Abstract
Substantial progress has been realized in the past several years in our understanding of the molecular mechanisms responsible for the expansions and deletions (genetic instabilities) of repeating tri-, tetra- and pentanucleotide repeating sequences associated with a number of hereditary neurological diseases. These instabilities occur by replication, recombination and repair processes, probably acting in concert, due to slippage of the DNA complementary strands relative to each other. The biophysical properties of the folded-back repeating sequence strands play a critical role in these instabilities. Non-B DNA structural elements (hairpins and slipped structures, DNA unwinding elements, tetraplexes, triplexes and sticky DNA) are described. The replication mechanisms are influenced by pausing of the replication fork, orientation of the repeat strands, location of the repeat sequences relative to replication origins and the flap endonuclease. Methyl-directed mismatch repair, nucleotide excision repair, and repair of damage caused by mutagens are discussed. Genetic recombination and double-strand break repair advances in Escherichia coli, yeast and mammalian models are reviewed. Furthermore, the newly discovered capacities of certain triplet repeat sequences to cause gross chromosomal rearrangements are discussed.
INTRODUCTION
Approximately twenty hereditary neurological diseases are linked to the expansions of triplet repeat sequences (TRSs). Since the early 1990s, substantial progress has been made in understanding the dynamic mutations involved in these processes. The diseases [including myotonic dystrophy (DM), Huntington's disease, fragile X syndrome (FRAX), and Friedreich's ataxia (FRDA)] have been reviewed (1–9) along with their inheritance patterns, chromosomal localizations, protein products and loci of the TRS. In some cases (type 2 diseases), the repeat expansions are massive (thousands of repeats) whereas in type 1 diseases, the TRS are in coding regions and elicit an expansion of a polyamino acid (usually glutamine) tract. The clinical observation of anticipation, the decrease in age of onset and increase in severity, is observed with many but not all of these diseases. In general, a more severe neurological syndrome is observed in patients with longer repeat tracts. The clinical observations and human genetic discoveries have been reviewed previously (4–8).
This review will focus on the molecular mechanisms of the genetic instabilities. Substantial work in the past fourteen years has demonstrated that the expansions and deletions are mediated by DNA replication, repair and recombination, probably acting in concert [reviewed in (1–3,9)]. The slippage of the repeating DNA complementary strands to form hairpin loops, or slipped conformations, with differing relative stabilities are important components in the mechanism. The inherent conformational properties of the repeating sequences, such as their high degree of flexibility, writhing and stability of hairpin formation, facilitate the strand slippage. The unusual DNA conformations affect DNA polymerase stalling, accentuating the disease-causing mutagenesis. A number of other genetic factors are likely to be involved including methyl-directed mismatch repair (MMR), nucleotide excision repair, single-strand DNA binding proteins, transcription and DNA polymerase proofreading (2,3). Studies have been conducted in bacterial cells, yeast, mammalian cell culture and transgenic mice (3). Clearly, the majority of the molecular mechanisms have been established first in well-defined genetic-biochemical systems (Escherichia coli and yeast) and then these results extrapolated to other experimental systems.
This review focuses on the molecular mechanisms of the expansion–deletion processes on work published within the past five years. Recent advances in our understanding of the DNA structural properties of the repeat sequences are described as related to genetic instabilities. Additionally, investigations on instabilities mediated by replication, followed by the involvement of DNA repair, and the influence of double-strand breaks (DSBs) and recombination (gene conversion) are discussed. The last section of this review describes the involvement of non-B DNA conformations formed by TRS in generating genomic rearrangements.
NON-B DNA STRUCTURES
The structural properties of repeating trinucleotide, tetranucleotide and pentanucleotide DNA sequences, associated with several hereditary neurological diseases, are proposed to be major contributing factors to the genetic instabilities observed in these diseases (1–3,10). Several DNA repeating sequences, namely CTG•CAG, CGG•CCG, GAA•TTC, GAC•GTC, CCTG•CAGG and ATTCT•AGAAT, have been shown to form non-B DNA structures including hairpins, slipped-strand DNA, DNA unwinding elements (DUEs), tetraplexes, triplexes, and more recently, the novel structure, sticky DNA (Figure 1). Almost all models involving replication, transcription, recombination and repair, hypothesize the formation of non-B DNA secondary structures at these DNA repeats (2,11–16). At least 10 non-B conformations (2,17) are formed at specific motifs as a result of negative supercoil density (15,18–20), replication and transcriptional polymerase pausing (14,21–23). Interestingly, these secondary structures have been shown to be targets for protein binding (24–26), including several enzymes involved in DNA repair (27–33).
Figure 1.
Non-B DNA conformations associated with repeating sequences and mechanisms of instability.
Hairpins and slipped structures
Hairpins and slipped structures occur at DNA sequences with direct repeat (DR) symmetry (17). The cellular processes of replication and transcription cause unwinding of the duplex that renders the DNA single-stranded, thus giving the repeat sequences the opportunity to fold back and form alternative base pairs within the same DNA strand. The repeat sequences that have been associated with neurological diseases, in particular, are prone to slipped-mispairing and hairpin formation during replication and transcription (2,34,35). The formation of these structures can presumably lead to expansions and deletions of the repeating tract because of the stability of the misaligned intermediates.
An order of hairpin stability, CGG > CTG > CAG > CCG, was established using CD, optical melting, differential scanning and calorimetry (36). The repeat sequence CTG•CAG associated with myotonic dystrophy type 1 (DM1) has been observed to form slipped structures and hairpins in a length and orientation-dependent manner under physiological conditions (34,37–39). The stability of hairpins in the CTG strand was attributed to the T•T mismatch that stacked more efficiently as opposed to the A•A mispair on the complementary CAG strand. Studies using chemical modification, P1 nuclease digestion and NMR have shown CGG•CCG repeats to form hairpin structures; the stability of which is dependent on the G–C content in each strand (2). More recently, the CCTG•CAGG tetranucleotide repeats, associated with myotonic dystrophy type 2 (DM2) (40), showed that the CAGG strand had a greater propensity to form a more stable hairpin-loop when compared to the CCTG strand.
Although the biological implications of hairpins and slipped structure formation are not completely understood, they have been shown to bind certain repair proteins such as MSH2 and UvrA (24,29) and are even hypothesized to be recognized by recombinational repair proteins in the event where structure formation at specific loci leads to DSBs (41). In fact, Wojciechowska et al. (41) showed a correlation between the propensity for CTG•CAG repeats to form non-B structures, such as hairpins, and the creation of DSBs near the sites of structure formation, leading to gross deletions. Furthermore, GAC•GTC repeats associated with skeletal dysplasias have been shown to undergo small expansion and deletion events due to slippage when transcription is inhibited and large expansion and deletion events due to the hairpin forming propensity of the GTC strand in presence of transcription (2,42).
DNA unwinding elements
DUEs are A + T rich sequences commonly associated with replication origins and chromosomal matrix attachment sites (17). The pentanucleotide repeat ATTCT•AGAAT associated with spinocerebellar ataxia 10 (SCA10) has been reported to be a DUE (12,22,43). Recent studies found the propensity for unwinding of this sequence allowed for accessibility to chemical probes within the region, as well as oligonucleotide hybridization, which led to aberrant DNA replication (12). The unscheduled DNA synthesis, as a result of the DUE was proposed to be a critical factor in the instability of the sequence. Also, a DUE was found at the AAT•ATT TRS in the surface glycoprotein gene of Trypanosoma brucei (44).
Tetraplexes
Tetraplexes (four-stranded DNA) assemble at G-rich DNA sequences forming a stable G-quartet. Although the ability to form tetrads has been commonly reported for single-stranded G-rich telomeric sequences (45), other residues were also observed to form tetrads. For example, studies showed the ability for base-pairing between hemi-protonated cytosines of one C-rich duplex with cytosine residues of a second duplex, forming a stable tetraplex structure, known as the i-motif (46,47). For this reason, the CGG•CCG repeats, associated with the FRAX, were suggested to form tetraplexes (either G-quartets or i-motifs) as demonstrated by circular dichroism, NMR and UV spectroscopy (46–50). Studies from several laboratories in the past decade on the biological conditions for tetraplex formation have concluded that different ionic and pH conditions, as well as repeat lengths, influence structural conformations within the CGG•CCG sequence (46,47,50). The stability of the G-quartet conformation has been implicated in the mediation of chromosomal degradation and condensation at telomeres (17,51), and has been suggested to play a role in blocking DNA replication (52,53) or inhibiting transcription (17,54).
Triplexes
Triplex DNA conformations have been studied for many decades. Since 1957, this structure has been observed both in vitro and in vivo in prokaryotes and eukaryotes by several laboratories (3,17,31,33,55,56). Long stretches of purine•pyrimidine (R•Y) mirror repeat sequence can readily form these three-stranded structures where the duplex DNA pairs with a third strand by Hoogsteen pairing with the purine strand of the duplex (17). Additional factors that influence the formation of triplexes include pH, binding of divalent metal ions and negative supercoiling (17,57,58). The TRS GAA•TTC has been observed to form both inter- and intramolecular triplexes (55,56,59,60). Potaman et al. (56) recently studied the propensity of these repeats to form an intramolecular triplex (H-DNA) or a bi-triplex structure at very long repeats. They proposed possible pathways to connect triplex formation with replication blockage and DNA template expansions. Other studies have also shown R•Y sequences can act as replication pause sites (17).
Several examples of R•Y sequences as protein binding sites to regulate gene expression have also been studied (33,61). Furthermore, triplex formation can affect nucleosome positioning (62–64). The structure formed by the GAA•TTC repeating sequence has been implicated to regulate gene expression (65–67) and genetic recombination (11).
Sticky DNA
A novel non-B DNA structure termed sticky DNA was discovered in 1999 by Sakamoto et al. (15). Sticky DNA is an intramolecular structure adopted by two long GAA•TTC repeating tracts in one DNA molecule (15,68) to give a dumbbell-shaped conformation in bacterial plasmids (Figure 1). It was first observed during studies on the genetic instability of the GAA•TTC repeats associated with Friedreich's ataxia in plasmid DNA in E.coli. Upon endonucleolytic cleavage of the plasmid external to the repeat tract, a retarded band was observed during insert analysis by gel electrophoresis (15). Since the DNA band ran with mobility much higher (up to 7-fold) compared to linear DNA, it indicated the presence of a stable alternative structure. Although the exact conformation of sticky DNA has yet to be elucidated, there are specific requirements for its formation, including the presence of two GAA•TTC tracts within the same molecule in a DR orientation, neutral pH, negative supercoiling and the presence of Mg2+ ions (15,68). Some of these conditions are similar to those required for an R•R•Y triplex, which may be indicative of the actual conformation. However, sticky DNA can only be formed by an intramolecular reaction and cannot be disassociated easily, even by heating to 80°C for 60 min (69). In situ nitrogen mustard crosslinking and electron microscopy of the digested plasmid, confirmed a stable interaction between the two tracts (15,18). Furthermore, sticky DNA has not been observed with GAA•TTC lengths <60 repeats. Long tracts of the GAA•TTC sequence have been shown to inhibit replication and transcription in E.coli, yeast and eukaryotic cells suggesting that sticky DNA may be a regulator of gene expression and DNA metabolism (21,67). Napierala et al. (11) observed a decrease in homologous recombination (HR) with increasing lengths of GAA•TTC. This decrease in frequency was attributed to sticky DNA formation in vivo. When novobiocin was used to decrease the negative supercoil density of plasmid DNA, which inhibits structure formation, the expected positive correlation between repeat tract length and recombination was restored.
Whereas conclusive proof of the role of sticky DNA in the etiology of FRDA remains to be demonstrated, substantial studies (11,15,18,21,67–72) are consistent with this concept.
REPLICATION
Replication slippage is generally considered to be a major factor influencing the genetic instabilities of the various TRS including CTG•CAG, GAA•TTC, CGG•CCG and GAC•GTC, which are implicated in several hereditary neurological diseases (1,3,38,42,73–78). More recently, replication was shown to influence the genetic instabilities of the CCTG•CAGG tetranucleotide repeats associated with DM2 (14). Although other factors such as repair and recombination (discussed below) also play an important role in these instabilities, replication is believed to be the first step in involving these other cellular processes.
Replication pausing
Numerous studies have shown the propensity of repeating sequences to fold back and form several non-B DNA structures including hairpins, triplexes, tetraplexes and sticky DNA (discussed above). It is believed that these secondary structures cause significant impediments for replication fork progression that leads to pausing of the DNA polymerase, finally causing replication fork collapse and involvement of the repair and recombination machinery to help restart replication. Both in vitro as well as in vivo studies have shown a variety of human, bacterial and phage replication polymerases to pause within the repeats in a length- and sequence-dependent manner (21,23,53,75,79–81). Recent 2D gel electrophoresis studies of replication intermediates from an in vivo yeast system showed a length- and sequence-dependent replication attenuation within the CTG•CAG, CGG•CCG and GAA•TTC repeats (21,82). The repeat expansions were proposed to occur when the replication fork attempts to escape from the pause site.
Orientation of repeat sequences
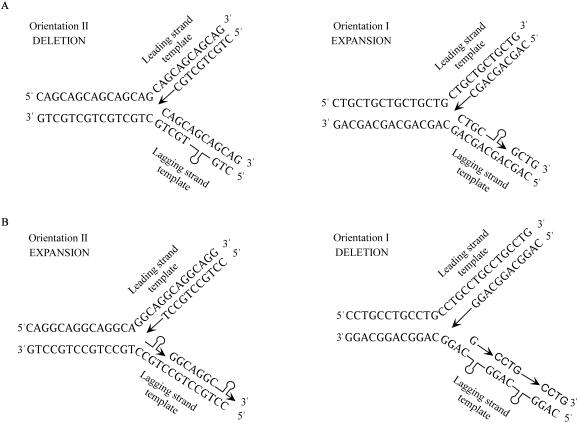
Studies on TRS show an orientation bias (relative to the replication origin) where repeats in one orientation are more stable than in the other. This orientation dependence is attributed to the ability of the repeat containing strand to form folded-back slipped structures. When the more stable structure is formed on the newly synthesized Okazaki fragment, expansions are generated. Alternatively, deletions are favored when the structure-prone strand is the template for replication (1,3,38,49,73,74,76,77,83,84). This is the case for all the TRS as well as the CCTG•CAGG tetranucleotide repeats (14). However, as shown in Figure 2 the essential difference between the DM1 CTG•CAG sequence and the DM2 CCTG•CAGG tetranucleotide repeats is that the CAGG strand forms a more stable hairpin-loop structure as compared its complementary CCTG strand (14). Alternatively, in the case of the triplet repeats, the CTG strand forms a more stable structure (36,38,39,85–88). This leads to a reversed behavior of the CCTG•CAGG sequence with respect to the orientation, where orientation I (CCTG on the leading strand template) is prone to delete and orientation II (CAGG on the leading strand template) expands (Figure 2). More interestingly, in contrast to the TRS where the orientation that yields deletions is considered to be unstable (deletions are the predominant products in most studies), the CCTG•CAGG repeats are unstable in the orientation prone to expand (14). Thus, the tetranucleotide repeats are different from the TRSs since they are better able to mimic the human instability behavior in the prokaryotic (E.coli) and eukaryotic (COS-7) model systems (14).
Figure 2.
Replication fork models to explain the orientation-dependent instability of the CTG•CAG and CCTG•CAGG repeats. (A) In the case of the CTG•CAG repeats, orientation I shows expansions due to the ability of the CTG strand to form a more stable hairpin-loop structure on the newly synthesized lagging strand, whereas the presence of the same structure on the lagging strand template in orientation II leads to deletions. (B) The CCTG•CAGG repeats exhibit a reversed behavior where orientation II gives expansions and orientation I leads to deletions. Although the orientation effect is reversed here, the principle, of one of the two strands forming a more stable fold-back structure, still remains the same.
Location of repeat sequences
In addition to the direction of replication, studies have also shown that the distance of the repeats from the origin of replication plays an important role in the amount of instability observed (14,38,74,77,89,90). The repeats cloned proximal to the replication origin are more prone to genetic instabilities as compared to the same repeats cloned distal to the origin. For the CCTG•CAGG repeats, we proposed that the initiation event occurring at the replication origin would render the repeats single-stranded for a sufficient period of time to give them the opportunity to form folded-back structures and thus lead to instability (14). Also, a model was suggested that the ability of the repeats to form non-B DNA structures would differ, depending on the location of the priming of the Okazaki fragment within the repeats, and this in turn would determine the amount and type of instability (77).
Flap endonuclease
Most models propose that the repeating tracts have a greater opportunity to form non-B DNA structures on the newly synthesized lagging strand, due to its single-stranded nature, than for the leading strand. Thus, Okazaki fragment maturation has been implicated in the expansions and contractions of these repeat sequences. One of the factors that has the most pronounced effect on genetic instability is the human flap endonuclease 1 (FEN-1) and Rad27 in yeast. FEN-1/Rad27 is involved in processing the 5′ ends of the Okazaki fragments (91–95). Previous studies have shown that mutations in FEN-1/Rad27 lead to destabilization of the repeating tracts resulting in expansions (76,96–101). This instability is attributed to the ability of the TRS to form folded structures in the 5′ flap of the Okazaki fragment, thus preventing processing of the flap and annealing of these flaps with the adjacent Okazaki fragments leading to expansions. Furthermore, the Rad27 mutants were shown recently to influence both the stability as well as the fragility of the CTG•CAG repeat tracts in a yeast system (102). It has been proposed that the inability of the Rad27 mutant cells to process the flaps of the Okazaki fragments causes breakage at the repeat sites, and their repair leads to genetic instability. Moreover, the same group has proposed a flap equilibration and endonuclease tracking model that attributes the stability of the repeats not to the cleavage activity of the endonuclease per se, but to the ability of the endonuclease to capture a cleavable flap among the equilibrating intermediates (103).
An in vitro study showed that the presence of the hFEN-1 was able to suppress repeat expansions of the GAA•TTC tracts during replication (104). However, if the hFEN-1 was added at a later stage during replication, it was unable to suppress these expansions. Most of the previously mentioned studies have been conducted in yeast, and not all of the features observed in yeast models are able to explain the expansions in human diseases. It has been shown that complete loss of FEN-1 in mammals is embryonically lethal (105). Transgenic mice were created that are either heterozygous or homozygous for FEN-1 and carry the Huntington disease causing repeat sequence CAG•CTG (106). The heterozygous mice showed an intergenerational expansion of the repeats when compared to the homozygous wild-type mice. Moreover, human cells that were deficient in FEN-1 also gave rise to instability further strengthening the protective role of FEN-1 in repeat tract expansions.
Polymerase switching in both prokaryotes and eukaryotes is believed to contribute to the genetic instabilities of the repeats associated with neurological diseases (14,23,107,108). Moreover, studies with mutants of other replication proteins such as Dna2, RNase HI and DNA ligase that are thought to work cooperatively with FEN-1/Rad27 had only a modest effect on repeat stability (102). Recently, the Srs2 DNA helicase was identified in a genetic screen for inhibitors of expansions in Saccharomyces cerevisiae and shown to block triplet repeat expansion through its helicase activity in conjunction with polymerase delta (109). The Bloom protein (BLM) was recently identified as interacting with FEN-1 and suppressed genomic instability by aiding FEN-1 cleavage of structure-containing flaps (110).
DNA REPAIR
The molecular pathways involved in DNA repair exist to protect the genomic integrity of the DNA sequence by preventing the permanent incorporation or loss of nucleotides within the genome, which would potentially create a deleterious mutation for the organism. These normally protective pathways have been hypothesized to be involved in the genetic instabilities of several microsatellites including trinucleotide repeats. Studies have shown that several types of cancers display microsatellite instabilities attributed to the loss of the DNA repair machinery, a result which is consistent with a possible role for DNA repair during the expansion of trinucleotide repeats associated with hereditary neurological diseases (111). The secondary structural properties of TRS (see above) are probably substrates to be recognized by these repair proteins. Herein, we discuss the ongoing research to determine the potential role of these repair pathways as sources for trinucleotide repeat instability.
Mismatch repair
Illegitimate incorporation of nucleotides during DNA replication resulting in a mismatch within the DNA sequence is a dangerous consequence of DNA polymerase infidelity. The MMR pathway is the main source for correcting these errors (112). Since several of the secondary structures formed by TRS result in the mismatching of bases within the structure or in the flanking regions, the MMR pathway is likely to recognize and act on these mismatches.
Jaworski et al. (30) first proposed MMR as a potential source of instability; plasmids harboring expanded CTG•CAG repeats in E.coli were more stable when grown in methyl-directed MMR-deficient strains than in the parental background. The authors hypothesized that the MMR proteins recognize three-base loops formed during replication. Repair of the loops generated gaps in the DNA, which were bypassed by DNA polymerase during resynthesis of the DNA. This process results in deletion of part of the triplet repeat sequence. Experiments with human MSH2 confirmed the preferential binding of MMR proteins to looped-out secondary structures formed by CTG•CAG repeats (29). The role of MMR was later refined by experiments which showed that active mismatch repair stabilized small instabilities (>8 repeats), but increased the occurrence of large deletions (35,107,113,114). However, experiments with human cell lines showed no increase in instability, at the DM1 and FRAXA loci, in two cell lines with mutations in different MMR proteins, MLH1 and MSH2 (115).
Studies in mouse model systems have shown mixed effects of mismatch repair on instability. Intergenerational instability of (CTG•CAG)84 in the human DM1 context was not stimulated in a MSH3- or MSH6-deficient background (116). However, somatic tissues showed a substantial increase in instability in the MSH6-deficient background. Interestingly, the somatic instability normally observed with this sequence was completely blocked in the MSH3-deficient background. This differential effect was hypothesized to be due to the competitive binding of MSH3 and MSH6 to MSH2 to form a functional complex. Somatic instability and germline expansions were found to be dependent upon MSH2 in Huntington disease transgenic mice (117–119). In experiments with DM1 transgenic mice with >300 repeats, a strong dependence was found on MSH2 for contractions during germline transmission and in spermatogonia (120,121). Most recently, a MutL homologue, Pms2, was determined to increase the somatic mosaicism of CTG•CAG repeats (122). These results support the hypothesis that mismatch repair is a key player in genetic instabilities, possibly through MSH2 or other downstream proteins, but its precise role that results in the expansions observed in the human diseases is still unclear.
Nucleotide excision repair (NER)
Nucleotide excision repair recognizes helical distortions in DNA generally created by bulky adducts such as pyrimidine dimers (123). The ability to recognize helical distortions also results in the recognition and removal of DNA loops often associated with the secondary structures formed by trinucleotide repeats (see above). Mistakes that result during the repair of the DNA gap naturally generated through the repair process have been hypothesized to be a potential source of expansions in human diseases.
Experiments in E.coli revealed that mutations in UvrA dramatically increased the instability of the (CTG•CAG)175 repeats harbored on the plasmid (124). Transcription through the repeat region stimulated deletions which were further enhanced by mutations in uvrA. The authors hypothesized that UvrA binds to secondary structures possibly formed on the single-stranded DNA generated during transcription, subsequently preventing replication bypass of the structure, which would have resulted in a deletion. In contrast to the above observations, experiments using a genetic selection system for deletions showed a decrease in deletion rates in UvrA-deficient strains (24). Additionally, the authors found that the UvrA protein preferentially bound to repeat loops of 1, 2 or 17 CAG repeats. This suggested that UvrA may stimulate NER repair of structures formed by trinucleotide repeats resulting in genomic instability. Experiments in yeast have shown that deletion of Rad1, which is partially involved in NER, does not stimulate the instability of CTG•CAG repeats (76). Hence, further work will be required to clarify the influence of NER on trinucleotide repeat instability.
Repair of damage caused by mutagens
The repair of DNA damage caused by chemical, radiation and environmental mutagens and their influence on the genetic instability of trinucleotide repeats has recently been explored. Due to the propensity for trinucleotide repeats to adopt secondary structures, certain non-paired nucleotides may be highly susceptible to damage-inducing agents (125–127). These nucleotide-adducts may stimulate a series of molecular pathways that increase instability. Additionally, this susceptibility to damage-inducing agents could be used as a potential basis for developing therapeutic strategies to reduce the size of the trinucleotide repeats within the affected gene, thus alleviating the deleterious effects of the expanded repeat on normal gene function.
The first experiments on this concept were conducted to ascertain if abasic sites, an intermediate during base excision repair, could induce expansions during DNA replication (128). The authors found that a single abasic site located at the 5′ end of the template strand induced dramatic trinucleotide repeat expansions in their in vitro primer extension assay. This instability was reduced when the site was relocated within the repeat region and was completely absent when in the primer region. These experiments implied that DNA damage can play a role in generating expansion. This hypothesis was later confirmed by Pineiro et al. (129) who found that the influence of mutagenic stress caused by exposure to mitomycin C increased expansions in a lymphoblastoid cell line at the DM1 locus. However, experiments conducted by Hashem et al. (130) found that mitomycin C, ethylmethanesulfonate (EMS), mitoxantrone and doxorubicin induced deletions of the CTG•CAG repeats at the DM1 locus.
Induction of deletions following treatment with DNA damaging agents has been shown by several laboratories. Along with the experiments conducted by Hashem et al. (130), transgenic mouse spermatocytes treated with cyclophosphomide or spermatocytes and stem cells treated with radiation, all induced deletions in the expanded DM1 locus (131). Additionally, spontaneous contractions were increased following treatment with aphidicolin, hydroxyurea and gamma radiation (132), observed using a selectable system for monitoring CTG•CAG repeats in mammalian cells. Chemically induced deletions were found using cytosine arabinoside, ethidium bromide, 5-azacytidine and aspirin in transgenic mice kidney cell lines (133). Surprisingly, caffeine was found to increase expansion rates by ∼60% in the same experiments. The mechanism for the reduction of the size of the expanded trinucleotide repeats is not entirely understood. However, these experiments represent initial attempts to delete expanded repeats as potential therapeutic strategies for the future.
GENETIC RECOMBINATION AND DOUBLE-STRAND BREAK REPAIR
Double-strand breaks are very potent instigators of genetic recombination (134). Proteins involved in the repair of the DSBs also participate in the recombination processes. It has been demonstrated, in different model systems, that TRS such as CTG•CAG and CGG•CCG efficiently induce DNA strand discontinuities that are repaired by the recombination machinery [reviewed in (27,78,135,136)]. Therefore, the role of DSB repair and recombination pathways in generating repeat instabilities will be reviewed together.
Replication slippage was the dominant model in the 1990s to explain microsatellite instability, since recombination (as defined by the reciprocal crossing-over exchange) was not revealed by studies of human cases (1). However, three important facts argue against reciprocal exchange as a mechanism of TRS instability in patients; first, the lack of evidence supporting the exchange of the flanking sequences, second, no corresponding length changes of the second allele during the expansion/deletion event occurring at the other allele, and third, a simple reciprocal exchange cannot explain extremely large expansions observed in some of the diseases. On the other hand, data from the patients pointed towards gene conversion (a non-reciprocal event) in the instability of CGG•CCG repeats in the FRAX and CTG•CAG tracts in the DM1 cases [reviewed in (135,136)]. Thus, gene conversion could be in principle, the recombination pathway involved in CTG•CAG, CGG•CCG repeat instabilities (Figure 3A). It should be pointed out that the small expansions in the polyalanine tracts (often encoded by different GCN triplets), recently shown to cause at least nine human diseases, are likely to arise from unequal crossing-over as the predominant mechanism (137–139). Since polyalanine tracts are usually encoded by imperfect trinucleotide repeats (i.e. by variants of the alanine codons), a simple replication slippage model cannot explain their instability (137–139).
Figure 3.
Inter-allelic and intra-allelic pathways in DSB repair leading to repeat instabilities. (A) Gene conversion without exchange of the flanking sequences. (B) Synthesis-dependent strand annealing (SDSA). (C) Non-homologous end joining (NHEJ). (D) Single-strand annealing (SSA). Only pathways which do not involve or do not alter the sequence of the second allele are presented. Note that strong likelihood exists for the repeats to form stable secondary structures (e.g. hairpins) at any stage during the processes of DNA synthesis and annealing of the single-stranded DNA ends. Thus, non-B DNA structures formed by tandem repeats, besides being an important cause of the DSB formation, are also a direct source of the repeat instabilities. Thicker red, grey and blue lines: repeat regions. The blue line designates the newly synthesized DNA tracts.
Recombination studies in E.coli
In model systems from bacteria to mammalian cells, recombination, including both gene conversion and crossing-over events, has been shown to be involved in TRS instability. Additionally, repetitive sequences also promoted HR in both prokaryotic and the eukaryotic systems presumably by virtue of forming unusual, non-B DNA structures. Different di-, tetra- and pentanucleotide repetitive sequences have previously been shown to stimulate recombination (140–144). The results of intermolecular and intramolecular studies in E.coli revealed that the frequency of crossing-over between long DM1 CTG•CAG repeats was significantly elevated when compared to the non-repeating controls (145,146). Stimulation of recombination was also observed for GAA•TTC repeats from the Friedreich's ataxia gene (11), however, the intramolecular process between long repeats was significantly hampered by formation of sticky DNA (see above). In the case of the CTG•CAG repeats, the recombination frequency was dependent on the orientation of the repeat tract relative to the unidirectional origin of replication. When the CTG repeats were present on the lagging strand template, the frequency of recombination was substantially higher in both inter- and intramolecular assays. The CTG•CAG tracts (as well as CGG•CCG and GAA•TTC) are known to arrest replication fork progression in vitro and in vivo (13,21,75,147), due to their capabilities to adopt non-B DNA conformations. In the case of CTG•CAG repeats, this occurs predominantly when the CTG strand is located on the lagging strand template for replication (75). In the model proposed to explain the orientation effect on recombination, stalling of the replication fork at the secondary structures led to the formation of nicks and/or DSB in the repeating tracts which stimulated their mutagenic repair via recombination (145,146). These studies also demonstrated a high level of TRS instability resulting from the recombination process in E.coli. A pronounced influence of DSB repair on TRS instability was also detected in the experiments, with transformation of break-containing plasmids into E.coli (148). Repair of the DSB located in the CTG•CAG and CGG•CCG repeats resulted in dramatic increase of TRS deletions. Recently, Hashem et al. (28) showed using a genetic system in bacteria that mutations in recA and recB, which decrease the rate of recombination, had a stabilizing effect on CTG•CAG repeats lowering the high rates of deletion seen in recombination proficient cells. Thus, the recombination proficiency also correlated with the high rates of genetic instability in the triplet repeats.
TRS instability during mitotic and meiotic recombination in yeast
Eukaryotic model systems, especially yeast, have been proven to be an excellent tool for the analysis of the involvement of recombination in the TRS instability, since mitotic and meiotic events can be analyzed separately in different genetic backgrounds (27,78,149). Independent analyses of the mitotic and meiotic processes may be crucial in order to understand the timing of the events leading to the TRS expansion in humans.
Recently, several studies in yeast have been aimed towards understanding the role of DSB repair and recombination in the instability of TRS tracts, primarily CTG•CAG repeats [reviewed in (27,78,135)]. Initial results obtained with relatively short TRS did not reveal a significant role of recombination in generating TRS instabilities (78). It has been speculated that short CTG•CAG tracts may not be very efficient in generating DSB in yeast or that the breaks induced in the shorter repeats are repaired by pathways other than HR (78,150). In addition, experiments with RAD52 mutants suggested that TRS instability is due to defects in replication rather than in recombination (78). However, elegant experiments with long CTG•CAG tracts (up to 250 repeats) definitively implicated DSBs and recombination as important mechanisms of the repeat instability (76). Freudenreich et al. (76) showed using both pulsed field electrophoresis of the yeast chromosomes and genetic assays that long CTG•CAG tracts (130–250 repeats) induce DSB in a length-dependent manner, and that these sequences have a high propensity for expansions during yeast transformation when the recombination event is initiated next to the repeat tract. The expansions of the CTG•CAG sequences in yeast were even more pronounced during meiosis when compared to the mitotic division (151–154). Jankowski et al. (153) attributed these instabilities to DSB-induced recombination. Sequences as short as (CTG•CAG)64 induced the spo11 dependent DSB formation during meiosis (154). Their repair resulted in deletions as well as in expansions of the CTG•CAG tract.
DSB were also artificially induced in yeast in vivo by use of the homing endonuclease (HO) (155). In a study with short CTG•CAG tracts, almost 20% of DSB-induced gene conversion events led to TRS deletions (almost exclusively in the recipient locus) (156). When longer repeats were used [(CTG•CAG)98], gene conversion resulted in frequent expansions (∼30% of events) (157). In the absence of the HO endonuclease, only contractions were observed. Interestingly, no expansions were detected when CTG•CAG repeats were replaced with the (CAA•TTG)87 tract, substantiating the role of non-B DNA structures in the instability processes since neither CAA nor TTG repeat tracts have been shown to form stable hairpin structures (85).
Recently, Richard et al. (149) proposed a unifying model for CTG•CAG instabilities observed during both mitotic and meiotic gene conversion in yeast. This model is based on the synthesis-dependent strand annealing (SDSA) pathway (158), modified for the specificity of the repetitive sequences. Four crucial considerations are accommodated into this model: (i) the initial formation of the DSB in one of the TRS tracts; (ii) the importance of the unusual DNA structures in generation of the repeat instability; (iii) the absence of evidence for crossing-over exchange; and (iv) no change in the sequence of the donor/template DNA. The initial event of the SDSA pathway is an invasion of one or both DNA strands of the processed DSB ends into the DNA template followed by DNA synthesis and dissociation of the newly synthesized strands from the template (Figure 3B). Out-of-register re-annealing of the unwound DNA strands together with hairpin structure formation on either of the strands results in the expansions or deletions of the TRS tract. Hence, the role of the unusual DNA structures in the recombinational instability of TRS tracts is not only limited to the initiation of the recombination event (via DSB induction), but it is also important at each of the subsequent synthesis and annealing steps, where slipped structures can be formed.
TRS recombination and DSB repair in mammalian models
The involvement of recombination and DSB repair in TRS instability has not been extensively studied in mammalian cells. Results of recent experiments in CHO cells demonstrated the influence of long CTG•CAG repeats (98 and 183 repeats) but not (CTG•CAG)17 on the recombination between two copies of the APRT gene (159). Meservy et al. (159) examined the changes in the CTG•CAG repeats initiated by HR between nearby APRT sequences. Long repeats underwent frequent large deletions (10-fold increase due to recombination). The frequency of the recombination-associated rearrangements extending outside of the CTG•CAG region was also increased over 50-fold. The presence of the CTG•CAG repeats also had a reciprocal effect on the types of the recombination events observed. In the cell lines harboring (CTG•CAG)183 repeats, the rate of the gene conversion events between the APRT loci was 3- to 4-fold lower and, in contrast, the rate of crossing-over was 2- to 3-fold higher when compared to the control cell lines lacking the repeats (159).
HR is a primary pathway of DSB repair in bacteria and lower eukaryota including yeast (160). In mammalian cells, non-homologous end joining (NHEJ) is the primary means of DSB repair (134,161). The influence of the mammalian DSB repair on the stability of CTG•CAG tracts was studied in COS1 cells (162). The DNA breaks were artificially introduced into the repeat region prior to transfection. The vast majority of the DSB repair events resulted in deletion of the TRS tracts, perhaps due to the structure formation at the repeat-containing DNA ends (Figure 3C). It would be interesting to analyze the TRS instability after in vivo induction of the DSB in mammalian cells, since different pathways are known to participate in the repair of the breaks generated in vitro compared to those induced in vivo (163).
Mouse genetic experiments support the involvement of DSB repair in CTG•CAG repeats instability. Savouret et al. (120) tested the influence of several genetic products in both HR and NHEJ (Rad52, Rad54 and DNA-PKcs) on intergenerational and somatic instability of CTG•CAG repeats in transgenic mice. No change in the repeat stability was observed in Rad54 and DNA-PKcs knockouts eliminating DSBR–HR as a likely mechanism of TRS expansions in their system. However, lack of Rad52 led to a significant decrease in the size of the expansions during intergenerational transmission. This implicated the contribution of the single-strand annealing (SSA, Figure 3D) pathway in the CTG•CAG repeats instability in mice.
In summary, substantial evidence has accumulated to support the following general model of DSB/recombination-mediated TRS instability. Structures formed by TRS related to the human neurological diseases are capable of blocking DNA replication. They can also be recognized and subjected to repair by endonuclease excision. These processes (arrest of the replication fork progression as well as nucleolytic repair of the ‘structural lesion’) may induce DNA strand discontinuities (nicks/breaks), which are very efficient substrates for recombinational repair. The repair of the DSB by the intra-allelic as well the inter-allelic (or ectopic) processes can lead to substantial TRS instability. It will be interesting to learn in the future how these processes are conducted in humans.
NON-B DNA CONFORMATIONS, REPEAT SEQUENCE MOTIFS AND GROSS REARRANGEMENTS
This laboratory has recently discovered that certain types of microsatellite sequences invoke gross deletions of the microsatellite sequences as well as flanking regions. These results document a new type of mutation caused by TRS. Bacolla et al. (164) discovered that a long (2.5 kb) and highly asymmetric (95% C + T in one strand) segment which adopts unusual DNA conformations is recognized by NER. Next, we broadened our attention to gross rearrangements which are involved in human diseases. Genomic rearrangements are a frequent source of instability, but the mechanisms involved are poorly understood. The 2.5 kb poly R•Y sequence mentioned above induced long deletions and other instabilities in plasmids that were mediated by mismatch repair and, in some cases, transcription (165). The breakpoints occurred at predicted non-B DNA structures. Computer searches of the locations of these features indicated a significant proximity of alternating purine–pyrimidine and oligo (purine•pyrimidine) tracts to breakpoint junctions in 222 gross deletions and translocations, involved in human diseases. In 11 deletions analyzed, breakpoints were explicable by non-B DNA structure formation. We concluded that alternative DNA conformations trigger genomic rearrangements through recombination-repair activities. Hence, the genomic rearrangements were apparently caused by the presence of the non-B DNA structures. Therefore, the sequences per se in the right-handed B structure are not mutagenic; however, in the triplex, slipped structure, tetraplex or cruciform conformations, they are mutagenic.
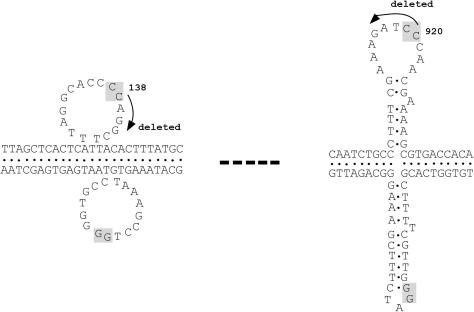
A representative pair of non-B DNA conformations that mediate a large deletion are shown in Figure 4. Note that the left DNA tract can be folded into a slipped structure whereas the right tract is accommodated in a cruciform conformation. The CC•GG region of homology is shown at position 138 and 920, respectively. Also, the deleted segment occurs precisely at the contorted regions (loops) of the slipped structure and cruciform (41). These discoveries on non-B DNA conformations, chromosomal rearrangements, triplet repeat and other inherited diseases, and related phenomena for class switch recombination have been recently reviewed (127).
Figure 4.
A representative pair of non-B DNA conformations that mediate a large deletion by recombination repair [see Figure 3 in (41)].
Other types of repeating DNA sequences which form non-B conformations are also effective in generating these types of mutations (41). The capacity of the long DM CTG•CAG and the Friedreich's ataxia GAA•TTC repeat tracts in plasmids to induce mutations in DNA flanking regions was evaluated in E.coli (41). Long (CTG•CAG)n (where n = 98 and 175) caused the deletion of most, or all, of the repeats and the flanking GFP gene. Deletions of 0.6–1.8 kb were found as well as inversions. Shorter repeat tracts (where n = 0 or 17) were essentially inert, as observed for the (GAA•TTC)176-containing plasmid. Under certain conditions, 30–50% of the products contained gross deletions. DNA sequence analyses of the breakpoint junctions of 47 deletions revealed the presence of 1–8 bp direct or inverted homologies in all cases. In addition, the presence of non-B DNA folded conformations (Figure 4) (i.e. slipped structures, cruciforms or triplexes) at or near the breakpoints was predicted for all rearrangements.
This genetic behavior, which was previously unrecognized for a TRS [reviewed in (127)], may provide the basis for a new type of instability of the myotonic dystrophy protein kinase (DMPK) gene in patients with a full mutation (41). Also, our discovery (159) that long CTG•CAG tracts from DM1 induce deletions and rearrangements during recombination at the APRT locus in CHO cells supports this concept.
These results are particularly exciting since they reveal a novel mechanism of mutagenesis elicited by certain TRS as well as demonstrate critical biological and/or medical functions for non-B DNA conformations. The latter has been an elusive goal for numerous laboratories for many years (1–3,17,26,39,52,57,61,111,127,160,162,166).
CONCLUDING REMARKS
Studies over the past fifteen years have provided remarkable advances in our knowledge of the molecular mechanisms involved in the genetic instabilities of repeating tri-, tetra- and pentanucleotide sequences involved in the etiology of hereditary neurological diseases. Whereas a number of factors including transcription, DNA repair, ligases, unwinding proteins and other factors, participate in the replication and recombination-mediated events, little is known about the temporal and spatial interrelationships of these factors. Also, we obviously want to understand these processes during different developmental phases in humans in order to comprehend the disease progressions. For the most part, these studies remain to be carried out in the future. The knowledge generated by the composite investigations will provide a basis for considering therapeutic strategies for ameliorating the devastating consequences of the diseases for the patients and their families.
The role of non-B DNA conformations in genetic instabilities has been an integral part of this field since its inception in the 1990s. Virtually, all workers have agreed that the capacity of the simple repeating sequences to adopt slipped structures, triplexes and other unusual conformations is an important component in mechanisms involved in expansions and deletions. Our inability to investigate these unusual conformations in living eukaryotic cells has been a substantial impediment for progress in this field. However, a major advance has been realized with the discovery that non-B DNA conformations serve as breakpoints for gross rearrangements (deletions, insertions, inversions and duplications) associated with a range of genetic diseases [reviewed in (127)]. Also, these studies (165) reveal a biological function for non-B DNA structures.
Due to the intrinsically interesting biological concepts involved and the medical implications of this field, a number of extremely talented and dedicated investigators have been attracted to generate important results. Thus, we have every reason to be optimistic that significant advances will be realized in the future.
Acknowledgments
This work was supported by a National Institutes of Health grant ES11347, the Robert A. Welch Foundation, the Friedreich's Ataxia Research Alliance and the Seek a Miracle Foundation. In addition, the authors thank our past and present co-workers for their numerous helpful suggestions. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wells R.D., Warren S.T., editors. Genetic Instabilities and Hereditary Neurological Diseases. San Diego: Academic Press; 1998. [Google Scholar]
- 2.Sinden R.R., Potaman V.N., Oussatcheva E.A., Pearson C.E., Lyubchenko Y.L., Shlyakhtenko L.S. Triplet repeat DNA structures and human genetic disease: dynamic mutations from dynamic DNA. J. Biosci. 2002;27:53–65. doi: 10.1007/BF02703683. [DOI] [PubMed] [Google Scholar]
- 3.Bowater R.P., Wells R.D. The intrinsically unstable life of DNA triplet repeats associated with human hereditary disorders. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:159–202. doi: 10.1016/s0079-6603(00)66029-4. [DOI] [PubMed] [Google Scholar]
- 4.Cummings C.J., Zoghbi H.Y. Fourteen and counting: unraveling trinucleotide repeat diseases. Hum. Mol. Genet. 2000;9:909–916. doi: 10.1093/hmg/9.6.909. [DOI] [PubMed] [Google Scholar]
- 5.Paulson H.L. Protein fate in neurodegenerative proteinopathies: polyglutamine diseases join the (mis)fold. Am. J. Hum. Genet. 1999;64:339–345. doi: 10.1086/302269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harper P.S. The Epidemiology of Huntington's Disease. London: Saunders; 1991. [Google Scholar]
- 7.Martin J.B. Molecular basis of the neurodegenerative disorders. N. Engl. J. Med. 1999;340:1970–1980. doi: 10.1056/NEJM199906243402507. [DOI] [PubMed] [Google Scholar]
- 8.Manto M.U., Pandolfo M., editors. The Cerebellum and its Disorders. Cambridge, UK: Cambridge University Press; 2002. [Google Scholar]
- 9.Rubinzstein D.C., Hayden M.R. Analysis of Triplet Repeat Disorders. Oxford: Bios Scientific Publishers; 1998. [Google Scholar]
- 10.Baldi P., Brunak S., Chauvin Y., Pedersen A.G. Structural basis for triplet repeat disorders: a computational analysis. Bioinformatics. 1999;15:918–929. doi: 10.1093/bioinformatics/15.11.918. [DOI] [PubMed] [Google Scholar]
- 11.Napierala M., Dere R., Vetcher A., Wells R.D. Structure-dependent recombination hot spot activity of GAA.TTC sequences from intron 1 of the Friedreich's ataxia gene. J. Biol. Chem. 2004;279:6444–6454. doi: 10.1074/jbc.M309596200. [DOI] [PubMed] [Google Scholar]
- 12.Potaman V.N., Bissler J.J., Hashem V.I., Oussatcheva E.A., Lu L., Shlyakhtenko L.S., Lyubchenko Y.L., Matsuura T., Ashizawa T., Leffak M., et al. Unpaired structures in SCA10 (ATTCT)n.(AGAAT)n repeats. J. Mol. Biol. 2003;326:1095–1111. doi: 10.1016/s0022-2836(03)00037-8. [DOI] [PubMed] [Google Scholar]
- 13.Siyanova E.I., Mirkin S.M. Expansion of trinucleotide repeats. Mol. Biol. 2001;35:208–223. [PubMed] [Google Scholar]
- 14.Dere R., Napierala M., Ranum L.P., Wells R.D. Hairpin structure-forming propensity of the (CCTG.CAGG) tetranucleotide repeats contributes to the genetic instability associated with myotonic dystrophy type 2. J. Biol. Chem. 2004;279:41715–41726. doi: 10.1074/jbc.M406415200. [DOI] [PubMed] [Google Scholar]
- 15.Sakamoto N., Chastain P.D., Parniewski P., Ohshima K., Pandolfo M., Griffith J.D., Wells R.D. Sticky DNA: self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedreich's ataxia. Mol. Cell. 1999;3:465–475. doi: 10.1016/s1097-2765(00)80474-8. [DOI] [PubMed] [Google Scholar]
- 16.Darlow J.M., Leach D.R.F. Secondary structures in d(CGG) and d(CCG): repeat tracts. J. Mol. Biol. 1998;275:3–16. doi: 10.1006/jmbi.1997.1453. [DOI] [PubMed] [Google Scholar]
- 17.Sinden R.R. DNA Structure and Function. San Diego: Academic Press; 1994. [Google Scholar]
- 18.Vetcher A.A., Wells R.D. Sticky DNA formation in vivo alters the plasmid dimer/monomer ratio. J. Biol. Chem. 2004;279:6434–6443. doi: 10.1074/jbc.M309595200. [DOI] [PubMed] [Google Scholar]
- 19.Shlyakhtenko L.S., Potaman V.N., Sinden R.R., Lyubchenko Y.L. Structure and dynamics of supercoil-stabilized DNA cruciforms. J. Mol. Biol. 1998;280:61–72. doi: 10.1006/jmbi.1998.1855. [DOI] [PubMed] [Google Scholar]
- 20.Rahmouni A.R., Wells R.D. Stabilization of Z DNA in vivo by localized supercoiling. Science. 1989;246:358–363. doi: 10.1126/science.2678475. [DOI] [PubMed] [Google Scholar]
- 21.Krasilnikova M.M., Mirkin S.M. Replication stalling at Friedreich's ataxia (GAA)n repeats in vivo. Mol. Cell. Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuura T., Fang P., Lin X., Khajavi M., Tsuji K., Rasmussen A., Grewal R.P., Achari M., Alonso M.E., Pulst S.M., et al. Somatic and germline instability of the ATTCT repeat in spinocerebellar ataxia type 10. Am. J. Hum. Genet. 2004;74:1216–1224. doi: 10.1086/421526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang S., Ohshima K., Shimizu M., Amirhaeri S., Wells R.D. Pausing of DNA synthesis in vitro at specific loci in CTG and CGG triplet repeats from human hereditary disease genes. J. Biol. Chem. 1995;270:27014–27021. doi: 10.1074/jbc.270.45.27014. [DOI] [PubMed] [Google Scholar]
- 24.Oussatcheva E.A., Hashem V.I., Zou Y., Sinden R.R., Potaman V.N. Involvement of the nucleotide excision repair protein UvrA in instability of CAG.CTG repeat sequences in Escherichia coli. J. Biol. Chem. 2001;276:30878–30884. doi: 10.1074/jbc.M104697200. [DOI] [PubMed] [Google Scholar]
- 25.Connelly J.C., Kirkham L.A., Leach D.R.F. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl Acad. Sci. USA. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guieysse A.L., Praseuth D., Helene C. Identification of a triplex DNA-binding protein from human cells. J. Mol. Biol. 1997;267:289–298. doi: 10.1006/jmbi.1997.0884. [DOI] [PubMed] [Google Scholar]
- 27.Lahue R.S., Slater D.L. DNA repair and trinucleotide repeat instability. Front. Biosci. 2003;8:653–665. doi: 10.2741/1107. [DOI] [PubMed] [Google Scholar]
- 28.Hashem V.I., Rosche W.A., Sinden R.R. Genetic recombination destabilizes (CTG)n.(CAG)n repeats in E. coli. Mutat. Res. 2004;554:95–109. doi: 10.1016/j.mrfmmm.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 29.Pearson C.E., Ewel A., Acharya S., Fishel R.A., Sinden R.R. Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum. Mol. Genet. 1997;6:1117–1123. doi: 10.1093/hmg/6.7.1117. [DOI] [PubMed] [Google Scholar]
- 30.Jaworski A., Rosche W.A., Gellibolian R., Kang S., Shimizu M., Bowater R.P., Sinden R.R., Wells R.D. Mismatch repair in Escherichia coli enhances instability of (CTG)n triplet repeats from human hereditary diseases. Proc. Natl Acad. Sci. USA. 1995;92:11019–11023. doi: 10.1073/pnas.92.24.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faruqi A.F., Datta H.J., Carroll D., Seidman M.M., Glazer P.M. Triple-helix formation induces recombination in mammalian cells via a nucleotide excision repair-dependent pathway. Mol. Cell. Biol. 2000;20:990–1000. doi: 10.1128/mcb.20.3.990-1000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shigemori Y., Oishi M. Specific cleavage of DNA molecules at RecA-mediated triple-strand structure. Nucleic Acids Res. 2004;32:e4. doi: 10.1093/nar/gnh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasquez K.M., Christensen J., Li L., Finch R.A., Glazer P.M. Human XPA and RPA DNA repair proteins participate in specific recognition of triplex-induced helical distortions. Proc. Natl Acad Sci. USA. 2002;99:5848–5853. doi: 10.1073/pnas.082193799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tam M., Erin Montgomery S., Kekis M., David Stollar B., Price G.B., Pearson C.E. Slipped (CTG).(CAG) repeats of the myotonic dystrophy locus: surface probing with anti-DNA antibodies. J. Mol. Biol. 2003;332:585–600. doi: 10.1016/s0022-2836(03)00880-5. [DOI] [PubMed] [Google Scholar]
- 35.Wells R.D., Parniewski P., Pluciennik A., Bacolla A., Gellibolian R., Jaworski A. Small slipped register genetic instabilities in Escherichia coli in triplet repeat sequences associated with hereditary neurological diseases. J. Biol. Chem. 1998;273:19532–19541. doi: 10.1074/jbc.273.31.19532. [DOI] [PubMed] [Google Scholar]
- 36.Paiva A.M., Sheardy R.D. Influence of sequence context and length on the structure and stability of triplet repeat DNA oligomers. Biochemistry. 2004;43:14218–14227. doi: 10.1021/bi0494368. [DOI] [PubMed] [Google Scholar]
- 37.Pearson C.E., Tam M., Wang Y.H., Montgomery S.E., Dar A.C., Cleary J.D., Nichol K. Slipped-strand DNAs formed by long (CAG).(CTG) repeats: slipped-out repeats and slip-out junctions. Nucleic Acids Res. 2002;30:4534–4547. doi: 10.1093/nar/gkf572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang S., Jaworski A., Ohshima K., Wells R.D. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nature Genet. 1995;10:213–218. doi: 10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- 39.Mitas M. Trinucleotide repeats associated with human disease. Nucleic Acids Res. 1997;25:2245–2254. doi: 10.1093/nar/25.12.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liquori C.L., Ricker K., Moseley M.L., Jacobsen J.F., Kress W., Naylor S.L., Day J.W., Ranum L.P. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 41.Wojciechowska M., Bacolla A., Larson J.E., Wells R.D. The myotonic dystrophy type 1 triplet repeat sequence induces gross deletions and inversions. J. Biol. Chem. 2005;280:941–952. doi: 10.1074/jbc.M410427200. [DOI] [PubMed] [Google Scholar]
- 42.Mochmann L.H., Wells R.D. Transcription influences the types of deletion and expansion products in an orientation-dependent manner from GAC.GTC repeats. Nucleic Acids Res. 2004;32:4469–4479. doi: 10.1093/nar/gkh787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin X., Ashizawa T. SCA10 and ATTCT repeat expansion: clinical features and molecular aspects. Cytogenet. Genome Res. 2003;100:184–188. doi: 10.1159/000072853. [DOI] [PubMed] [Google Scholar]
- 44.Ohshima K., Kang S., Larson J.E., Wells R.D. TTA.TAA triplet repeats in plasmids form a non-H bonded structure. J. Biol. Chem. 1996;271:16784–16791. doi: 10.1074/jbc.271.28.16784. [DOI] [PubMed] [Google Scholar]
- 45.Neidle S., Parkinson G.N. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003;13:275–283. doi: 10.1016/s0959-440x(03)00072-1. [DOI] [PubMed] [Google Scholar]
- 46.Mills M., Lacroix L., Arimondo P.B., Leroy J.L., Francois J.C., Klump H., Mergny J.L. Unusual DNA conformations: implications for telomeres. Curr. Med. Chem. Anti-Canc. Agents. 2002;2:627–644. doi: 10.2174/1568011023353877. [DOI] [PubMed] [Google Scholar]
- 47.Fojtik P., Vorlickova M. The fragile X chromosome (GCC) repeat folds into a DNA tetraplex at neutral pH. Nucleic Acids Res. 2001;29:4684–4890. doi: 10.1093/nar/29.22.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saha T., Usdin K. Tetraplex formation by the progressive myoclonus epilepsy type-1 repeat: implications for instability in the repeat expansion diseases. FEBS Lett. 2001;491:184–187. doi: 10.1016/s0014-5793(01)02190-1. [DOI] [PubMed] [Google Scholar]
- 49.Balakumaran B.S., Freudenreich C.H., Zakian V.A. CGG.CCG repeats exhibit orientation-dependent instability and orientation-independent fragility in Saccharomyces cerevisiae. Hum. Mol. Genet. 2000;9:93–100. doi: 10.1093/hmg/9.1.93. [DOI] [PubMed] [Google Scholar]
- 50.Fry M., Loeb L.A. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He Y., Neumann R.D., Panyutin I.G. Intramolecular quadruplex conformation of human telomeric DNA assessed with 125I-radioprobing. Nucleic Acids Res. 2004;32:5359–5367. doi: 10.1093/nar/gkh875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Usdin K. NGG-triplet repeats form similar intrastrand structures: implications for the triplet expansion diseases. Nucleic Acids Res. 1998;26:4078–4085. doi: 10.1093/nar/26.17.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Usdin K., Woodford K.J. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 1995;23:4202–4209. doi: 10.1093/nar/23.20.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 55.Tiner W.J., Sr, Potaman V.N., Sinden R.R., Lyubchenko Y.L. The structure of intramolecular triplex DNA: atomic force microscopy study. J. Mol. Biol. 2001;314:353–357. doi: 10.1006/jmbi.2001.5174. [DOI] [PubMed] [Google Scholar]
- 56.Potaman V.N., Oussatcheva E.A., Lyubchenko Y.L., Shlyakhtenko L.S., Bidichandani S.I., Ashizawa T., Sinden R.R. Length-dependent structure formation in Friedreich ataxia (GAA)n.(TTC)n repeats at neutral pH. Nucleic Acids Res. 2004;32:1224–1231. doi: 10.1093/nar/gkh274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mirkin S.M., Frank-Kamenetskii M.D. H-DNA and related structures. Annu. Rev. Biophys. Biomol. Struct. 1994;23:541–576. doi: 10.1146/annurev.bb.23.060194.002545. [DOI] [PubMed] [Google Scholar]
- 58.James P.L., Brown T., Fox K.R. Thermodynamic and kinetic stability of intermolecular triple helices containing different proportions of C+*GC and T*AT triplets. Nucleic Acids Res. 2003;31:5598–5606. doi: 10.1093/nar/gkg782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mariappan S.V., Catasti P., Silks L.A., III, Bradbury E.M., Gupta G. The high-resolution structure of the triplex formed by the GAA.TTC triplet repeat associated with Friedreich's ataxia. J. Mol. Biol. 1999;285:2035–2052. doi: 10.1006/jmbi.1998.2435. [DOI] [PubMed] [Google Scholar]
- 60.Shimizu M., Hanvey J.C., Wells R.D. Multiple non-B-DNA conformations of polypurine.polypyrimidine sequences in plasmids. Biochemistry. 1990;29:4704–4713. doi: 10.1021/bi00471a027. [DOI] [PubMed] [Google Scholar]
- 61.Kohwi Y., Kohwi-Shigematsu T. Altered gene expression correlates with DNA structure. Genes Dev. 1991;5:2547–2554. doi: 10.1101/gad.5.12b.2547. [DOI] [PubMed] [Google Scholar]
- 62.Espinas M.L., Jimenez-Garcia E., Martinez-Balbas A., Azorin F. Formation of triple-stranded DNA at d(GA.TC)n sequences prevents nucleosome assembly and is hindered by nucleosomes. J. Biol. Chem. 1996;271:31807–31812. doi: 10.1074/jbc.271.50.31807. [DOI] [PubMed] [Google Scholar]
- 63.Westin L., Blomquist P., Milligan J.F., Wrange O. Triple helix DNA alters nucleosomal histone–DNA interactions and acts as a nucleosome barrier. Nucleic Acids Res. 1995;23:2184–2191. doi: 10.1093/nar/23.12.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown P.M., Fox K.R. DNA triple-helix formation on nucleosome-bound poly(dA).poly(dT) tracts. Biochem. J. 1998;333:259–267. doi: 10.1042/bj3330259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bidichandani S.I., Ashizawa T., Patel P.I. The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am. J. Hum. Genet. 1998;62:111–121. doi: 10.1086/301680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grabczyk E., Usdin K. The GAA.TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohshima K., Montermini L., Wells R.D., Pandolfo M. Inhibitory effects of expanded GAA.TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo. J. Biol. Chem. 1998;275:14588–14595. doi: 10.1074/jbc.273.23.14588. [DOI] [PubMed] [Google Scholar]
- 68.Vetcher A.A., Napierala M., Wells R.D. Sticky DNA: effect of the polypurine.polypyrimidine sequence. J. Biol. Chem. 2002;277:39228–39234. doi: 10.1074/jbc.M205210200. [DOI] [PubMed] [Google Scholar]
- 69.Vetcher A.A., Napierala M., Iyer R.R., Chastain P.D., Griffith J.D., Wells R.D. Sticky DNA, a long GAA.GAA.TTC triplex that is formed intramolecularly, in the sequence of intron 1 of the frataxin gene. J. Biol. Chem. 2002;277:39217–39227. doi: 10.1074/jbc.M205209200. [DOI] [PubMed] [Google Scholar]
- 70.Sakamoto N., Ohshima K., Montermini L., Pandolfo M., Wells R.D. Sticky DNA, a self-associated complex formed at long GAA*TTC repeats in intron 1 of the frataxin gene, inhibits transcription. J. Biol. Chem. 2001;276:27171–27177. doi: 10.1074/jbc.M101879200. [DOI] [PubMed] [Google Scholar]
- 71.Ohshima K., Sakamoto N., Labuda M., Poirier J., Moseley M.L., Montermini L., Ranum L.P., Wells R.D., Pandolfo M. A nonpathogenic GAAGGA repeat in the Friedreich gene: implications for pathogenesis. Neurology. 1999;53:1854–1857. doi: 10.1212/wnl.53.8.1854. [DOI] [PubMed] [Google Scholar]
- 72.Sakamoto N., Larson J.E., Iyer R.R., Montermini L., Pandolfo M., Wells R.D. GGA*TCC-interrupted triplets in long GAA*TTC repeats inhibit the formation of triplex and sticky DNA structures, alleviate transcription inhibition, and reduce genetic instabilities. J. Biol. Chem. 2001;276:27178–27187. doi: 10.1074/jbc.M101852200. [DOI] [PubMed] [Google Scholar]
- 73.Shimizu M., Gellibolian R., Oostra B.A., Wells R.D. Cloning, characterization and properties of plasmids containing CGG triplet repeats from the FMR-1 gene. J. Mol. Biol. 1996;258:614–626. doi: 10.1006/jmbi.1996.0273. [DOI] [PubMed] [Google Scholar]
- 74.Maurer D.J., O'Callaghan B.L., Livingston D.M. Orientation dependence of trinucleotide CAG repeat instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:6617–6622. doi: 10.1128/mcb.16.12.6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Samadashwily G.M., Raca G., Mirkin S.M. Trinucleotide repeats affect DNA replication in vivo. Nature Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 76.Freudenreich C.H., Kantrow S.M., Zakian V.A. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 77.Cleary J.D., Nichol K., Wang Y.H., Pearson C.E. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nature Genet. 2002;31:37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- 78.Lenzmeier B.A., Freudenreich C.H. Trinucleotide repeat instability: a hairpin curve at the crossroads of replication, recombination, and repair. Cytogenet. Genome Res. 2003;100:7–24. doi: 10.1159/000072836. [DOI] [PubMed] [Google Scholar]
- 79.Krasilnikov A.S., Panyutin I.G., Samadashwily G.M., Cox R., Lazurkin Y.S., Mirkin S.M. Mechanisms of triplex-caused polymerization arrest. Nucleic Acids Res. 1997;25:1339–1346. doi: 10.1093/nar/25.7.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ohshima K., Wells R.D. Hairpin formation during DNA synthesis primer realignment in vitro in triplet repeat sequences from human hereditary disease genes. J. Biol. Chem. 1997;272:16798–16806. doi: 10.1074/jbc.272.27.16798. [DOI] [PubMed] [Google Scholar]
- 81.Viguera E., Canceill D., Ehrlich S.D. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 2001;20:2587–2595. doi: 10.1093/emboj/20.10.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pelletier R., Krasilnikova M.M., Samadashwily G.M., Lahue R., Mirkin S.M. Replication and expansion of trinucleotide repeats in yeast. Mol. Cell. Biol. 2003;23:1349–1357. doi: 10.1128/MCB.23.4.1349-1357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miret J.J., Pessoa-Brandao L., Lahue R.S. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1998;95:12438–12443. doi: 10.1073/pnas.95.21.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bichara M., Pinet I., Schumacher S., Fuchs R.P. Mechanisms of dinucleotide repeat instability in Escherichia coli. Genetics. 2000;154:533–542. doi: 10.1093/genetics/154.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gacy A.M., Goellner G., Juranic N., Macura S., McMurray C.T. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 86.Sarkar P.S., Chang H.C., Boudi F.B., Reddy S. CTG repeats show bimodal amplification in E. coli. Cell. 1998;95:531–540. doi: 10.1016/s0092-8674(00)81620-7. [DOI] [PubMed] [Google Scholar]
- 87.Moore H., Greenwell P.W., Liu C., Arnheim N., Petes T.D. Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl Acad. Sci. USA. 1999;96:1504–1509. doi: 10.1073/pnas.96.4.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chi L.M., Lam S.L. Structural roles of CTG repeats in slippage expansion during DNA replication. Nucleic Acids Res. 2005;33:1604–1617. doi: 10.1093/nar/gki307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nenguke T., Aladjem M.I., Gusella J.F., Wexler N.S., Arnheim N. Candidate DNA replication initiation regions at human trinucleotide repeat disease loci. Hum. Mol. Genet. 2003;12:1021–1028. doi: 10.1093/hmg/ddg111. [DOI] [PubMed] [Google Scholar]
- 90.Nichol Edamura K., Leonard M.R., Pearson C.E. Role of replication and CpG methylation in fragile X syndrome CGG deletions in primate cells. Am. J. Hum. Genet. 2005;76:302–311. doi: 10.1086/427928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ishimi Y., Claude A., Bullock P., Hurwitz J. Complete enzymatic synthesis of DNA containing the SV40 origin of replication. J. Biol. Chem. 1988;263:19723–19733. [PubMed] [Google Scholar]
- 92.Waga S., Bauer G., Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J. Biol. Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 93.Bambara R.A., Murante R.S., Henricksen L.A. Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 94.Gary R., Park M.S., Nolan J.P., Cornelius H.L., Kozyreva O.G., Tran H.T., Lobachev K.S., Resnick M.A., Gordenin D.A. A novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA and implications for genetic risk. Mol. Cell. Biol. 1999;19:5373–5382. doi: 10.1128/mcb.19.8.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Henricksen L.A., Veeraraghavan J., Chafin D.R., Bambara R.A. DNA ligase I competes with FEN1 to expand repetitive DNA sequences in vitro. J. Biol. Chem. 2002;277:22361–22369. doi: 10.1074/jbc.M201765200. [DOI] [PubMed] [Google Scholar]
- 96.Gordenin D.A., Kunkel T.A., Resnick M.A. Repeat expansion—all in a flap? Nature Genet. 1997;16:116–118. doi: 10.1038/ng0697-116. [DOI] [PubMed] [Google Scholar]
- 97.Kokoska R.J., Stefanovic L., Tran H.T., Resnick M.A., Gordenin D.A., Petes T.D. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase delta (pol3-t) Mol. Cell. Biol. 1998;18:2779–2788. doi: 10.1128/mcb.18.5.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schweitzer J.K., Livingston D.M. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum. Mol. Genet. 1998;7:69–74. doi: 10.1093/hmg/7.1.69. [DOI] [PubMed] [Google Scholar]
- 99.Spiro C., Pelletier R., Rolfsmeier M.L., Dixon M.J., Lahue R.S., Gupta G., Park M.S., Chen X., Mariappan S.V., McMurray C.T. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol. Cell. 1999;4:1079–1085. doi: 10.1016/s1097-2765(00)80236-1. [DOI] [PubMed] [Google Scholar]
- 100.White P.J., Borts R.H., Hirst M.C. Stability of the human fragile X (CGG)(n) triplet repeat array in Saccharomyces cerevisiae deficient in aspects of DNA metabolism. Mol. Cell. Biol. 1999;19:5675–5684. doi: 10.1128/mcb.19.8.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee S., Park M.S. Human FEN-1 can process the 5′-flap DNA of CTG/CAG triplet repeat derived from human genetic diseases by length and sequence dependent manner. Exp. Mol. Med. 2002;34:313–317. doi: 10.1038/emm.2002.44. [DOI] [PubMed] [Google Scholar]
- 102.Callahan J.L., Andrews K.J., Zakian V.A., Freudenreich C.H. Mutations in yeast replication proteins that increase CAG/CTG expansions also increase repeat fragility. Mol. Cell. Biol. 2003;23:7849–7860. doi: 10.1128/MCB.23.21.7849-7860.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu Y., Zhang H., Veeraraghavan J., Bambara R.A., Freudenreich C.H. Saccharomyces cerevisiae flap endonuclease 1 uses flap equilibration to maintain triplet repeat stability. Mol. Cell. Biol. 2004;24:4049–4064. doi: 10.1128/MCB.24.9.4049-4064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ruggiero B.L., Topal M.D. Triplet repeat expansion generated by DNA slippage is suppressed by human flap endonuclease 1. J. Biol. Chem. 2004;279:23088–23097. doi: 10.1074/jbc.M313170200. [DOI] [PubMed] [Google Scholar]
- 105.Kucherlapati M., Yang K., Kuraguchi M., Zhao J., Lia M., Heyer J., Kane M.F., Fan K., Russell R., Brown A.M., et al. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc. Natl Acad. Sci. USA. 2002;99:9924–9929. doi: 10.1073/pnas.152321699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Spiro C., McMurray C.T. Nuclease-deficient FEN-1 blocks Rad51/BRCA1-mediated repair and causes trinucleotide repeat instability. Mol. Cell. Biol. 2003;23:6063–6074. doi: 10.1128/MCB.23.17.6063-6074.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schumacher S., Fuchs R.P., Bichara M. Expansion of CTG repeats from human disease genes is dependent upon replication mechanisms in Escherichia coli: the effect of long patch mismatch repair revisited. J. Mol. Biol. 1998;279:1101–1110. doi: 10.1006/jmbi.1998.1827. [DOI] [PubMed] [Google Scholar]
- 108.Waga S., Stillman B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 109.Bhattacharyya S., Lahue R.S. Saccharomyces cerevisiae Srs2 DNA helicase selectively blocks expansions of trinucleotide repeats. Mol. Cell. Biol. 2004;24:7324–7330. doi: 10.1128/MCB.24.17.7324-7330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang W., Bambara R.A. Human bloom protein stimulates flap endonuclease 1 activity by resolving DNA secondary structure. J. Biol. Chem. 2005;280:5391–5399. doi: 10.1074/jbc.M412359200. [DOI] [PubMed] [Google Scholar]
- 111.Modrich P., Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 112.Lahue R.S., Au K.G., Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 113.Schmidt K.H., Abbott C.M., Leach D.R. Two opposing effects of mismatch repair on CTG repeat instability in Escherichia coli. Mol. Microbiol. 2000;35:463–471. doi: 10.1046/j.1365-2958.2000.01727.x. [DOI] [PubMed] [Google Scholar]
- 114.Parniewski P., Jaworski A., Wells R.D., Bowater R.P. Length of CTG.CAG repeats determines the influence of mismatch repair on genetic instability. J. Mol. Biol. 2000;299:865–874. doi: 10.1006/jmbi.2000.3796. [DOI] [PubMed] [Google Scholar]
- 115.Kramer P.R., Pearson C.E., Sinden R.R. Stability of triplet repeats of myotonic dystrophy and fragile X loci in human mutator mismatch repair cell lines. Hum. Genet. 1996;98:151–157. doi: 10.1007/s004390050179. [DOI] [PubMed] [Google Scholar]
- 116.van den Broek W.J., Nelen M.R., Wansink D.G., Coerwinkel M.M., te Riele H., Groenen P.J., Wieringa B. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 117.Manley K., Shirley T.L., Flaherty L., Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nature Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 118.Kovtun I.V., McMurray C.T. Trinucleotide expansion in haploid germ cells by gap repair. Nature Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 119.Wheeler V.C., Lebel L.A., Vrbanac V., Teed A., te Riele H., MacDonald M.E. Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Hum. Mol. Genet. 2003;12:273–281. doi: 10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- 120.Savouret C., Brisson E., Essers J., Kanaar R., Pastink A., te Riele H., Junien C., Gourdon G. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Savouret C., Garcia-Cordier C., Megret J., te Riele H., Junien C., Gourdon G. MSH2-dependent germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Mol. Cell. Biol. 2004;24:629–637. doi: 10.1128/MCB.24.2.629-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gomes-Pereira M., Fortune M.T., Ingram L., McAbney J.P., Monckton D.G. Pms2 is a genetic enhancer of trinucleotide (CAG.CTG) repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum. Mol. Genet. 2004;13:1815–1825. doi: 10.1093/hmg/ddh186. [DOI] [PubMed] [Google Scholar]
- 123.Nickoloff J.A., Hoekstra M.F., editors. Volume I: DNA Repair and Prokaryotes and Lower Eukaryotes. Totowa: Humana Press; 1998. [Google Scholar]
- 124.Parniewski P., Bacolla A., Jaworski A., Wells R.D. Nucleotide excision repair affects the stability of long transcribed (CTG.CAG) tracts in an orientation-dependent manner in Escherichia coli. Nucleic Acids Res. 1999;27:616–623. doi: 10.1093/nar/27.2.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wright B.E., Schmidt K.H., Minnick M.F. Mechanisms by which transcription can regulate somatic hypermutation. Genes Immun. 2004;5:176–182. doi: 10.1038/sj.gene.6364053. [DOI] [PubMed] [Google Scholar]
- 126.Wright B.E., Reimers J.M., Schmidt K.H., Reschke D.K. Hypermutable bases in the p53 cancer gene are at vulnerable positions in DNA secondary structures. Cancer Res. 2002;62:5641–5644. [PubMed] [Google Scholar]
- 127.Bacolla A., Wells R.D. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- 128.Lyons-Darden T., Topal M.D. Abasic sites induce triplet-repeat expansion during DNA replication in vitro. J. Biol. Chem. 1999;274:25975–25978. doi: 10.1074/jbc.274.37.25975. [DOI] [PubMed] [Google Scholar]
- 129.Pineiro E., Fernandez-Lopez L., Gamez J., Marcos R., Surralles J., Velazquez A. Mutagenic stress modulates the dynamics of CTG repeat instability associated with myotonic dystrophy type 1. Nucleic Acids Res. 2003;31:6733–6740. doi: 10.1093/nar/gkg898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hashem V.I., Pytlos M.J., Klysik E.A., Tsuji K., Khajavi M., Ashizawa T., Sinden R.R. Chemotherapeutic deletion of CTG repeats in lymphoblast cells from DM1 patients. Nucleic Acids Res. 2004;32:6334–6346. doi: 10.1093/nar/gkh976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang Y., Monckton D.G., Siciliano M.J., Connor T.H., Meistrich M.L. Detection of radiation and cyclophosphamide-induced mutations in individual mouse sperm at a human expanded trinucleotide repeat locus transgene. Mutat. Res. 2002;516:121–138. doi: 10.1016/s1383-5718(02)00035-9. [DOI] [PubMed] [Google Scholar]
- 132.Gorbunova V., Seluanov A., Dion V., Sandor Z., Meservy J.L., Wilson J.H. Selectable system for monitoring the instability of CTG.CAG triplet repeats in mammalian cells. Mol. Cell. Biol. 2003;23:4485–4493. doi: 10.1128/MCB.23.13.4485-4493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gomes-Pereira M., Monckton D.G. Chemically induced increases and decreases in the rate of expansion of a CAG.CTG triplet repeat. Nucleic Acids Res. 2004;32:2865–2872. doi: 10.1093/nar/gkh612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bishop A.J., Schiestl R.H. Homologous recombination as a mechanism for genome rearrangements: environmental and genetic effects. Hum. Mol. Genet. 2000;9:2427–2334. doi: 10.1093/hmg/9.16.2427. [DOI] [PubMed] [Google Scholar]
- 135.Jakupciak J.P., Wells R.D. Genetic instabilities of triplet repeat sequences by recombination. IUBMB Life. 2000;50:355–359. doi: 10.1080/713803749. [DOI] [PubMed] [Google Scholar]
- 136.Cleary J.D., Pearson C.E. The contribution of cis-elements to disease-associated repeat instability: clinical and experimental evidence. Cytogenet. Genome Res. 2003;100:25–55. doi: 10.1159/000072837. [DOI] [PubMed] [Google Scholar]
- 137.Warren S.T. Polyalanine expansion in synpolydactyly might result from unequal crossing-over of HOXD13. Science. 1997;275:408–409. doi: 10.1126/science.275.5298.408. [DOI] [PubMed] [Google Scholar]
- 138.Amiel J., Trochet D., Clement-Ziza M., Munnich A., Lyonnet S. Polyalanine expansions in human. Hum. Mol. Genet. 2004;13:235–243. doi: 10.1093/hmg/ddh251. [DOI] [PubMed] [Google Scholar]
- 139.Brown L.Y., Brown S.A. Alanine tracts: the expanding story of human illness and trinucleotide repeats. Trends Genet. 2004;20:51–58. doi: 10.1016/j.tig.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 140.Sargent R.G., Merrihew R.V., Nairn R., Adair G., Meuth M., Wilson J.H. The influence of a (GT)29 microsatellite sequence on homologous recombination in the hamster APRT gene. Nucleic Acids Res. 1996;24:746–753. doi: 10.1093/nar/24.4.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Majewski J., Ott J. GT repeats are associated with recombination on human chromosome 22. Genome Res. 2000;10:1108–1114. doi: 10.1101/gr.10.8.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Benet A., Molla G., Azorin F. d(GA x TC)(n) microsatellite DNA sequences enhance homologous DNA recombination in SV40 minichromosomes. Nucleic Acids Res. 2000;28:4617–4622. doi: 10.1093/nar/28.23.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shiroishi T., Sagai T., Moriwaki K. Hotspots of meiotic recombination in the mouse major histocompatibility complex. Genetica. 1993;88:187–196. doi: 10.1007/BF02424475. [DOI] [PubMed] [Google Scholar]
- 144.Kirkpatrick D.T., Wang Y.H., Dominska M., Griffith J.D., Petes T.D. Control of meiotic recombination and gene expression in yeast by a simple repetitive DNA sequence that excludes nucleosomes. Mol. Cell. Biol. 1999;19:7661–7671. doi: 10.1128/mcb.19.11.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Napierala M., Parniewski P.P., Pluciennik A., Wells R.D. Long CTG CAG repeat sequences markedly stimulate intramolecular recombination. J. Biol. Chem. 2002;277:34087–34100. doi: 10.1074/jbc.M202128200. [DOI] [PubMed] [Google Scholar]
- 146.Pluciennik A., Iyer R.R., Napierala M., Larson J.E., Filutowicz M., Wells R.D. Long CTG.CAG repeats from myotonic dystrophy are preferred sites for intermolecular recombination. J. Biol. Chem. 2002;277:34074–34086. doi: 10.1074/jbc.M202127200. [DOI] [PubMed] [Google Scholar]
- 147.Ohshima K., Kang S., Larson J.E., Wells R.D. Cloning, characterization, and properties of seven triplet repeat DNA sequences. J. Biol. Chem. 1996;271:16773–16783. doi: 10.1074/jbc.271.28.16773. [DOI] [PubMed] [Google Scholar]
- 148.Hebert M.L., Spitz L.A., Wells R.D. DNA double-strand breaks induce deletion of CTG.CAG repeats in an orientation-dependent manner in Escherichia coli. J. Mol. Biol. 2004;336:655–672. doi: 10.1016/j.jmb.2003.12.038. [DOI] [PubMed] [Google Scholar]
- 149.Richard G.F., Cyncynatus C., Dujon B. Contractions and expansions of CAG.CTG trinucleotide repeats occur during ectopic gene conversion in yeast, by a MUS81-independent mechanism. J. Mol. Biol. 2003;326:769–782. doi: 10.1016/s0022-2836(02)01405-5. [DOI] [PubMed] [Google Scholar]
- 150.Freudenreich C.H., Stavenhagen J.B., Zakian V.A. Stability of a CTG.CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol. Cell. Biol. 1997;17:2090–2098. doi: 10.1128/mcb.17.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Arai N., Akiyama R., Niimi N., Nakatsubo H., Inoue T. Meiotic contraction of CAG repeats in Saccharomyces cerevisiae. Genes Genet. Syst. 1999;74:159–167. doi: 10.1266/ggs.74.159. [DOI] [PubMed] [Google Scholar]
- 152.Cohen H., Sears D.D., Zenvirth D., Hieter P., Simchen G. Increased instability of human CTG repeat tracts on yeast artificial chromosomes during gametogenesis. Mol. Cell. Biol. 1999;19:4153–4158. doi: 10.1128/mcb.19.6.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jankowski C., Nasar F., Nag D.K. Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc. Natl Acad. Sci. USA. 2000;97:2134–2139. doi: 10.1073/pnas.040460297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jankowski C., Nag D.K. Most meiotic CAG repeat tract-length alterations in yeast are SPO11 dependent. Mol. Genet. Genom. 2002;267:64–70. doi: 10.1007/s00438-001-0635-4. [DOI] [PubMed] [Google Scholar]
- 155.Richard G.F., Paques F. Mini- and microsatellite expansions: the recombination connection. EMBO Rep. 2000;1:122–126. doi: 10.1093/embo-reports/kvd031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Richard G.F., Dujon B., Haber J.E. Double-strand break repair can lead to high frequencies of deletions within short CAG.CTG trinucleotide repeats. Mol. Gen. Genet. 1999;261:871–882. doi: 10.1007/s004380050031. [DOI] [PubMed] [Google Scholar]
- 157.Richard G.F., Goellner G.M., McMurray C.T., Haber J.E. Recombination-induced CAG trinucleotide repeat expansions in yeast involve the MRE11-RAD50-XRS2 complex. EMBO J. 2000;19:2381–2390. doi: 10.1093/emboj/19.10.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nassif N., Penney J., Pal S., Engels W.R., Gloor G.B. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Meservy J.L., Sargent R.G., Iyer R.R., Chan F., McKenzie G.J., Wells R.D., Wilson J.H. Long CTG tracts from the myotonic dystrophy gene induce deletions and rearrangements during recombination at the APRT locus in CHO cells. Mol. Cell. Biol. 2003;23:3152–3162. doi: 10.1128/MCB.23.9.3152-3162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lieber M.R., Ma Y., Pannicke U., Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nature Rev. Mol. Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 161.Kanaar R., Hoeijmakers J.H., van Gent D.C. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 162.Marcadier J.L., Pearson C.E. Fidelity of primate cell repair of a double-strand break within a (CTG).(CAG) tract. Effect of slipped DNA structures. J. Biol. Chem. 2003;278:33848–33856. doi: 10.1074/jbc.M304284200. [DOI] [PubMed] [Google Scholar]
- 163.van Heemst D., Brugmans L., Verkaik N.S., van Gent D.C. End-joining of blunt DNA double-strand breaks in mammalian fibroblasts is precise and requires DNA-PK and XRCC4. DNA Repair (Amst.) 2004;3:43–50. doi: 10.1016/j.dnarep.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 164.Bacolla A., Jaworski A., Connors T.D., Wells R.D. PKD1 unusual DNA conformations are recognized by nucleotide excision repair. J. Biol. Chem. 2001;276:18597–18604. doi: 10.1074/jbc.M100845200. [DOI] [PubMed] [Google Scholar]
- 165.Bacolla A., Jaworski A., Larson J.E., Jakupciak J.P., Chuzhanova N., Abeysinghe S.S., O'Connell C.D., Cooper D.N., Wells R.D. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl Acad. Sci. USA. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wells R.D., Ashizawa T., editors. Genetic Instabilities and Neurological Diseases, 2nd edn. Elsevier-Academic Press, Inc.; 2006. in press. [Google Scholar]