Abstract
We have developed new surface to ensure a proper spacing between immobilized biomolecules. While DNA microarray on this surface provided each probe DNA with ample space for hybridization with incoming target DNAs, the microarray showed enhanced discrimination efficiency for various types of single nucleotide polymorphism. The high discrimination efficiency holds for all tested cases (100:<1 for internal mismatched cases; 100:<28 for terminal mismatched ones). In addition, by investigating influence of hybridization temperature and washing condition on the fluorescence intensity and the discrimination efficiency with and without controlled mesospacing, it was observed that the nanoscale-controlled surface showed good discrimination efficiency in a wide range of temperature (37–50°C), and hybridization behavior on the surface was in agreement with the solution one. Intriguingly, it was found that washing process after the hybridization was critical for the high discrimination efficiency. For the particular case, washing process was so efficient that only 30 s washing was sufficient to reach the optimal discrimination ratio.
INTRODUCTION
Immobilized biomolecules on surface have found application in various fields such as a biosensor, a biochip, an affinity chromatography, an ELISA, a bead assay, an SPR spectroscopy, etc. While all the biomolecules have inherent activity and function in aqueous phase, a part of its property is distorted frequently through interaction with the surface (1–3). Undesirable orientation, reduced degree of freedom, and significant interaction with non-innocent surface are main causes of the distortion. In addition, poorly controlled packing of biomolecules on surface disturbs pristine biomolecular interaction due to severe electrostatic disturbance and/or steric hindrance. These problems can generally be alleviated through proper surface design and rational conjugation approach.
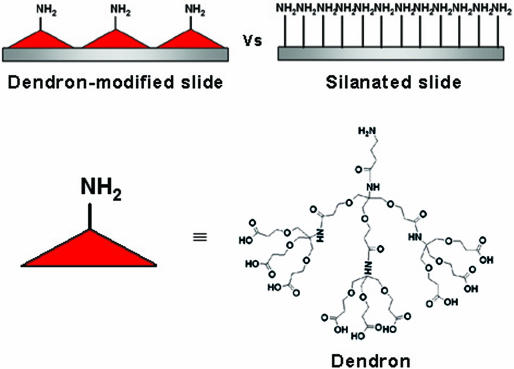
Immobilization of biomolecules with precise control has been studied extensively since early 1990s (4–9), while the importance of the lateral spacing between the biomolecules was recognized only recently. Until now, mixed self-assembled monolayers have been generally utilized to control the lateral spacing between biomolecules on surface (10–16). However, statistics governs distribution of the spacing for the case. Usually the situation gets worse, because molecules with the similar functional group are inclined to associate closely to form aggregates. On the other hand, a cone-shaped dendrimer molecule, ‘dendron’ ensures the regular distance between biomolecules on surface when its termini are glued to the surface (Scheme 1) (17–19). A dendron, which is a word to call one branch of a dendrimer, has uniform size and molecular weight as well as well-defined structure unlike a typical polymer (20–22). Because it is possible to control its size and structure precisely, we can regulate the lateral spacing between each dendron molecule on a surface. Previously, we reported preparation of DNA microarray on the second generation dendron-modified surface and beneficial influence of the mesospacing (18,19). Here we report, examination of the microarray for detecting various mutant types and investigation on the performance in several hybridization and washing conditions. For comparison, DNA microarrays were fabricated also on silanated slides which have high surface density of reactive amine functionality.
Scheme 1.
Dendron-modified slide, silanated slide and the employed dendron molecule.
MATERIALS AND METHODS
Materials
All chemicals and solvents for synthesis of the dendron were of reagent grade from Sigma-Aldrich and Mallinckrodt Laboratory Chemicals. The silane coupling reagent (3-glycidoxypropyl)methyldiethoxysilane was purchased from Gelest, Inc. and all other chemicals for surface reaction were of reagent grade from Sigma-Aldrich. Reaction solvents for the silylation are anhydrous ones in Sure/Seal bottles from Aldrich. All washing solvents for the substrates are of HPLC grade from Mallinckrodt Laboratory Chemicals. Glass slides (2.5 cm × 7.5 cm) were purchased from Corning Co. Silanated slides purchased from Telechem International, Inc. are amine-modified ones. All of the oligonucleotides were purchased from Metabion. Ultrapure water (18 M Ω/cm) was obtained from a Milli-Q purification system (Millipore).
Instruments
Oligonucleotides were spotted using MicroSys5100 microarrayer (Cartesian Technologies, Inc.) or Piezorray (PerkinElmer LAS). Hybridization was performed with GeneTAC™ HybStation (Genomic Solutions, Inc.) or HS4800 (TECAN). The fluorescence signal of the microarray was measured with both ScanArray Lite (GSI Lumonics) and ArrayWorx (Applied Precision, Inc.), and the images were analyzed with an Imagene 4.0 software (Biodiscovery).
Preparation of the dendron-modified slides
The dendron-modified slide was prepared according to the procedure as described previously (19) and a detailed procedure is included in Supplementary Material.
Arraying probe oligonucleotides on the dendron-modified slides
The deprotected dendron-modified slides were incubated in the acetonitrile solution dissolving di(N-succinimidyl)carbonate (25 mM) and a catalytic amount of DIPEA. After 4 h reaction under nitrogen atmosphere, the slides were placed in DMF with stirring for 30 min and washed with methanol. After being dried in a vacuum chamber, probe oligonucleotides (20 μM) in a spotting buffer solution [25 mM NaHCO3, 5.0 mM MgCl2 and 10% DMSO (pH 8.5)] were printed on the surface with a microarrayer, and the spots were arranged to make a 4 × 4 format. The microarrays were incubated in a humidity chamber (80% humidity) for half a day to give the amine-tethered oligonucleotides sufficient reaction time. The slides were then placed in a stirred hybridization buffer solution [2× SSPE buffer (pH 7.4) dissolving 7.0 mM SDS] at 37°C for 30 min and subsequently in boiling water for 5 min to remove non-specifically bound oligonucleotides. Finally, the DNA-functionalized microarrays were dried under a stream of nitrogen and stored at 4°C for the next step.
Arraying probe oligonucleotides on the silanated slides
Purchased silanated slides were incubated in a mixed solvent [DMF/pyridine 90:10 (v/v)] dissolving 1,4-phenylenediisothiocyanate (PDITC) linker (12.5 mM). After reaction for 2 h, the plates were sonicated three times each in methanol and acetone in a sequential manner. The washed plates were dried in a vacuum chamber (30–40 mTorr). Probe oligonucleotides dissolved in sodium carbonate/bicarbonate buffer solution (100 mM, pH 9.0) were printed on the PDITC-modified slides with a microarrayer, and the spots were arranged to make a 4 × 4 format. The slides were incubated in a saturated humidity chamber at 37°C for 1 h to immobilize the amine-tethered oligonucleotides. The slides were then soaked in 1% NH4OH aqueous solution for 3 min. Finally, the DNA-functionalized slides were washed with deionized water several times and dried under a stream of nitrogen.
Hybridization
A hybridization buffer solution dissolving a target oligonucleotide tagged with a Cy3 fluorescent dye was allowed to contact with the microarray using an automatic hybridization machine. After hybridization, the microarrays were washed with the hybridization buffer solution, and then with deionized water at 20°C for 1–2 s. For each hybridization experiment, 100 μl volume of a target DNA solution was utilized.
RESULTS AND DISCUSSION
Surface density and spacing of a probe oligonucleotide
Previously, surface density of the active amino group on the dendron-modified surface was measured by introducing a gold nanoparticle onto the apex of the dendron molecule. In this research, a direct measurement of the probe oligonucleotide was carried out. The surface density of a probe oligonucleotide on both slides was obtained from calibration graphs of fluorescence intensity versus surface loading of an oligonucleotide tethering a Cy3. The density of the probe oligonucleotide on the dendron slide turned out to be 0.03 Ea/nm2, which was slightly smaller than previously reported dendron density (0.05 Ea/nm2) (19). Decrease of the surface density could be from steric hindrance and/or charge repulsion between oligonucleotides. While fluorescence intensity of the tagged probe on the silanated slide is three times higher than that on the dendron slide, actual surface density of the probe oligonucleotide on the silanated slide should be >0.09 Ea/nm2, because the calibration curve showed that fluorescence was quenched to a certain degree at this range. Therefore, it is reasonable to assume each probe oligonucleotide on the dendron-modified surface occupies 33 nm2 in average, and is away from neighboring one with ∼6 nm separation.
Signal intensity versus concentration of a probe oligonucleotide
To investigate dependence of fluorescence signal intensity upon the concentration of a probe oligonucleotide, the amine-tethered probe oligonucleotide (P1) of various concentrations (2.0, 20, 50, 100 and 200 μM) was microarrayed on the dendron-modified slide and the silanated one. The oligonucleotide was immobilized on the silanated slides as described by Guo et al. (23) with slight modification. Use of di(N-succinimidyl)carbonate linker on the silanated slide resulted in low fluorescence intensity and non-uniform intensity profile within a spot. On the other hand, PDITC linker assured strong fluorescence signal and uniform profile (19). Also, the linker-modified surface gave rise to low non-specific binding of target DNA as little as that of the dendron-modified surface. For hybridization, a complementary oligonucleotide (T1) at 1.0 μM concentration was utilized to expose the DNA microarray with sufficient amount of the target oligonucleotide. After hybridization with the target tethering a Cy3 dye at 45°C for 1 h, the microarray was washed with the hybridization buffer solution at 37°C for 1 min, and then with deionized water briefly. Because salts and SDS left on the surface prevent the slides from manifesting homogeneous signal profile and low background, it is essential to wash salts and SDS from the slides with water. The fluorescence signal on the dendron-modified slide increased sharply from 2.0 to 20 μM probe concentration, and it was constant ∼20 μM concentration, whereas that on the silanated slide saturated at 200 μM concentration (Figure 1). Rather low signal intensity of the dendron-modified slide reflects lower surface density of the probe. The data show that it is not necessary to use probe concentration >20 μM concentration to maximize the signal intensity for the latter case. The average value and error bar were obtained from 16 spots on a slide.
Figure 1.
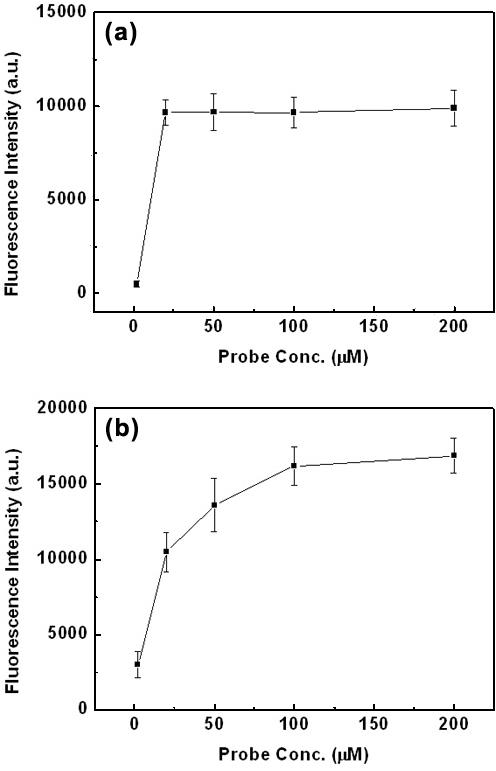
Fluorescence signal intensity versus probe oligonucleotide concentration (a) on the dendron-modified slide and (b) on the silanated slide. Target concentration is 1.0 μM. The average value at each condition was obtained from 16 spots on a slide and an error bar represents its SD. Laser power and photomultiplier tube (PMT) gain of a laser scanner were adjusted for the optimal reliability, and fixed for a set of experiments.
Signal intensity versus concentration of a target oligonucleotide
To investigate the correlation between fluorescence intensity and concentration of a target oligonucleotide, the probe oligonucleotide (P1) of 20 μM was spotted on the dendron-modified slide, whereas that of 200 μM on the silanated one. Thus-formed DNA microarrays were allowed for hybridization with the complementary counterpart (T1) at various concentrations (100 pM, 1.0 nM, 10 nM, 100 nM and 1.0 μM) (Figure 2). After hybridization at 45°C for 1 h, the microarray was washed with the hybridization buffer at 37°C for 1 min. The fluorescence intensity on the dendron-modified slide increased with the concentration of the target oligonucleotide continuously and saturated at a concentration ∼10 nM, while that on the silanated slide at a concentration ∼100 nM. Because the saturation point is a function of surface density of the probe oligonucleotide and hybridization efficiency, behavior of the microarray on the dendron-modified surface is quite understandable. In other words, early saturation and smaller signal intensity reflect higher hybridization efficiency and lower surface density (19). This is in harmony with previously observed surface nature of the dendron-modified surface. It is worth noting sensitive signal dependence on the target concentration between 100 pM and 1.0 nM for the dendron-modified case.
Figure 2.
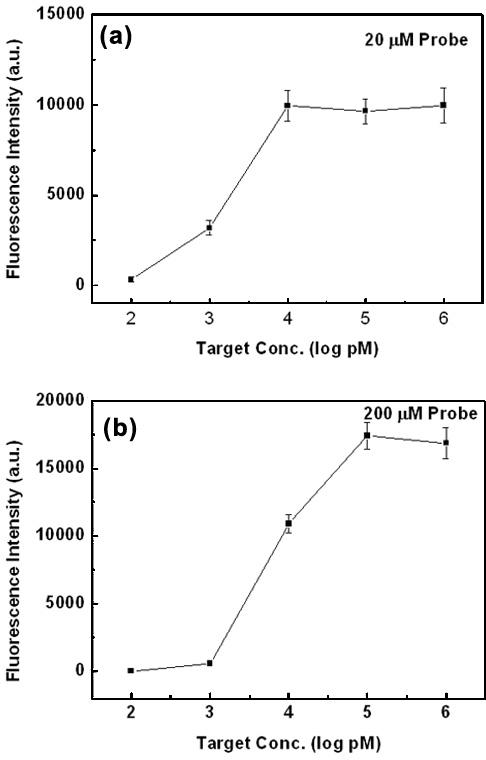
Fluorescence signal intensity versus target oligonucleotide concentration (a) on the dendron-modified slide and (b) on the silanated slide. Probe oligonucleotide concentration is 20 μM for the former case, and 200 μM for the latter case.
SNP discrimination efficiency of DNA microarrays
To examine single nucleotide polymorphism (SNP) discrimination efficiency of DNA microarrays on the dendron-modified slide and the silanated one, various types of mismatches including internal mismatch, insertion and deletion were tested. Probe and target oligonucleotides utilized in this work were listed in Table 1. For fair comparison, identical oligonucleotide in a solution was used for both slides, and all probe oligonucleotides were spotted on a single slide. Hybridization process was performed at 45°C for 1 h, followed by rinsing the microarrays with the hybridization buffer at 50°C for 1 min. Because the discrimination efficiency depends on washing condition and hybridization temperature, the optimal condition should be employed for the highest efficiency. For the both microarrays under scrutiny, the above condition was optimal coincidentally (vide infra).
Table 1.
Sequences and nomenclature of probe and target oligonucleotides
| Designation | Sequence |
|---|---|
| P1 | 5′-NH2-C6-CATTCCGAGTGTCCA-3′ |
| P2 | 5′-NH2-C6-CATTCCGTGTGTCCA-3′ |
| P3 | 5′-NH2-C6-CATTCCGGGTGTCCA-3′ |
| P4 | 5′-NH2-C6-CATTCCGCGTGTCCA-3′ |
| P5 | 5′-NH2-C6-CATTCCGAGTGTCAA-3′ |
| P6 | 5′-NH2-C6-CATTCCGAGTGTCTA-3′ |
| P7 | 5′-NH2-C6-CATTCCGAGTGTCGA-3′ |
| P8 | 5′-NH2-C6-CATTCCGAAGTGTCCA-3′ |
| P9 | 5′-NH2-C6-CATTCCG_GTGTCCA-3′ |
| P10 | 5′-NH2-C6-CATTCCGAGTGTC_A-3′ |
| P11 | 5′-NH2-C6-CATTCCGAGTGTCC_-3′ |
| P12 | 5′-NH2-C6-CATTCCGAGTGTCCT-3′ |
| P13 | 5′-NH2-C6-CATTCCGAGTGTCCG-3′ |
| P14 | 5′-NH2-C6-CATTCCGAGTGTCCC-3′ |
| P15 | 5′-NH2-C6-ACAAGCACAGTTAGG-3′ |
| T1 | 5′-Cy3-TGGACACTCGGAATG-3′ |
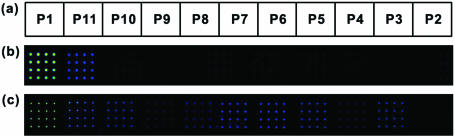
Figure 3a shows position of the various oligonucleotides spotted on a single slide. P1 is the complementary probe oligonucleotide, P2–P4 the oligonucleotides with one base mismatch at the middle position, and P5–P7 those with one base mismatch at the second last position from the 3′ end. P8 oligonucleotide has extra one base at the middle position, and P9–P11 designate oligonucleotides with deletion at three different positions. For the DNA microarray on the dendron-modified slide, all mismatched cases showed discrimination ratio (i.e. the fluorescence intensity of the mismatched duplex divided by that of the complementary one) <1%, except one terminal deletion case giving 10% (Figure 3b and Table 2). In general, it is difficult to discriminate the DNA duplex with one base deletion at the terminal site from the complementary one due to the tiny difference of Gibbs free energy (ΔG) and melting temperature (Tm). On the other hand, the DNA microarray on the silanated slide showed inferior discrimination efficiency: Most of the ratios are much larger than 1% except P2 case (Figure 3c and Table 2). Our previous study showed that the dendron-modified surface discriminated internal mismatched cases efficiently as if the oligonucleotides were in solution phase (19). Here, we demonstrated that the dendron-modified surface could ensure efficient discrimination for many other mismatched cases, and the lateral spacing between probe oligonucleotides must be one of the key factors to be considered for the good discrimination efficiency.
Figure 3.
Fluorescence images after the hybridization for various mutant types. (a) Each position of probe oligonucleotides is shown. Image (b) was obtained from the microarray on the dendron-modified surface, and image (c) from one on the silanated surface. Fluorescence images were obtained after the hybridization with T1 target oligonucleotide. Laser power and PMT gain of a laser scanner were adjusted to record the reasonable signals of mismatched pairs while avoiding saturation of the signal of the matched one.
Table 2.
Single nucleotide polymorphism (SNP) discrimination ratio
| Discrimination ratioa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | P11 | P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | |
| Dendron-modified slide | 100 | 10 | 0.4 | 0.1 | 0.4 | 0.4 | 0.3 | 0.4 | 0 | 0.5 | 0 |
| Silanated slide | 100 | 30 | 15 | 4 | 9 | 22 | 21 | 22 | 6 | 22 | 0.3 |
aThe Discrimination ratio represents the fluorescence intensity of the mismatched pair divided by that of the complementary one in percentage.
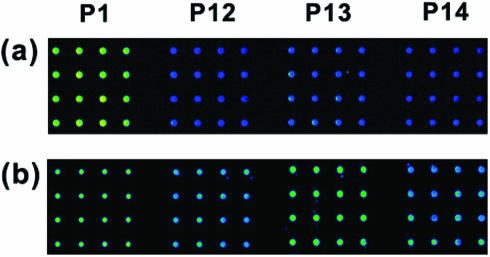
To understand characteristics of the dendron-modified slide further, we also tested single base mismatches at the terminal site. It is challenging to discriminate single terminal mismatches with a DNA microarray. The nearest neighbor model shows that difference in ΔG and Tm in solution is only 0.45–0.87 kcal/mol and 1.2–2.4°C, which is too small in comparison with those of the internal mismatched cases (1.97–3.26 kcal/mol and 5.7–9.8°C) (http://ozone2.chem.wayne.edu.) With three oligonucleotides (P12–P14), the observed discrimination ratios were 16, 28 and 14% (T:T, G:T and C:T) (Figure 4a). On the other hand, the DNA microarray fabricated on the silanated slide showed poor discrimination ratios, i.e. 66, 96 and 61% (T:T, G:T and C:T) (Figure 4b). For the G:T mismatched pair, that signal is almost indistinguishable with that of the matched case. Here, capability to discriminate even the most challenging terminal mismatched case is demonstrated for the DNA microarrays on the mesospaced surface.
Figure 4.
Fluorescence images for the terminal mutant types. Image (a) was obtained from the microarray on the dendron-modified slide, and image (b) from that on the silanated slide.
Signal intensity and discrimination efficiency in various conditions: washing time, washing temperature and hybridization temperature
We have measured the signal intensity and the discrimination efficiency in various conditions, such as washing time and washing/hybridization temperature. In this study, P1–P4 and T1 oligonucleotides were employed, and the silanated slide was utilized for comparative purpose.
Washing time
After hybridization at 45°C for 1 h, the microarray was washed with the hybridization buffer solution at 37°C. Figure 5 shows change of the fluorescence intensity and the discrimination ratio with the washing time. For both DNA microarrays on the dendron-modified slide and the silanated one, the signal intensity decreased as the washing time increased, but slope of the decrease and saturation behavior are quite different. In the case of DNA microarray on the dendron-modified slide, the intensity decreased moderately for the matched pair, while steep decrease during the first 10 s was observed for three mismatched ones. For T:G mismatched one, the discrimination ratio dropped from 90 to 15% during the time. It was observed that the short washing time was sufficient to achieve the saturation value of the discrimination efficiency. The sharp initial decrease for the mismatched cases is a peculiarity of the dendron-modified surface, and the phenomenon seems to be associated with facile dissociation of the target oligonucleotide weakly associated through abnormal fashion, of which nature has yet to be understood. Because melting points of the duplex for the mismatched cases (40–45°C) are slightly over the washing temperature, steeper decrease of intensity is what expected. However, the initial decrease during the first 10 s is too fast to explain with the melting points. This is the reason why a working hypothesis involving an unusual and weak binding mode is proposed.
Figure 5.
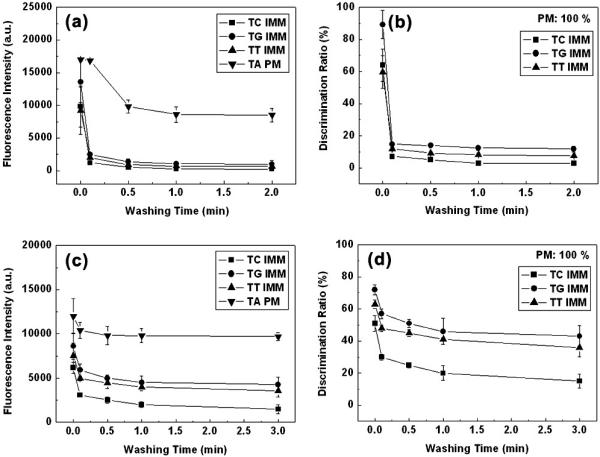
Effect of washing time on the fluorescence signal intensity and the discrimination ratio (a and b) on the dendron-modified slide and (c and d) on the silanated one. (a and c) Fluorescence signal intensity versus washing time and (b and d) discrimination ratio versus washing time. PM signifies perfect match and IMM one base internal mismatch. The discrimination ratio represents the fluorescence intensity of the mismatched pair divided by that of the complementary one.
On the other hand, DNA microarray on the silanated slide showed relatively slow signal decrease for the mismatched pairs as well as the matched one. After the initial decrease, the signal kept decreasing until 3 min. Also, the initial decrease for the mismatched cases is not as steep as the dendron-modified slide. While the elongated washing time up to 3 min enhanced the discrimination efficiency marginally, its values never reached those of the dendron-modified surface. As a result, it is observed that the washing process for DNA microarrays on the dendron-modified surface is so effective that only short washing time is required to reach the saturation point. It is interesting to note that the discrimination efficiency for both slides is enhanced during the washing step. This observation suggests a washing step should be implemented for even continuous monitoring systems, such as some electrochemical devices if high discrimination efficiency is an important goal to achieve.
Washing temperature
We studied the effect of the washing temperature on the signal intensity and the discrimination efficiency. After hybridization with the target oligonucleotide at 45°C for 1 h, the microarray was washed with the hybridization buffer solution at various temperatures for 1 min.
For DNA microarray fabricated on the dendron-modified slide, the discrimination efficiency for all three mismatched pairs was enhanced up to 50°C, and the best discrimination (<1%) was obtained at 50°C (Figure 6b). Because calculated Tm values of each duplex are 50.2°C (complementary), 42.2°C (T:T mismatch), 44.5°C (T:G mismatch) and 40.4°C (T:C mismatch) in solution (http://ozone2.chem.wayne.edu.), the above result is in harmony with the expectation. At elevated temperature such as 55°C, tarnished discrimination was observed, mainly because the temperature was high enough to dissociate even matched target oligonucleotide. This is evident in Figure 6a, where the signal intensity for all four cases decreases to almost zero. Moderately good discrimination efficiency is maintained at 37 and 45°C as well, and this suggests the efficiency is not hyper-sensitive to washing temperature. The latter characteristic is quite beneficial, because for a certain application >10000 different oligonucleotides were spotted in a single slide. For the last case, DNA duplexes with a rather broad range of Tm values should be treated at a fixed condition, and the tolerance should be of help to maintain the high discrimination efficiency. For a comparison, DNA microarrays on the silanated slide were tested. It was also true that the DNA microarray showed the best discrimination efficiency at 50°C (Figure 6d). In addition, its discrimination ratio rebounded over 50°C as the previous case, but its discrimination efficiency in the entire range could not reach that of the former DNA microarray.
Figure 6.
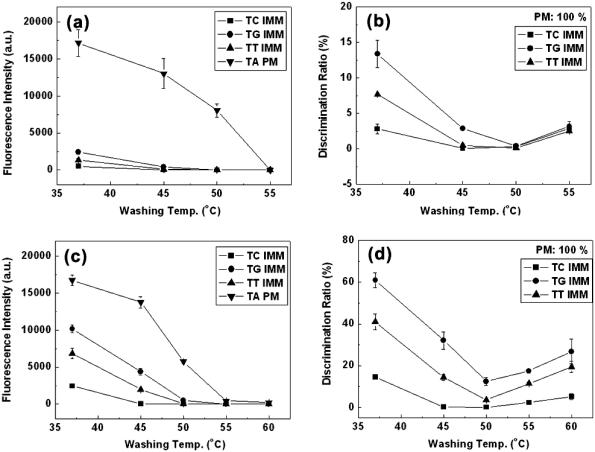
Effect of washing temperature on the fluorescence signal intensity and the discrimination ratio (a and b) on the dendron-modified slide and (c and d) on the silanated one. (a and c) Fluorescence signal intensity versus washing temperature and (b and d) discrimination ratio versus washing temperature.
Hybridization temperature
Hybridization temperature has been a key factor under scrutiny for enhanced performance of DNA microarrays. It is expected that the temperature should be strongly associated with range and distribution of melting points of DNA duplexes of the entire pixels. In this study, hybridization was allowed at various temperatures (37, 45, 50 and 60°C), and the slide was washed with hybridization buffer solution at 37°C for 1 min.
Figure 7 shows change of the fluorescence intensity and the discrimination ratio with the hybridization temperature. In the case of DNA microarray on the dendron-modified slide, the discrimination ratio of all three mismatched pairs decreased as the hybridization temperature increased up to 45°C, but rebounded over 50°C (Figure 7b), mostly because the fluorescence intensity of the matched one was constant up to 45°C, but dropped drastically over 45°C (Figure 7a). While hybridization at 45°C or 50°C can provide the highest discrimination efficiency, the fluorescence intensity at 45°C is twice as high as that at 50°C. As far as the discrimination efficiency is concerned, the tolerance holds again.
Figure 7.
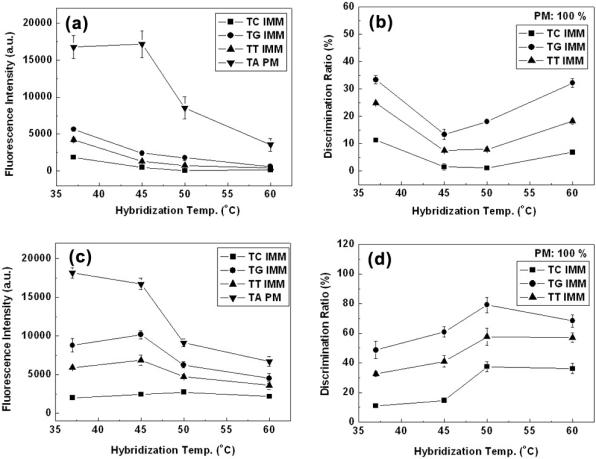
Effect of hybridization temperature on the fluorescence signal intensity and the discrimination ratio (a and b) on the dendron-modified slide and (c and d) on the silanated one. (a and c) Fluorescence signal intensity versus hybridization temperature and (b and d) discrimination ratio versus hybridization temperature.
This optimal hybridization condition depends absolutely on Tm values of the DNA duplexes utilized for this particular DNA microarray. Considering calculated Tm values of the DNA duplexes in the hybridization salt condition used in this work, we may say the above results agrees very well with the general concept that the discrimination in solution between a matched duplex and a mismatched one becomes the highest at the temperature near or right below Tm of the complementary duplex (24). Also, this obedient behavior is another evidence that mesopaced surface realized through the dendron allows unhindered hybridization. In the previous work, we showed that DNA microarray on the dendron-modified slide could achieve the same discrimination efficiency as that in solution. Therefore, these evidences support that the dendron-modified surface could establish an environment similar to the solution phase on the two dimensional space.
On the other hand, the DNA microarray fabricated on the silanated slide did not show any enhancement of the discrimination efficiency as hybridization temperature increased from 37 to 60°C (Figure 7d). In other words, change of its discrimination ratio does not coincide with the above general concept. It is not straightforward to explain why fluorescence intensity of the mismatched cases increases when the hybridization temperature increases from 37 to 45°C. This unexpected increase results in the deterioration of the discrimination efficiency at elevated temperatures. We may say this ought to be associated with a complicated environment far from the ideal in the silanated surface. In addition, its highest discrimination efficiency is far inferior to that of the DNA microarray on the dendron-modified slide. Overall, it can be said that effect of the hybridization temperature on the discrimination efficiency is very dependent on the nature of the surface, and sufficient spacing on surface provides DNA microarrays that are predictable with a model calculation.
CONCLUSION
This nanoscale-controlled surface provided DNA microarrays close to ideal, in which each probe DNA strand was given ample space for the incoming target DNA resulting in far superior SNP discrimination efficiency to that of the DNA microarray on the surface where spacing was poorly controlled. Detailed investigation showed that the washing process was an essential step for the effective discrimination, and its efficacy depends critically on the nature of the surface. In addition, the DNA microarray fabricated on the dendron-modified slide showed excellent discrimination efficiency in a wide range of hybridization and washing temperature. Moreover, its optimal condition for the discrimination efficiency and the signal intensity is in harmony with the melting points of the duplexes calculated with a model calculation. Its predictability is very important to design diagnosing DNA microarrays that require high reliability and reproducibility.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
Student fellowships of the Brain Korea 21 are greatly acknowledged, and also the work is supported by the Korea Science and Engineering Foundation (KOSEF) through Center for Integrated Molecular Systems. Funding to pay the Open Access publication charges for this article was provided by Pohang University of Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Southern E., Mir K., Shchepinov M. Molecular interactions on microarrays. Nature Genet. 1999;21:5–9. doi: 10.1038/4429. [DOI] [PubMed] [Google Scholar]
- 2.Schäferling M., Kambhampati D. Protein microarray surface chemistry and coupling schemes. In: Kambhampati D, editor. Protein Microarray Technology. Weinheim: Wiley-VCH Verlang GmbH & Co. KgaA; 2004. pp. 11–38. [Google Scholar]
- 3.Angenendt P., Glökler J., Murphy D., Lehrach H., Cahill D.J. Toward optimized antibody microarrays: a comparison of current microarray support materials. Anal. Biochem. 2002;309:253–260. doi: 10.1016/s0003-2697(02)00257-9. [DOI] [PubMed] [Google Scholar]
- 4.Prime K.L., Whitesides G.M. Self-assembled organic monolayers: model systems for studying adsorption of proteins at surfaces. Science. 1991;252:1164–1167. doi: 10.1126/science.252.5009.1164. [DOI] [PubMed] [Google Scholar]
- 5.Prime K.L., Whitesides G.M. Adsorption of proteins onto surfaces containing end-attached oligio(ethylene oxide): a model system using self-assembled monolayers. J. Am. Chem. Soc. 1993;115:10714–10721. [Google Scholar]
- 6.Rabinow B.E., Ding Y.S., Qin C., McHalsky M.L., Schneider J.H., Ashline K.A., Shelbourn T.L., Albrecht R.M. Biomaterials with permanent hydrophilic surfaces and low protein adsorption properties. J. Biomater. Sci. Polym. Ed. 1994;6:91–109. doi: 10.1163/156856295x00788. [DOI] [PubMed] [Google Scholar]
- 7.Chapman R.G., Ostuni E., Liang M.N., Meluleni G., Kim E., Yan L., Pier G., Warren H.S., Whitesides G.M. Polymeric thin films that resist the adsorption of proteins and the adhesion of bacteria. Langmuir. 2001;17:1225–1233. [Google Scholar]
- 8.Holmlin R.E., Chen X., Chapman R., Takayama S., Whitesides G.M. Zwitterionic SAMs that resist nonspecific adsorption of protein from aqueous buffer. Langmuir. 2001;17:2841–2850. doi: 10.1021/la0015258. [DOI] [PubMed] [Google Scholar]
- 9.Luk Y., Kato M., Mrksich M. Self-assembled monolayers of alkanethiolates presenting mannitol groups are inert to protein adsorption and cell attachment. Langmuir. 2000;16:9604–9608. [Google Scholar]
- 10.Roberts C., Chen C.S., Mrksich M., Martichonok V., Ingber D.E., Whitesides G.M. Using mixed self-assembled monolayers presenting RGD and (EG)3OH groups to characterize long-term attachment of bovine capillary endothelial cells to surfaces. J. Am. Chem. Soc. 1998;120:6548–6555. [Google Scholar]
- 11.Lahiri J., Isaacs L., Grzybowski B., Carbeck J.D., Whitesides G.M. Biospecific binding of carbonic anhydrase to mixed SAMs presenting benzenesulfonamide ligands: a model system for studying lateral steric effects. Langmuir. 1999;15:7186–7198. [Google Scholar]
- 12.Lahiri J., Isaacs L., Tien J., Whitesides G.M. A strategy for the generation of surfaces presenting ligands for studies of binding based on an active ester as a common reactive intermediate: a surface plasmon resonance study. Anal. Chem. 1999;71:777–790. doi: 10.1021/ac980959t. [DOI] [PubMed] [Google Scholar]
- 13.Peterson A.W., Wolf L.K., Georgiadis R.M. Hybridization of mismatched or partially matched DNA at surfaces. J. Am. Chem. Soc. 2002;124:14601–14607. doi: 10.1021/ja0279996. [DOI] [PubMed] [Google Scholar]
- 14.Yeo W., Yousaf M.N., Mrksich M. Dynamic interfaces between cells and surfaces: electroactive substrates that sequentially release and attach cells. J. Am. Chem. Soc. 2004;125:14994–14995. doi: 10.1021/ja038265b. [DOI] [PubMed] [Google Scholar]
- 15.Oh S.J., Cho S.J., Kim C.O., Park J.W. Characteristics of DNA microarrays fabricated on various aminosilane layers. Langmuir. 2002;18:1764–1769. [Google Scholar]
- 16.Shchepinov M.S., Case-Green S.C., Southern E.M. Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acid Res. 1997;25:1155–1161. doi: 10.1093/nar/25.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong B.J., Shim J.Y., Oh S.J., Park J.W. Self-assembly of a dendron through multiple ionic interaction to give mesospacing between reactive amine groups on the surface. Langmuir. 2003;19:2357–2365. [Google Scholar]
- 18.Choi Y., Yoon C.W., Lee H.D., Park M., Park J.W. Efficient protein-ligand interaction by guaranteeing mesospacing between immobilized biotins. Chem. Commun. 2004;11:1316–1317. doi: 10.1039/b403797a. [DOI] [PubMed] [Google Scholar]
- 19.Hong B.J., Oh S.J., Youn T.O., Kwon S.H., Park J.W. Nanoscale-controlled spacing provides DNA microarrays with the SNP discrimination efficiency in solution phase. Langmuir. 2005;21:4257–4261. doi: 10.1021/la046951y. [DOI] [PubMed] [Google Scholar]
- 20.DeMattei C.R., Huang B., Tomalia D.A. Designed dendrimer syntheses by self-assembly of single-site, ssDNA functionalized dendrons. Nano Lett. 2004;4:771–777. [Google Scholar]
- 21.Zeng F., Zimmerman S.C. Dendrimers in supramolecular chemistry: from molecular recognition to self-assembly. Chem. Rev. 1997;97:1681–1712. doi: 10.1021/cr9603892. [DOI] [PubMed] [Google Scholar]
- 22.Bosman A.W., Janssen H.M., Meijer E.W. About dendrimers: structure, physical properties and applications. Chem. Rev. 1999;99:1665–1688. doi: 10.1021/cr970069y. [DOI] [PubMed] [Google Scholar]
- 23.Guo A., Guilfoyle R.A., Thiel A.J., Wang R., Smith L.M. Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res. 1994;22:5456–5465. doi: 10.1093/nar/22.24.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cantor C.R., Smith C.L. Genomics: The Science and Technology Behind The Human Genome Project. NY: John Wiley & Sons:; 1999. pp. 71–77. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.