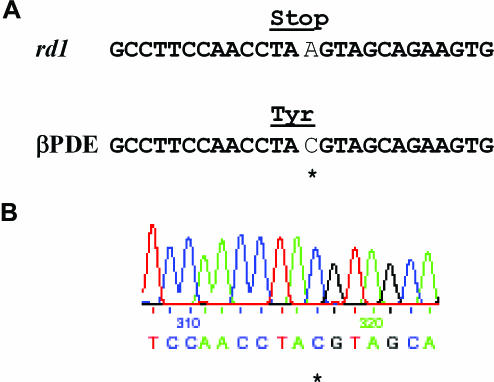
Furthermore, as in studies of the eBFP target, clones were expanded from individual green-positive cells isolated following transfection with 25-LNA-PDEas, and gene correction was demonstrated by sequence analysis (Figure 6). In contrast, no clones could be expanded from 293T-rd1-eGFP* cells transfected with 25-PS-PDEas ON.
Figure 6.
DNA sequence analyses of corrected clones. (A) rd1 and wt-βPDE sequences. (B) Sequence of chromosomal rd1-eGFP* target in a GFP-positive clone of 293T-rd1-eGFP* cells treated 25-LNA-PDEas. The coding sequence was amplified by PCR from genomic DNA and directly sequenced, which confirmed that the rd1 sequence had been corrected into wt-βPDE. The asterisk indicates the position of the modified base.
Improved photoreceptor cell survival in rd1 mice treated with specific ONs with flanking LNAs
Because the rd1 mutation could be corrected by single-stranded ONs with flanking LNAs when integrated in the genomic DNA of 293T cells, we investigated whether this mutation could also be targeted in vivo in rd1 mice. Using a technique that we developed for ON transfer into mouse retina, we were recently able to detect a modest improvement of photoreceptor survival in rd1 mice following transfer of 25-PS-PDEs ON, which was correlated with a low-level of gene correction (C. Andrieu-Soler et al., manuscript submitted). Therefore, LNA and phosphorothioate-modified ONs were transfected in retina of rd1 mice at postnatal days 4, 6 and 8. We then examined photoreceptor cells in the ONL on eye sections at PN28, a late degenerative stage in rd1 mice (19).
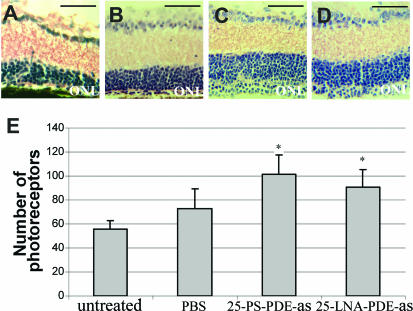
While a single row of cells was observed at PN28 in the ONL of untreated rd1 retina (Figure 7A), numerous areas containing three to four rows of cells were scattered over the ONL of mice treated with 25-LNA-PDEas and 25-PS-PDEas ONs (examples are shown in Figure 7C and 7D). Areas of ONL showing no improvement of photoreceptor survival may correspond to lower levels of ON transfection. In control PBS-treated eyes, a thickening of ONL could be observed which was small but present over the entire retina (Figure 7B), a phenomenon which is consistently observed upon PBS injection (20,21). In order to quantitate the effect of ON transfection on photoreceptor survival, ONL cell numbers were counted in retinas from treated and untreated rd1 mice. In 25-PS-PDEas ON- and 25-LNA-PDEas ON-treated retinas, there was a modest but significant increase in ONL cell numbers when compared with PBS-treated retinas or to the untreated retinas from rd1 mice (Figure 7D). Results of a preliminary screen had previously suggested that 25-PDEas ON was inactive; the difference with the present study could be due to synthesis or purification defects in the previous ON batches used. As a comparison, ONL cell numbers measured here in rd1 retinas treated with 25-PS-PDEas and 25-LNA-PDEas ONs were roughly equivalent to those obtained previously with 25-PDEs ON (C. Andrieu-Soler et al., manuscript submitted). These results suggest that ONs with flanking LNAs could stimulate sequence-specific modification in vivo. However, in contrast to the results obtained in cultured cells, there was no benefit of LNA over phosphorothioate modifications in this in vivo assay, and the number of photoreceptor cells remained insufficient to obtain clinical improvement.
Figure 7.
Eye sections of control and treated rd1 mice at PN28 and photoreceptor cell numbers in the outer nuclear layer (ONL). Hematoxylin–eosin stained sections of eyes of (A) untreated, (B) PBS-treated, (C) 25-PS-PDEas ON-treated and (D) 25-LNA-PDEas ON-treated rd1 mice. A and B are representative of the entire ONL of the corresponding retinas, whereas (C and D) are selected are as showing markedly increased cell numbers in the ONL of ON-treated rd1 mice. Scale bars: 100 μm. (E) Counting of cells in the ONL showed a modest but significant overall increase of cells in 25-PS-PDEas or 25-LNA-PDEas ON-treated compared to PBS-treated eyes (P < 0.0001) or untreated eyes (P < 0.0001) (asterisk). Results were expressed as means ± SD, represented as vertical bars, and compared using the non-parametric Mann–Whitney test.
DISCUSSION
Testing activity of ONs on single copy model target genes
In order to measure gene editing at the single cell level, we have used assays depending on detecting GFP following targeted modification of GFP mutant cDNAs consisting either of eBFP or a fusion between rd1-βPDE and GFP cDNAs. Because we applied Flp recombinase methodology, we could easily obtain cell lines containing the transgenes integrated as transcriptionally active single copies. As a consequence, GFP-fluorescence indicates the successful modification of the target gene and the number of GFP-positive cells provides a measure of target gene modification. Indeed, we could rigorously validate the assay by directly showing that in GFP-positive cells, the desired gene modification had been successfully introduced and stably propagated after transfection with sequence-specific ONs with flanking LNAs. An asset of our experimental approach is that further use of Flp recombinase methodology will allow introducing different model gene targets at identical genomic sites and it should therefore be invaluable to analyze the importance of the target DNA sequence in gene editing. It should also facilitate the characterization of complex ONs, such as RNA/DNA ONs, whose gene editing efficiency can vary from one synthesis to another (3,22,23). The strategy we have used to test correction of the rd1 mutation, relying on detecting correction of an inactive GFP fusion cDNA, could be easily applied to test the possibility of correcting other stop codons involved in human or animal diseases.
Strand-bias in ON-mediated mutagenesis is dependent on target sequence
Targeted gene modification by ONs takes place in two steps: pairing to the target, which likely involves protein complexes that usually take part in homologous recombination, and gene modification per se, which may involve mismatch repair or other mechanisms (3). For the eBFP sequence, we have found here that targeting the transcribed strand was much more efficient than targeting the non-transcribed strand (15- to 30-fold), as reported in most studies (3). In contrast, for the rd1-eGFP* sequence, there was no significant difference. The two targets tested, eBFP and rd1-eGFP*, were integrated at the same genomic site of 293T-FRT cells and transcribed from the same CMV promoter. Therefore, our results indicate that the target sequence may by itself determine a strand-bias in targeting efficiency, perhaps as a result of different abilities to reach and pair to their target. A similar conclusion was recently proposed following studies of an episomal target (24). Such a conclusion does not exclude; however, that genomic context (for example, determining orientation and timing of replication) or the promoter of the target sequence play an additional role in strand-bias. Until the molecular mechanisms of gene editing by ONs are better understood, it therefore remains important to test the activity of the two complementary sequences in order to achieve the best efficiency of gene editing.
Properties of the ON vectors
Using our assays of episomal and integrated eBFP targets, we have compared the activity of several types of single-stranded ONs as well as a branched ON and a double-stranded fragment. It appeared that short single-stranded ONs with flanking LNAs were efficient vectors, allowing gene editing of 0.2% integrated eBFP target sequences. Importantly, similar efficiencies were observed for the second target tested here, the rd1 stop mutation, as well as for a third unrelated sequence (C. Andrieu-Soler et al., manuscript in preparation), suggesting that short ONs with flanking LNAs constitute a simple and robust ON design for gene editing.
When comparing single-stranded vectors targeting the eBFP sequence, several observations could be made. First, we found that 25 nt ONs end-capped with single LNAs or with six phosphorothioate linkages were 30- to 60-fold more efficient than their non-modified counterparts, most likely due to increased resistance to nucleases and as a consequence a higher concentration of effective ON reaching the nucleus after transfection. Second, we also found that 25 nt ONs end-capped with single LNAs or with six phosphorothioate linkages were consistently superior to their longer counterparts. Shorter ONs may mediate more efficient gene editing per se, for instance as a result of higher affinity to proteins facilitating recognition or invasion of the target double-stranded sequence. Longer ONs may also be more prone to binding to non-specific sites, thereby diluting effective ON concentration at the targeted sequence. Third, we found that chemically modified ONs of 45 nt were not superior to the ON of standard chemistry. This observation was unexpected and may perhaps be explained by the ability of the 45 nt sequence to adopt structures conferring resistance to nucleases. In any case, we must be cautious in interpreting differences between ON activities. Many steps are necessary to achieve gene editing: transfection, nucleocytoplasmic transport, binding to molecular machines responsible for DNA editing, annealing to target DNA, degradation by nucleases…, all of which may be differentially affected by ON chemistry and sequence and impact on final ON activity.
Surprisingly, the 430-ds-eGFP exhibited limited activity on integrated targets even though it was relatively efficient on episomal eBFP cDNA. We tested extensive transfection conditions but no increase in conversion of chromosomal eBFP could be observed (data not shown). Differences in transfection conditions may be involved but are unlikely to entirely explain the very low activity of 430-ds-eGFP on integrated targets. In our model, the sequence homologous to 430-ds-eGFP starts immediately downstream of the CMV transcription initiation site. In consequence, association of 430-ds-eGFP to its homologous sequence or recruitment of the recombination machinery may be limited by the transcription initiation complex. Considering the high efficiency of small homologous fragments reported in other studies (18), our finding suggests that this method is extremely target-dependent. It nevertheless deserves further investigation, because it may allow performing more extensive mutations than with shorter ONs.
We have also found that a branched ON was roughly equivalent to the best single-stranded vectors on both episomal and chromosomal targets. This is the second example of gene targeting with branched ONs, which were originally designed by Olsen et al. (5), and it therefore proves to be an efficient vector. The branched ON structure is reminiscent of results obtained by Parekh-Olmedo et al. (11), who showed that in yeast, a hybrid between a long ON and a shorter complementary ON could exhibit higher activity than either ON taken alone. However, in our hands, such hybrids did not significantly change gene editing levels, whether annealing of ONs was performed prior to transfection or not (data not shown).
Stable transmission of targeted gene modification by ONs with flanking LNAs
LNAs are proving to be versatile reagents in many aspects of gene control and manipulation (10). Insertion of LNAs into an ON was originally designed to increase binding affinity toward complementary DNA and RNA. LNAs are therefore important tools in diagnostic and analytical applications involving nucleic acid hybridization. Additional important benefits were found in LNAs, including resistance to nucleases (25,26). In our study, an ON with a single LNA at its 5′ end only was as efficient as an ON flanked by LNA at both ends on the eBFP sequence target and an ON with a single LNA at its 3′ end only was as efficient as an ON flanked by LNA at both ends on the second rd1-eGFP sequence target. Therefore, a single protection may be sufficient but no general rule can be made: the differences observed between ONs with a single LNA at either 5′ or 3′ end are gene dependent. LNA modification at both ON ends therefore appears to be the optimal chemistry to use for gene editing. A second important feature of ONs incorporating LNAs is their low toxicity in vivo (25,26). The latter property likely explains why LNA-modified ONs were superior to the branched and PS-modified ONs in making it possible to isolate clones of cells bearing the desired sequence-specific modification. Branched ONs have been previously shown to be cytotoxic (5), apoptosis being triggered in corrected cells. Cytotoxicity is also a well-known caveat of phosphorothioate ONs that likely explains why growth of corrected cells appeared severely impaired. Other studies of gene editing by phosphorothioate-modified ONs have either described normal growth of corrected cells (16) or impaired cell cycle progression (27). Cytotoxicity of phosphorothioate-modified ONs is known to be sequence-dependent (28) and this could explain such variability in the growth of corrected cells. Other experimental differences may also be involved, such as cell lines and ON transfection reagents used. Variable success has similarly been reported for RNA/DNA ONs, which either did (29) or did not (30) allow growth and isolation of corrected cells. Possibly, further studies of such ONs may allow reducing their cytotoxicity without compromising their ability to mediate gene modification. However, in contrast, the use of 25 nt ONs with flanking LNAs appears to provide a straightforward and robust design for obtaining cell lines containing stable transmission of the desired gene modification. In addition to the two targets described here, we have recently obtained 293T cell lines with modification of several other model sequences, further validating the power of ONs with flanking LNAs. The next step will hence consist in demonstrating targeting of endogenous genes. The mutagenesis rates measured here, in the range of 0.1–0.5%, appear too low to envisage direct isolation of modified cells. Progressive isolation from pools of treated cells by PCR (15) or the use of a selection strategy, as suggested by Yoon and co-workers (31), will therefore be necessary to obtain cells with targeted mutations in endogenous genes. Mutagenesis rates may also be increased as a result of better understanding of the mechanisms of gene editing directed by ONs (32) or by the simultaneous use of triplex-forming ONs (6).
Finally, we compared the activity of LNA- and phosphorothioate-containing ONs in the rd1 mouse model. Following transfer of ONs into the retina of rd1 mouse mutant, an ON with flanking LNAs led to a modest but significant increase in photoreceptor survival, which was slightly lower than that obtained with phosphorothioate ONs. This result is consistent with the finding that the ON with flanking LNAs was less active on the single copy rd1-eGFP* transgene than the phosphorothioate ON at 3 days after transfection. Given that photoreceptor cells stop dividing after postnatal day 6, i.e. during the period when ON treatments were performed in rd1 mice, it may not be so surprising that the differences observed on growth of cultured cells in vitro do not translate into a detectable advantage when treating rd1 photoreceptor cells. One may expect to observe an advantage of LNA- over phosphorothioate-modified ONs in treatment of diseases involving proliferating cells or when correction rates achieved lead to therapeutic benefit and differences in toxicity between the two chemistries then become relevant. The comparison we have performed of targeted mutagenesis directed by LNA- or phosphorothioate-modified ONs on different targets illustrates the need to carefully tailor ON design to the specific gene target and experimental purpose. Our finding that ONs with flanking LNAs allow to reliably isolate cell lines containing the desired gene modifications may help in further applications of ON-mediated targeted mutagenesis in functional genomics and gene therapy.
Acknowledgments
C.A. is funded by Optis France and the French Ministry of Research and Education. This work was supported by the GenHomme program from the French Ministry for Research and Education (grants 2001 number 01 H 0203, 2002 number 01 H 0204), NIH NEI R01 EY12514 and R03 EY13986, the Foundation Fighting Blindness, the Research to Prevent Blindness association and AFM. The authors thank S. Haffar and P. Eog-Eog at Institut Cochin for technical help and Y. Courtois and M.-F. Blanchet-Tournier for critical reading of the manuscript. Clontech is acknowledged for their kind help in getting eBFP cDNA, which is no longer commercially available. Funding to pay the Open Access publication charges for this article was provided by INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vasquez K.M., Marburger K., Intody Z., Wilson J.H. Manipulating the mammalian genome by homologous recombination. Proc. Natl Acad. Sci. USA. 2001;98:8403–8410. doi: 10.1073/pnas.111009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell C.R., Keown W., Lowe L., Kirschling D., Kucherlapati R. Homologous recombination involving small single-stranded oligonucleotides in human cells. New Biol. 1989;1:223–227. [PubMed] [Google Scholar]
- 3.Igoucheva O., Alexeev V., Yoon K. Oligonucleotide-directed mutagenesis and targeted gene correction: a mechanistic point of view. Curr. Mol. Med. 2004;4:445–463. doi: 10.2174/1566524043360465. [DOI] [PubMed] [Google Scholar]
- 4.Liu L., Parekh-Olmedo H., Kmiec E.B. The development and regulation of gene repair. Nature Rev. Genet. 2003;4:679–689. doi: 10.1038/nrg1156. [DOI] [PubMed] [Google Scholar]
- 5.Olsen P.A., McKeen C., Krauss S. Branched oligonucleotides induce in vivo gene conversion of a mutated EGFP reporter. Gene Ther. 2003;10:1830–1840. doi: 10.1038/sj.gt.3302079. [DOI] [PubMed] [Google Scholar]
- 6.Seidman M.M., Glazer P.M. The potential for gene repair via triple helix formation. J. Clin. Invest. 2003;112:487–494. doi: 10.1172/JCI19552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taubes G. Gene therapy. The strange case of chimeraplasty. Science. 2002;298:2116–2120. doi: 10.1126/science.298.5601.2116. [DOI] [PubMed] [Google Scholar]
- 8.van der Steege G., Schuilenga-Hut P.H., Buys C.H., Scheffer H., Pas H.H., Jonkman M.F. Persistent failures in gene repair. Nat. Biotechnol. 2001;19:305–306. doi: 10.1038/86664. [DOI] [PubMed] [Google Scholar]
- 9.De Semir D., Aran J.M. Misleading gene conversion frequencies due to a PCR artifact using small fragment homologous replacement. Oligonucleotides. 2003;13:261–269. doi: 10.1089/154545703322460630. [DOI] [PubMed] [Google Scholar]
- 10.Jepsen J.S., Sorensen M.D., Wengel J. Locked nucleic acid: a potent nucleic acid analog in therapeutics and biotechnology. Oligonucleotides. 2004;14:130–146. doi: 10.1089/1545457041526317. [DOI] [PubMed] [Google Scholar]
- 11.Parekh-Olmedo H., Drury M., Kmiec E.B. Targeted nucleotide exchange in Saccharomyces cerevisiae directed by short oligonucleotides containing locked nucleic acids. Chem. Biol. 2002;9:1073–1084. doi: 10.1016/s1074-5521(02)00236-3. [DOI] [PubMed] [Google Scholar]
- 12.Pittler S.J., Baehr W. Identification of a nonsense mutation in the rod photoreceptor cGMP phosphodiesterase beta-subunit gene of the rd mouse. Proc. Natl Acad. Sci. USA. 1991;88:8322–8326. doi: 10.1073/pnas.88.19.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Gorman S., Fox D.T., Wahl G.M. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science. 1991;251:1351–1355. doi: 10.1126/science.1900642. [DOI] [PubMed] [Google Scholar]
- 14.Tsien R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 15.Dekker M., Brouwers C., te Riele H. Targeted gene modification in mismatch-repair-deficient embryonic stem cells by single-stranded DNA oligonucleotides. Nucleic Acids Res. 2003;31:e27. doi: 10.1093/nar/gng027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nickerson H.D., Colledge W.H. A comparison of gene repair strategies in cell culture using a lacZ reporter system. Gene Ther. 2003;10:1584–1591. doi: 10.1038/sj.gt.3302049. [DOI] [PubMed] [Google Scholar]
- 17.Igoucheva O., Alexeev V., Yoon K. Targeted gene correction by small single-stranded oligonucleotides in mammalian cells. Gene Ther. 2001;8:391–399. doi: 10.1038/sj.gt.3301414. [DOI] [PubMed] [Google Scholar]
- 18.Gruenert D.C., Bruscia E., Novelli G., Colosimo A., Dallapiccola B., Sangiuolo F., Goncz K.K. Sequence-specific modification of genomic DNA by small DNA fragments. J. Clin. Invest. 2003;112:637–641. doi: 10.1172/JCI19773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter-Dawson L.D., LaVail M.M., Sidman R.L. Differential effect of the rd mutation on rods and cones in the mouse retina. Invest. Ophthalmol. Vis. Sci. 1978;17:489–498. [PubMed] [Google Scholar]
- 20.Frasson M., Picaud S., Leveillard T., Simonutti M., Mohand-Said S., Dreyfus H., Hicks D., Sabel J. Glial cell line-derived neurotrophic factor induces histologic and functional protection of rod photoreceptors in the rd/rd mouse. Invest. Ophthalmol. Vis. Sci. 1999;40:2724–2734. [PubMed] [Google Scholar]
- 21.Silverman M.S., Hughes S.E. Photoreceptor rescue in the RCS rat without pigment epithelium transplantation. Curr. Eye Res. 1990;9:183–191. doi: 10.3109/02713689008995205. [DOI] [PubMed] [Google Scholar]
- 22.Graham I.R., Manzano A., Tagalakis A.D., Mohri Z., Sperber G., Hill V., Beattie S., Schepelmann S., Dickson G., Owen J.S. Gene repair validation. Nat. Biotechnol. 2001;19:507–508. doi: 10.1038/89209. [DOI] [PubMed] [Google Scholar]
- 23.Manzano A., Mohri Z., Sperber G., Ogris M., Graham I., Dickson G., Owen J.S. Failure to generate atheroprotective apolipoprotein AI phenotypes using synthetic RNA/DNA oligonucleotides (chimeraplasts) J. Gene. Med. 2003;5:795–802. doi: 10.1002/jgm.403. [DOI] [PubMed] [Google Scholar]
- 24.Sorensen C.B., Krogsdam A.M., Andersen M.S., Kristiansen K., Bolund L., Jensen T.G. Site-specific strand bias in gene correction using single-stranded oligonucleotides. J. Mol. Med. 2005;83:39–49. doi: 10.1007/s00109-004-0592-6. [DOI] [PubMed] [Google Scholar]
- 25.Kurreck J., Wyszko E., Gillen C., Erdmann V.A. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30:1911–1918. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wahlestedt C., Salmi P., Good L., Kela J., Johnsson T., Hokfelt T., Broberger C., Porreca F., Lai J., Ren K., et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl Acad. Sci. USA. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsen P.A., Randol M., Krauss S. Implications of cell cycle progression on functional sequence correction by short single-stranded DNA oligonucleotides. Gene Ther. 2005;12:546–551. doi: 10.1038/sj.gt.3302454. [DOI] [PubMed] [Google Scholar]
- 28.Drygin D., Barone S., Bennett C.F. Sequence-dependent cytotoxicity of second-generation oligonucleotides. Nucleic Acids Res. 2004;32:6585–6594. doi: 10.1093/nar/gkh997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexeev V., Yoon K. Stable and inheritable changes in genotype and phenotype of albino melanocytes induced by an RNA–DNA oligonucleotide. Nat. Biotechnol. 1998;16:1343–1346. doi: 10.1038/4322. [DOI] [PubMed] [Google Scholar]
- 30.Tagalakis A.D., Dickson J.G., Owen J.S., Simons J.P. Correction of the neuropathogenic human apolipoprotein E4 (APOE4) gene to APOE3 in vitro using synthetic RNA/DNA oligonucleotides (chimeraplasts) J. Mol. Neurosci. 2005;25:95–104. doi: 10.1385/JMN:25:1:095. [DOI] [PubMed] [Google Scholar]
- 31.Alexeev V., Igoucheva O., Yoon K. Simultaneous targeted alteration of the tyrosinase and c-kit genes by single-stranded oligonucleotides. Gene Ther. 2002;9:1667–1675. doi: 10.1038/sj.gt.3301862. [DOI] [PubMed] [Google Scholar]
- 32.Wu X.S., Xin L., Yin W.X., Shang X.Y., Lu L., Watt R.M., Cheah K.S., Huang J.D., Liu D.P., Liang C.C. Increased efficiency of oligonucleotide-mediated gene repair through slowing replication fork progression. Proc. Natl Acad. Sci. USA. 2005;102:2508–2513. doi: 10.1073/pnas.0406991102. [DOI] [PMC free article] [PubMed] [Google Scholar]