Abstract
Enhancers are DNA sequences that can activate gene transcription from remote positions. In yeast, regulatory sequences that are functionally equivalent to the metazoan enhancers are called upstream activating sequences (UASs). UASs show a lower degree of flexibility than their metazoan counterparts, but can nevertheless activate transcription from a distance of >1000 bp from the promoter. One of several models for the mechanism of action of transcriptional enhancers proposes that enhancer-bound activating proteins contact promoter-bound transcription factors and thereby get in close proximity to the promoter region with concomitant looping of the intervening DNA. We tested the mode of enhancer activity in yeast. A polymerase II-transcribed gene was paired with a remote, inducible enhancer. An independent reporter system was inserted next to the promoter to monitor the potential modes of enhancer activity. Our results show that the enhancer activated the reporter system only in the presence of a functional promoter. We also demonstrate that the heterologous expression of GAGA, a factor known to facilitate DNA loop formation, allows enhancer action in yeast over a distance of 3000 bp.
INTRODUCTION
In metazoans, enhancers can influence gene expression independent of their orientation and from remote locations, upstream, downstream or even in introns of the corresponding transcription unit (1–4). Yeast enhancers, which are called upstream activating sequences (UASs) are less flexible in terms of distance and position relative to the regulated promoter compared with their metazoan counterparts, but can nevertheless work from a distance of up to 1200 bp from the promoter (5–7). One unresolved question is how regulatory proteins bound to enhancer sequences can influence the activity of another protein, RNA polymerase II (pol II), at a distance. Several models have been proposed. The three most popular models have been dubbed ‘scanning’, ‘linking’ and ‘looping’ (8–14).
The scanning model (also known as sliding or tracking) proposes that enhancer-bound activators have a high affinity for the RNA pol II complex, or parts of it, and recruit it to DNA. Starting from the enhancer, the recruited complex subsequently scans the DNA until it reaches a promoter sequence, where it initiates transcription. The ability of boundary elements to block the action of an enhancer is consistently explained by a scanning mechanism. A boundary element would act as a roadblock for complexes scanning along DNA and thereby prevent initiation at the promoter. In the case of the bacteriophage T4 late genes that are activated by a distantly located enhancer element, it was shown that an endonuclease-defective EcoRI bound between promoter and enhancer was efficiently blocking enhancer activity (8,15). The linking model (also known as oozing) was proposed as a mechanism by which the Locus Control Region (LCR) in the β-globin locus regulates the activation of the different globin genes during development. Activators bound to enhancer sequences recruit enhancer facilitators and initiate the formation of a protein structure that extends from the enhancer along the chromatin fiber until it reaches proteins bound to a transcriptionally competent promoter. The activated promoter acts as a boundary to the further spread of the chain, and initiation of transcription by RNA pol II at the promoter is stimulated (11). The looping model proposes that the contacts between enhancer-bound activating proteins and transcription factors working at promoters bring the enhancer and promoter elements into close proximity (direct communication through physical apposition), with concomitant looping out of the intervening DNA to facilitate the reaction (9). Such DNA looping might be facilitated by a variety of protein–protein and protein–DNA interactions, including those that affect the chromatin structure (9,10,12,16–18). As it has been pointed out by others (10,13), DNA loops could also be generated by ‘scanning’ or ‘linking’ activities that, in their movement away from the enhancer, carry along this DNA segment. Thus, the most salient mechanistic distinction between the DNA looping model and the other models described above is that contact between activators and the core transcriptional machinery causes physical apposition of enhancers and promoters, and consequently DNA loop formation. In this case, DNA looping is expected to require the presence of both the enhancer and the promoter elements.
Ptashne and collaborators have demonstrated that the DNA loop formed by a telomere in yeast can bring a remote enhancer close to a promoter and thereby stimulate activation at a distance. These results clearly indicate that DNA looping can permit enhancer–promoter communication over distances that are too large for such interactions to occur autonomously in yeast. However, they do not show that activation at a distance must work by a looping mechanism that is controlled by transcription factors. For instance, forcing an enhancer close to a promoter by introducing a telomeric DNA loop might allow the enhancer-tethered transcription complex to directly contact (‘jump to’) the promoter, even though, in a natural situation, this complex would scan the DNA until it reaches the promoter (19–21). Similar data have been presented to indicate that phenomena such as transvection or enhancer–promoter functional interactions between concatenated plasmids are consistent with DNA looping, but do not show it (11).
The results presented here provide genetic evidence for DNA looping between an enhancer and a promoter that is mediated by a transcriptional activator in yeast. Implications for the mechanism of action of transcriptional enhancers in higher eukaryotes are discussed.
MATERIALS AND METHODS
Strains and plasmids
Deletion of the endogenous SNR6 gene in the Saccharomyces cerevisiae strain NLY2 (MATα ura3-52 leu2-2 his3Δ200 Δ trp1 Δlys2 Δgal4 Δgal80) containing a URA3-marked centromeric plasmid bearing the SNR6-6 allele was carried out by using a kanamycin knockout construct (22). The resulting strain was called MPy26. The SNR6-6 allele, which lacks 6 nt, can fully rescue the lethal snr6 phenotype (M. Petrascheck and A. Barberis, unpublished data). The yeast reporter constructs were derived from pDE200 (7), an integrating HIS3-marked lacZ reporter gene controlled by six LexA-binding sites (LexA BS), each separated by a unique restriction site. The second LexA BS was replaced by a DNA fragment containing the SNR6 gene under the control of five Gal4p-binding sites (UASG) (23), thus generating pDE251, the reporter construct with the oppositely oriented SNR6 and lacZ genes. This reporter construct was digested with XhoI and SalI to excise the HIS3 ORF and part of its 3′-UTR, which eliminated the PstI site in the 3′-UTR. The resulting plasmid was designated pDE251ΔXS. The distance between the SNR6 gene and the UASG element was increased by inserting 100, 200 and 400 bp spacer DNA fragments that were derived from the β-globin ORF by PCR. These fragments were inserted into the unique PstI site of pDE251ΔXS located downstream of the SNR6 gene resulting in pDE251/100ΔXS, for the 100 bp spacer, and pDE251/200ΔXS, for the 200 bp spacer. The SalI–XhoI HIS3 gene fragment was inserted into pDE251/100ΔXS and pDE251/200ΔXS to generate pDE251/100 and pDE251/200, respectively. The enhancer-deleted reporter construct pMP107 was generated by cutting out the four distal LexA-binding sites from pDE251/200 by SalI/NotI digestion, treating the DNA fragments with the Klenow enzyme and religating the plasmid backbone. The promoter-deleted reporter construct pMP108 lacking the pol II promoter, the proximal LexA site and part of the lacZ gene was obtained by cutting pDE251/200 with EcoRI and religating the plasmid backbone. The reporter constructs pDE251, pDE251/100, pDE251/200, pMP107 and pMP108 were linearized at the XhoI site in the HIS3 3′-UTR, and integrated into MPy26 to generate MPy254/no spacer, MPy254/100, MPy254/200, MPy254/-enh and MPy254/-prom, respectively.
The lacZ-GAGA reporter (pMP204A) was a two-step cloning. First a blunt-ended UASG-GAGA fragment cut out of pTM3 (24) using BamHI/HindIII was inserted into the SmaI site located 3′ of the lacZ gene of pJP158. In a second step, the GAGA sites containing SalI/XbaI fragment of pTM2 (24) was inserted upstream of the lacZ gene promoter into the corresponding sites. The resulting construct was designated pMP204A and inserted into the URA3 locus of NLY2 to obtain the reporter strain MPy204A. To express the GAGA factor, pMP190 was constructed by performing PCR with the following primers 5′-GTCCCCCGGGTCGCTGCCAATGAATTCGCTGTATTC-3′ and 5′-GCGCTCTAGACTACTGCGGCTGCGGCTGTTGCTG-3′ using pTM5 (24) as a template. The PCR product was cut by SmaI/XbaI and inserted into the vector pJP156 that was first digested with NcoI, blunt-ended with Klenow enzyme and after purification digested using XbaI.
Gene expression assays
For the growth assays to indirectly monitor expression of the SNR6 reporter gene, cells were picked from the original transformation plate, grown to an OD600 between 1 and 2 in 5 ml of synthetic drop-out (SD) liquid medium, and then adjusted to an OD600 of 0.5. Ten microliter aliquots of these cultures containing equal amounts of cells were diluted and spotted in parallel on SD control plates or on SD plates supplemented to 0.1% 5-fluoro-orotic acid (5-FOA). These plates were incubated at 30°C for 2–4 days. For each experiment, at least three different trials were carried out using different yeast reporter clones and colony isolates. Measurements of lacZ expression were performed as previously described (7). RNA mapping was performed by growing 20 ml of cells to early log phase (OD600 = 0.4) and preparing total RNA (BIO101 kit) for quantification of SNR6 expression by primer-extension according to standard protocols (U6 primer sequence: 5′-GCA GGG GAA CTC ATC ATC TCT G-3′).
RESULTS
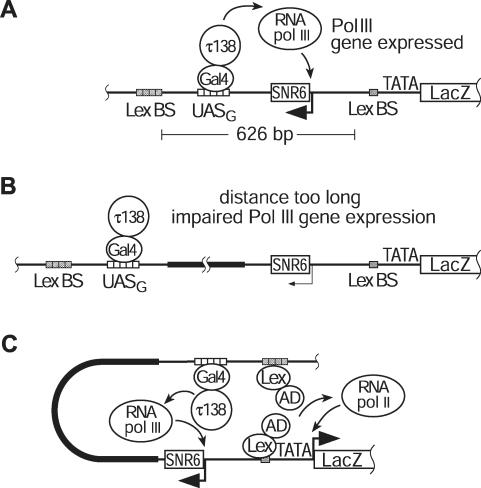
A hallmark of pol II enhancers is that they can influence gene expression from a remote position. In contrast, regulatory sequences of genes transcribed by RNA polymerase III (pol III) are normally located in close proximity to the gene, and cannot work over long distances (25). In agreement with this observation, our studies on the regulation of UASG-SNR6, a yeast pol III gene that is activated by tethering the TFIIIC subunit τ138 to the UASG sequence via the fusion with the Gal4p DNA-binding domain (DBD) (26,27), indicate that splitting this UASG-SNR6 gene such as to shift the UASG element to a more distant location significantly reduces its expression (see below). Such functional differences between pol II and pol III regulatory sequences provided the basis for the system that we established to investigate the mode of enhancer activity in vivo. For this system, we considered the possibility that the mechanism of action of a pol II enhancer (whatever that is, i.e. scanning, linking or looping) located next to the dislocated artificial pol III regulatory element might help to restore communication between this element (UASG) and the pol III promoter and thus allow activation of the SNR6 pol III gene at a distance. Figure 1 schematically depicts the molecular basis of this system. The SNR6 gene and a lacZ reporter gene transcribed from a pol II promoter were ligated in divergent orientations. Downstream of SNR6, we positioned five Gal4p-binding sites (UASG) and, adjacently, four binding sites (BS) for LexA as the pol II enhancer. A chimeric protein bearing the activation domain of Gal4p fused to LexA (Lex-AD) was used as the pol II-specific activator. To mimic the promoter architecture of metazoans more closely, a single LexA-binding site was inserted proximal to the GAL1 TATA box to facilitate the action of the distant enhancer (7) (Figure 1). This pol II–pol III reporter construct was integrated into the genome of a yeast strain that bears a deletion of the endogenous SNR6 gene. Since this knockout is lethal, we provided a functional SNR6 allele containing a 6 bp deletion (SNR6-6) on an episomal plasmid marked with the URA3 gene that fully complements depletion of the endogenous wild-type U6 snRNA and is not affected by Gal4DBD-τ138. This marker gene allowed us to monitor expression of the UASG-SNR6 reporter gene by selecting for the loss of the SNR6-6 episomal plasmid upon cell growth on 5-FOA media (26,28). The efficiency of cell growth on this selective medium correlates with the level of expression of the UASG-SNR6 gene (26) (see below). In addition, the shorter U6 snRNA product of the SNR6-6 allele carried by the URA3-marked centromeric plasmid allows monitoring the expression of both SNR6 alleles in a single primer-extension assay, since the SNR6-6 is easily distinguishable from the product of the SNR6 reporter gene by gel electrophoresis. In our experimental set up, the three models described above make different predictions of the requirements for reactivation of the split UASG-SNR6 pol III reporter gene by the pol II enhancer. These predictions were tested (see below).
Figure 1.
Reporter systems to test enhancer action in vivo. (A) The divergently oriented SNR6 and lacZ genes, transcribed by RNA polymerase III and II, respectively, are integrated into the yeast genome. The lacZ gene is under the control of the GAL1 promoter (TATA), one proximal and four distal LexA-binding sites (Lex BS). The SNR6 gene is activated by tethering τ138 as a Gal4p DNA-binding domain fusion (Gal4DBD-τ138) to five Gal4p-binding sites (UASG). When the UASG is inserted at the original B block position, Gal4DBD-τ138 activates SNR6 (bent arrow) by RNA polymerase III (RNA pol III). The distances (in bp) between the various sequence motifs are in scale. (B) The UASG is shifted further downstream of the SNR6 gene by inserting spacer DNA (thick line). As a consequence, Gal4DBD-τ138 only weakly activates transcription from this split SNR6 gene (thin, bent arrow). (C) Binding of the pol II activator proteins (Lex-AD) to the remote and proximal LexA-binding sites and their interactions with the promoter-binding pol II transcription complex might induce DNA looping and, by that, bring Gal4DBD-τ138 closer to the SNR6 transcriptional start site, which would result in stronger SNR6 activation. Note: Thick bent arrows at the 5′ end of a gene indicate strong expression; thin bent arrows indicate weak expression; if the gene is not expressed in the set up shown, the arrow has been omitted.
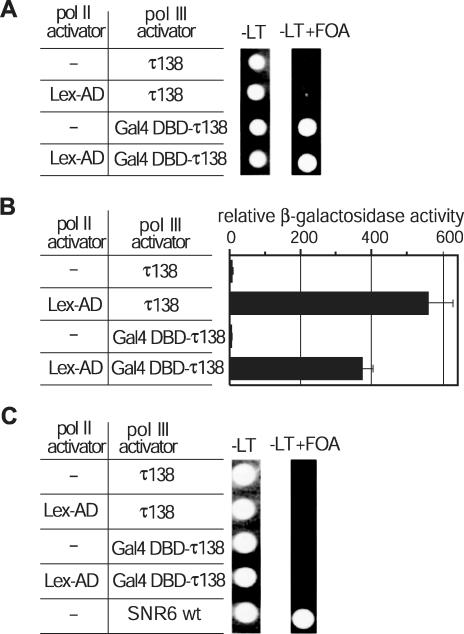
The yeast strain bearing the reporter constructs schematically depicted in Figure 1 was transformed with plasmids expressing the pol II and pol III transcription factors indicated in Figure 2. Activation of the SNR6 reporter gene was monitored by spotting aliquots of these yeast cultures on permissive (−Leu −Trp SD) as well as non-permissive (−Leu −Trp SD + 5-FOA) plates in order to select for those cells that could lose the episomal copy of SNR6. Expression of the pol III transcription factor τ138 lacking the Gal4p DBD moiety did not cause activation of either the UASG-SNR6 (Figure 2A) or of the lacZ (Figure 2B) reporter genes. Expression of the pol II transcriptional activator Lex-AD strongly activated lacZ gene expression (Figure 2B), but did not activate expression of the UASG-SNR6 gene (Figure 2A). Expression of the Gal4DBD-τ138 fusion protein activated transcription of the UASG-SNR6 gene (Figure 2A), but not of the lacZ reporter gene (Figure 2B). Combining the pol II (Lex-AD) and the pol III (Gal4DBD-τ138) activators had a slight but reproducible negative effect on their activities. However, they were capable of inducing expression of their respective reporter genes also when simultaneously present on the reporter DNA (Figure 2A and B). In a control experiment, we monitored growth on 5-FOA plates of an isogenic strain that carries a reporter construct lacking the UASG-SNR6 gene, but otherwise bearing all other sequences described above. Figure 2C shows that none of the expressed transcription factors stimulated growth of this yeast strain on plates containing 5-FOA, thus indicating that selective growth of cells carrying the complete reporter construct was a direct effect of the activation of the reporter SNR6 gene. These results show that the divergently oriented genes are functional, even when both polymerases are activated, and that the pol II activator did not directly influence gene expression of the pol III reporter gene and vice versa.
Figure 2.
The divergent RNA polymerase II and III genes are functional and exclusively activated by their specific transcription factors. The reporter construct shown in Figure 1A was integrated into the genome and the activity of the divergent reporter genes was tested (A) Equal amounts of transformed cells were spotted on minimal drop-out plates (−LT) and on plates containing 5-fluoro-orotic acid (−LT + FOA) to monitor expression of the UASG-SNR6 reporter gene. (B) Quantitative β-galactosidase assay to determine the relative expression levels of the lacZ gene. (C) The SNR6 gene and its regulatory sequences were omitted in the reporter construct integrated in the yeast cells tested here. None of the effector plasmids allowed growth on 5-FOA plates, while the presence of a TRP1-marked plasmid bearing the wild-type SNR6 gene (SNR6 wt) allowed growth on 5-FOA plates.
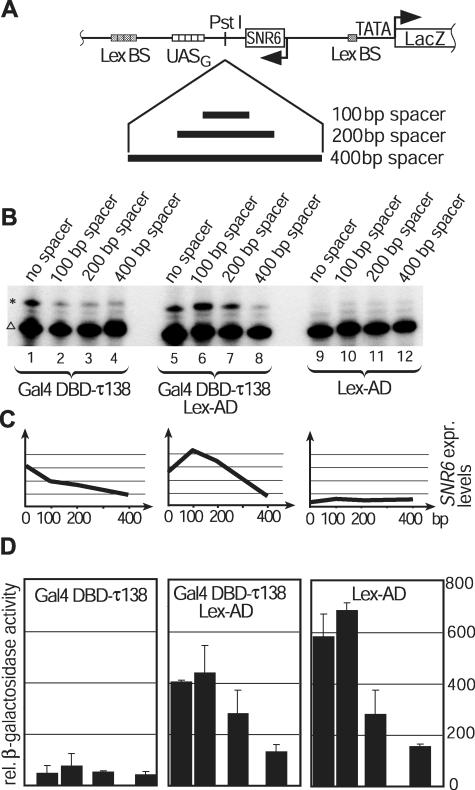
The distance between the Gal4DBD-τ138-binding UASG element and the SNR6 gene on the basic reporter construct (Figure 1) was increased in a stepwise manner by introducing either a 100, 200 or a 400 bp DNA fragment between these two sequences (Figure 3A). We then tested the level of SNR6 and β-galactosidase transcriptional activation of these pol III reporter constructs in the presence of Gal4DBD-τ138, Lex-AD or the combination of Gal4DBD-τ138 and Lex-AD, and compared it to the activation level of the spacer-less UASG-SNR6. The stepwise increase of the distance between the UASG element and the SNR6 gene led to a rapid decline of SNR6 expression as monitored by primer-extension assays (Figure 3B and C). Activation of the lacZ gene by the pol II activator Lex-AD was only reduced to ∼60% of the original activity by the insertion of the 200 bp spacer DNA (Figure 3D, middle and right panel). We tested whether the reduced SNR6 expression, caused by moving the UASG element further downstream of SNR6, could be rescued by a pol II activator bound to the remote enhancer. We observed that cells bearing the 100 and 200 bp spacer reporter constructs, but not the cells carrying the spacer-less construct, showed higher (reporter) SNR6 transcription levels when co-transformed with Gal4DBD-τ138 and the pol II activator Lex-AD than when transformed with Gal4DBD-τ138 alone (compare ‘no spacer’ in Figure 3B lanes 1 and 5, ‘100 bp spacer’ lanes 2 and 6, ‘200 bp spacer’ lanes 3 and 7, and Figure 4B). Figure 3C plots the SNR6 expression as a function of distance between the reporter SNR6 and the UASG element. The co-expression of Gal4DBD-τ138 and Lex-AD increased SNR6 expression only at distances (100 and 200 bp) at which Lex-AD was capable of activating the lacZ gene. These results show that the helping effect of the pol II activator Lex-AD on the action of Gal4DBD-τ138 only occurred in the case of the increased distance between the binding sites for this protein and the SNR6 sequence. The helping effect was not observed when the binding sites for Gal4DBD-τ138 were proximal to the pol III gene.
Figure 3.
Reduced expression of the split pol III genes can be rescued by a pol II activator working from a remote enhancer position to activate a pol II promoter. (A) Spacer sequences of 100, 200 and 400 bp were inserted between the SNR6 gene and the UASG sequence of the reporter construct depicted in Figure 1A. (B) Yeast reporter strains with the indicated spacer insertions were transformed with Gal4DBD-τ138 and either Lex-AD or empty plasmids. SNR6 expression was monitored by Primer-extension analysis of the U6 snRNA expressed from the reporter SNR6 constructs (indicated by an asterisk). The lower band (indicated by a triangle) represents the shorter U6 snRNA that is constitutively expressed from the wild-type pol III promoter. (C) Quantification of SNR6 reporter gene expression levels. The U6snRNA levels expressed from the reporter constructs (indicated by an asterisk) were normalized against the constitutively expressed U6snRNA (SNR6-6) levels driven by the wild-type pol III promoter (triangle) using imagequant software. Co-expression of both activators increased reporter SNR6 expression in constructs with distantly positioned enhancers, but fails to increase reporter expression at proximal enhancer positions or positions too remote for lacZ reporter activation (arbitrary units). (D) Quantitative β-galactosidase assay to determine the relative expression levels of the lacZ reporter genes.
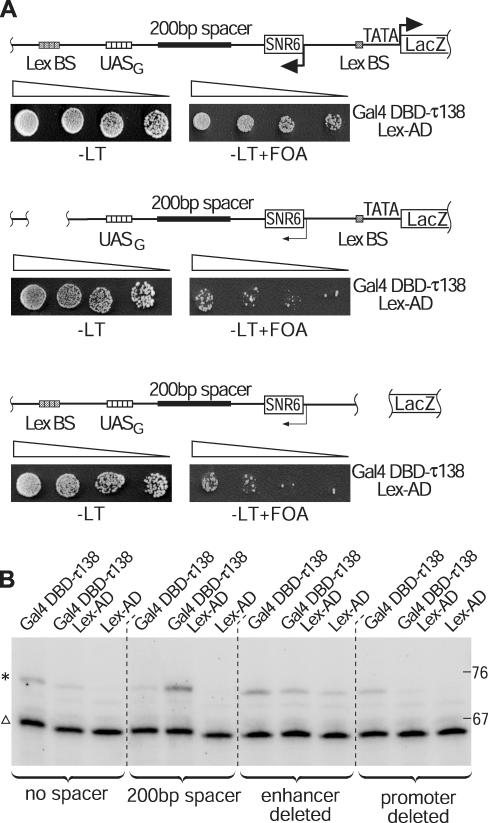
Figure 4.
Deletion of either the pol II enhancer or the pol II promoter sequence abolishes the rescue of the reduced expression of the split pol III genes by the pol II activator. (A) 3-fold stepwise dilutions of yeast cells containing one of the three depicted reporter constructs (top: complete; middle: enhancer deleted; bottom: promoter deleted) were spotted on non-selective (−LT) and selective plates (−LT + 0.1%FOA). All three strains expressed Gal4DBD-τ138 in combination with the pol II activator Lex-AD. (B) Primer-extension analysis of the U6 snRNA expressed from the reporter SNR6 constructs (asterisk). The lower band (triangle) represents the shorter U6 snRNA (SNR6-6) that is constitutively expressed from the wild-type pol III promoter. The integrated reporter constructs of the individual yeast strains are indicated with curly braces while the effectors expressed in each strain are indicated at the top of each lane.
We therefore asked whether this helping effect of the pol II activator is also dependent on the presence of the pol II promoter and the pol II enhancer. As shown in Figure 4, such a helping effect of the pol II activator required the presence of both the pol II enhancer and the promoter sequences, since deletion of either one of these elements abolished stimulation of SNR6 gene expression. The decreased SNR6 expression from the ‘no-spacer’, ‘enhancer-deleted’ and ‘promoter-deleted’ reporter strains in the presence of both the pol II and pol III activators, as compared to the presence of the pol III activator alone, is probably due to the same non-specific inhibitory effect (squelching) seen in the control experiments. Consistent with this explanation, a slight growth-rate reduction in the presence of the over-expressed pol II activator was also observed for all tested yeast strains under non-selective conditions (data not shown). These results show that the simultaneous presence of pol II enhancer and promoter is necessary for the helping effect of the pol II activator Lex-AD on the action of Gal4DBD-τ138 at an increased distance between the binding sites for this protein and the SNR6 sequence.
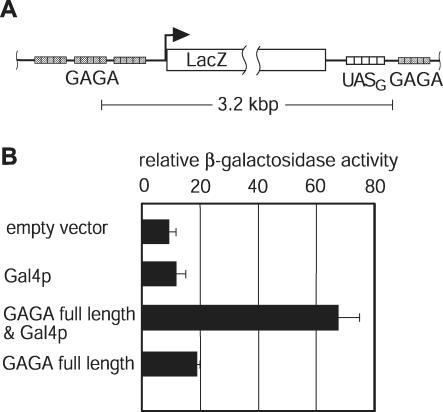
We reasoned that the reduced SNR6 expression, caused by an increased distance between the UASG element and the SNR6 promoter can be rescued by the formation of a loop that brings the distantly located bound activator Gal4DBD-τ138 in close proximity to the SNR6 promoter. Pol II enhancer activation in yeast is limited to a distance of ∼1200 bp (5–7). A yeast pol II enhancer that is unable to activate a reporter gene due to its large distance to the respective promoter (>1200 bp) might therefore be rescued in a manner similar to the rescue of the SNR6 expression. Reactivation of a distantly located enhancer would therefore require a protein that facilitates enhancer–promoter communication by bringing the enhancer into close proximity of the regulated promoter thereby looping out the intervening DNA. GAGA, a protein encoded by the essential Trithorax-like (Trl) gene of Drosophila melanogaster, has been shown to perform a multitude of chromosomal tasks and to be required for the expression of a wide range of different genes (29–32). In vitro GAGA facilitates transcription of distantly located enhancers by linking them to their cognate promoter through oligomerization when bound to DNA (24,33,34). Therefore, a factor like GAGA that is able to link distantly located enhancers to its respective promoter in vitro should enable enhancers to activate transcription from larger enhancer–promoter distances than naturally occurring in yeast. A lacZ reporter gene flanked by several GAGA elements containing four USAG-binding sites between the 3′ end of the lacZ gene and the downstream GAGA elements was integrated into the genome (Figure 5A). As expected from previous works, neither the classical activator Gal4p nor GAGA, shown to act as a desilencer rather than a classical activator, did activate transcription of the lacZ reporter gene (35). Co-expression, however, resulted in a low but robust expression of the reporter. This is not only remarkable because of the unprecedented distance but also because classical pol II activators in yeast have been shown not to work from positions downstream of the TATA box (36,37). We therefore conclude that Gal4p is recruited to a region upstream of the TATA box of the Gal1 promoter by the multimerizing GAGA factors bound to the corresponding sites downstream and upstream of the lacZ gene.
Figure 5.
GAGA-mediated enhancer recruitment to a promoter allows long distance activation in yeast. (A) Structure of the reporter system to monitor GAGA activity in yeast. The lacZ gene is under the control of the GAL1 promoter. Four Gal4p-binding sites (UASG) were inserted at the 3′ end of the lacZ gene. Upstream of the GAL1 promoter and adjacent to the (UASG) sites GAGA sites were introduced. (B) Expression of GAGA promotes transcriptional activation at large distances. The reporter construct depicted in (A) was integrated into the genome. The resulting reporter strain was transformed with Gal4p and/or GAGA factor expressing or empty plasmids. Expression of the lacZ reporter was quantified by β-galactosidase assays.
DISCUSSION
Our results show that the reduced SNR6 expression, which is caused by the increased distance between the DNA-tethered Gal4DBD-τ138 and this gene, can be rescued by the binding of a pol II activator to the enhancer sequence that is located adjacent to the binding site for Gal4DBD-τ138 (see Figures 3 and 4). Since binding of the pol II activators to the enhancer strongly activated the lacZ gene, but did not activate SNR6 expression in the absence of the Gal4DBD-τ138 fusion protein (Figures 2, 3 and 4), it is excluded that the pol II activator directly influences the SNR6 expression. This conclusion is in agreement with published results showing that pol II activators do not activate the SNR6 gene (26,27).
Our results are consistent with mechanisms that require the formation of a DNA loop to allow the physical apposition of enhancer and promoter sequences. Our results are inconsistent with mechanisms by which enhancers would essentially send out a signal along the DNA that is received and interpreted by distant promoters. If such an enhancer signal, which might be carried by ‘scanning’ or ‘linking’ activities, could stimulate expression of the SNR6 pol III reporter gene, e.g. by altering the chromatin structure, it would not require the simultaneous presence of the pol II promoter. Moreover, its effect would not give rise to the observed correlation with the increasing distance between the enhancer and the SNR6 sequence. The level of SNR6 expression stimulated by such a signal is actually expected to be either unaffected or diminished by the increased distance between the pol II enhancer and the SNR6 sequence applied in our experiments. Neither a ‘scanning’ nor a ‘linking’ mechanism predicts a gain of transcriptional activity at an increased distance between the regulatory elements. DNA loop formation in vivo was shown to be inefficient at distances <80 bp and is optimal ∼200 bp, which is in good correlation with our data. Looping is dependent on length and stiffness of DNA, and shows a ‘bell-shaped’ graph when plotted as a function of distance (38) similar to the graph obtained when SNR6 transcription was plotted as a function of distance in the presence of both activators (Figure 3C). Ineffective loop formation due to the stiffness of DNA explains the higher SNR6 activation seen in the 100 and 200 bp spacer constructs compared with the no spacer constructs.
Recently, it was shown that yeast genes form loops that bring terminator and promoter regions in close spatial proximity and that the formation of these loops is dependent on the phosphorylation of the C-terminal domain (CTD) of the large subunit of pol II (39). Although pol II and pol III share many subunits, the CTD, a domain that contains multiple heptapeptide repeats, is unique to pol II. This offers an attractive explanation for the fact that pol III activators are incapable of activating transcription at a distance. Due to the lack of a CTD domain, pol III complexes are unable to induce the formation of a loop and their ability to activate transcription is confined to short distances (40). If the physical apposition of promoter and enhancer sequences is not generated by random collisions but by an enhancer complex scanning along DNA, the role of CTD phosphorylation might be to stop the scanning enhancer complex by exchanging protein complexes, thereby anchoring the loop to the promoter. If so, our results suggest that the loops formed by scanning in the absence of a functional promoter must be transient or unstable since they were unable to rescue SNR6 expression at a distance (Figure 4).
The results of our genetic experiments strongly favor the view that signaling between the pol II enhancer and the promoter is direct and requires physical apposition of these elements, which can be accommodated by DNA looping. Such a DNA looping brought the UASG-tethered τ138 closer to the SNR6 gene located near the pol II promoter, thus stimulating the transcription of this gene. Therefore, we interpret our findings as evidence for DNA looping induced by a transcriptional enhancer in vivo. This, however, does not exclude preceding ‘scanning’ or ‘linking’ events that might favor the formation of a DNA loop.
Can enhancers in metazoan work by the same mechanism over much larger distances than in yeast? It is generally assumed that the interactions that have been shown to occur between enhancer-binding activators and transcription factors working at promoters do not suffice to overcome the entropy barriers to proper interactions between very distant enhancer and promoter elements with concomitant DNA loop formation (11). Several proteins and DNA elements have been identified that might facilitate communication between enhancers and promoters in metazoans (10,20,21,24,41–43). One of the models that have been suggested to explain their mechanism of action proposes that this class of factors might control formation of DNA loops between enhancers and promoters, such as to bring these elements de facto closer to each other. Since many transcriptional activators, as well as many components of the transcriptional machinery, are conserved between yeast and metazoans, it is plausible that the basic mechanism of enhancer–promoter apposition mediated by the interactions between activators and the transcription complex is also conserved in all eukaryotic cells. Metazoans require the intervention of additional factors (‘facilitators’) in order to allow promoter–enhancer communication over very large distances, perhaps by shrinking the actual distance in three-dimensional space between enhancers and promoters to the equivalent of a few hundred to a few thousand base pairs length. Some of these metazoan-specific chromatin remodeling factors have been shown to regulate the flexibility (‘bending properties’) of DNA (38,44), and might play an important role in regulating the access of the enhancer to its corresponding promoter during development and cell differentiation. Since in metazoan development many genes are regulated by several enhancers, it is interesting to speculate that long distance activation ‘facilitators’ might have been a pre-requirement for metazoan evolution. With the expression of the GAGA factor and the resulting long distance activation in yeast, we show that at least some of these mechanisms can be reconstituted in yeast.
Acknowledgments
We thank Drs K. Basler, R. Eckner, L. Martin, M. Rosenberg and members of the Barberis lab for valuable comments, and Drs M.-C. Marsolier and A. Sentenac for plasmids. This work was supported by the Kanton of Zürich, the Swiss National Science Foundation and the Helmut Horten Foundation. Funding to pay the Open Access publication charges for this article was provided by the State of Zurich.
Conflict of interest statement. None declared.
REFERENCES
- 1.Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 2.Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33:729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- 3.Moreau P., Hen R., Wasylyk B., Everett R., Gaub M.P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981;9:6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillies S.D., Morrison S.L., Oi V.T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983;33:717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- 5.Barberis A., Pearlberg J., Simkovich N., Farrell S., Reinagel P., Bamdad C., Sigal G., Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 6.Brand A.H., Micklem G., Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987;51:709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- 7.Escher D., Bodmer-Glavas M., Barberis A., Schaffner W. Conservation of glutamine-rich transactivation function between yeast and humans. Mol. Cell Biol. 2000;20:2774–2782. doi: 10.1128/mcb.20.8.2774-2782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herendeen D.R., Kassavetis G.A., Geiduschek E.P. A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science. 1992;256:1298–1303. doi: 10.1126/science.1598572. [DOI] [PubMed] [Google Scholar]
- 9.Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986;322:697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- 10.Dorsett D. Distant liaisons: long-range enhancer–promoter interactions in Drosophila. Curr. Opin. Genet. Dev. 1999;9:505–514. doi: 10.1016/s0959-437x(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 11.Bulger M., Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 12.Engel J.D., Tanimoto K. Looping, linking, and chromatin activity: new insights into beta-globin locus regulation. Cell. 2000;100:499–502. doi: 10.1016/s0092-8674(00)80686-8. [DOI] [PubMed] [Google Scholar]
- 13.Blackwood E.M., Kadonaga J.T. Going the distance: a current view of enhancer action. Science. 1998;281:61–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 14.Fraser P., Grosveld F. Locus control regions, chromatin activation and transcription. Curr. Opin. Cell Biol. 1998;10:361–365. doi: 10.1016/s0955-0674(98)80012-4. [DOI] [PubMed] [Google Scholar]
- 15.Hatzis P., Talianidis I. Dynamics of enhancer–promoter communication during differentiation-induced gene activation. Mol. Cell. 2002;10:1467–1477. doi: 10.1016/s1097-2765(02)00786-4. [DOI] [PubMed] [Google Scholar]
- 16.Dillon N., Trimborn T., Strouboulis J., Fraser P., Grosveld F. The effect of distance on long-range chromatin interactions. Mol. Cell. 1997;1:131–139. doi: 10.1016/s1097-2765(00)80014-3. [DOI] [PubMed] [Google Scholar]
- 17.Mueller-Storm H.P., Sogo J.M., Schaffner W. An enhancer stimulates transcription in trans when attached to the promoter via a protein bridge. Cell. 1989;58:767–777. doi: 10.1016/0092-8674(89)90110-4. [DOI] [PubMed] [Google Scholar]
- 18.Li R., Knight J.D., Jackson S.P., Tjian R., Botchan M.R. Direct interaction between Sp1 and the BPV enhancer E2 protein mediates synergistic activation of transcription. Cell. 1991;65:493–505. doi: 10.1016/0092-8674(91)90467-d. [DOI] [PubMed] [Google Scholar]
- 19.de Bruin D., Zaman Z., Liberatore R.A., Ptashne M. Telomere looping permits gene activation by a downstream UAS in yeast. Nature. 2001;409:109–113. doi: 10.1038/35051119. [DOI] [PubMed] [Google Scholar]
- 20.Carter D., Chakalova L., Osborne C.S., Dai Y.F., Fraser P. Long-range chromatin regulatory interactions in vivo. Nature Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 21.Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 22.Wach A., Brachat A., Pohlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 23.Marsolier M.C., Chaussivert N., Lefebvre O., Conesa C., Werner M., Sentenac A. Directing transcription of an RNA polymerase III gene via GAL4 sites. Proc. Natl Acad. Sci. USA. 1994;91:11938–11942. doi: 10.1073/pnas.91.25.11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahmoudi T., Katsani K.R., Verrijzer C.P. GAGA can mediate enhancer function in trans by linking two separate DNA molecules. EMBO J. 2002;21:1775–1781. doi: 10.1093/emboj/21.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaffner W. Enhancer. In: Creighton A.E, editor. The Encyclopedia of Molecular Biology. NY: John Wiley & Sons Inc.; 1999. pp. 823–828. [Google Scholar]
- 26.Marsolier M.C., Prioleau M.N., Sentenac A. A RNA polymerase III-based two-hybrid system to study RNA polymerase II transcriptional regulators. J. Mol. Biol. 1997;268:243–249. doi: 10.1006/jmbi.1997.0979. [DOI] [PubMed] [Google Scholar]
- 27.Petrascheck M., Castagna F., Barberis A. Two-hybrid selection assay to identify proteins interacting with polymerase II transcription factors and regulators. Biotechniques. 2001;30:296–298. doi: 10.2144/01302st02. 300, 302. [DOI] [PubMed] [Google Scholar]
- 28.Boeke J.D., LaCroute F., Fink G.R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 29.Farkas G., Gausz J., Galloni M., Reuter G., Gyurkovics H., Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 30.Farkas G., Leibovitch B.A., Elgin S.C. Chromatin organization and transcriptional control of gene expression in Drosophila. Gene. 2000;253:117–136. doi: 10.1016/s0378-1119(00)00240-7. [DOI] [PubMed] [Google Scholar]
- 31.Henikoff S., Vermaak D. Bugs on drugs go GAGAA. Cell. 2000;103:695–698. doi: 10.1016/s0092-8674(00)00172-0. [DOI] [PubMed] [Google Scholar]
- 32.Mahmoudi T., Verrijzer C.P. Chromatin silencing and activation by Polycomb and trithorax group proteins. Oncogene. 2001;20:3055–3066. doi: 10.1038/sj.onc.1204330. [DOI] [PubMed] [Google Scholar]
- 33.Katsani K.R., Hajibagheri M.A., Verrijzer C.P. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 1999;18:698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Espinas M.L., Jimenez-Garcia E., Vaquero A., Canudas S., Bernues J., Azorin F. The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. J. Biol. Chem. 1999;274:16461–16469. doi: 10.1074/jbc.274.23.16461. [DOI] [PubMed] [Google Scholar]
- 35.Ishii K., Laemmli U.K. Structural and dynamic functions establish chromatin domains. Mol. Cell. 2003;11:237–248. doi: 10.1016/s1097-2765(03)00010-8. [DOI] [PubMed] [Google Scholar]
- 36.Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc. Natl Acad. Sci. USA. 1984;81:7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guarente L., Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the ‘TATA box’. Proc. Natl Acad. Sci. USA. 1984;81:7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ringrose L., Chabanis S., Angrand P.O., Woodroofe C., Stewart A.F. Quantitative comparison of DNA looping in vitro and in vivo: chromatin increases effective DNA flexibility at short distances. EMBO J. 1999;18:6630–6641. doi: 10.1093/emboj/18.23.6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Sullivan J.M., Tan-Wong S.M., Morillon A., Lee B., Coles J., Mellor J., Proudfoot N.J. Gene loops juxtapose promoters and terminators in yeast. Nature Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 40.Gerber H.P., Hagmann M., Seipel K., Georgiev O., West M.A., Litingtung Y., Schaffner W., Corden J.L. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature. 1995;374:660–662. doi: 10.1038/374660a0. [DOI] [PubMed] [Google Scholar]
- 41.Georgiev P.G., Murav'eva E.E., Golovnin A.K., Gracheva E.M., Belen'kaia T. [Insulators and interaction between long-distance regulatory elements in higher eukaryotes] Genetika. 2000;36:1588–1597. [PubMed] [Google Scholar]
- 42.Cai H.N., Shen P. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science. 2001;291:493–495. doi: 10.1126/science.291.5503.493. [DOI] [PubMed] [Google Scholar]
- 43.Muravyova E., Golovnin A., Gracheva E., Parshikov A., Belenkaya T., Pirrotta V., Georgiev P. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science. 2001;291:495–498. doi: 10.1126/science.291.5503.495. [DOI] [PubMed] [Google Scholar]
- 44.Ross E.D., Hardwidge P.R., Maher L.J., III HMG proteins and DNA flexibility in transcription activation. Mol. Cell Biol. 2001;21:6598–6605. doi: 10.1128/MCB.21.19.6598-6605.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]